Abstract
Potential single nucleotide polymorphisms (SNPs) were detected between two chicken breeds (Kashmir favorella and broiler) using deep RNA sequencing. This was carried out to comprehend the coding area alterations, which cause variances in the immunological response to Salmonella infection. In the present study, we identified high impact SNPs from both chicken breeds in order to delineate different pathways that mediate disease resistant/susceptibility traits. Samples (liver and spleen) were collected from Salmonella resistant (K. favorella) and susceptible (broiler) chicken breeds. Salmonella resistance and susceptibility were checked by different pathological parameters post infection. To explore possible polymorphisms in genes linked with disease resistance, SNP identification analysis was performed utilizing RNA seq data from nine K. favorella and ten broiler chickens. A total of 1778 (1070 SNPs and 708 INDELs) and 1459 (859 SNPs and 600 INDELs) were found to be specific to K. favorella and broiler, respectively. Based on our results, we conclude that in broiler chickens the enriched pathways mostly included metabolic pathways like fatty acid metabolism, carbon metabolism and amino acid metabolism (Arginine and proline metabolism), while as in K. favorella genes with high impact SNPs were enriched in most of the immune-related pathways like MAPK signaling pathway, Wnt signaling pathway, NOD-like receptor signaling pathway, etc., which could be a possible resistance mechanism against salmonella infection. In K. favorella, protein–protein interaction analysis also shows some important hub nodes, which are important in providing defense against different infectious diseases. Phylogenomic analysis revealed that indigenous poultry breeds (resistant) are clearly separated from commercial breeds (susceptible). These findings will offer fresh perspectives on the genetic diversity in chicken breeds and will aid in the genomic selection of poultry birds.
1. Introduction
Salmonella enterica serovar typhimurium is a Gram-negative, facultative anaerobe, non-spore producing, motile bacillus in the Enterobacteriaceae family. It colonizes the digestive tracts of many vertebrates and cause severe intestinal pathology in young chicken [1,2]. Salmonellosis poses a serious socioeconomic hazard and is associated with significant human and animal mortality and morbidity [3]. Salmonella is one of the most prevalent bacteria, responsible for sporadic cases or outbreaks of gastroenteritis [4]. The annual economic cost of foodborne illness has been estimated to be as high as USD90 billion [5]. Globally, Salmonella is the most common cause of foodborne illnesses, with poultry and poultry-related products considered the primary causes of such outbreaks [6]. Salmonella infection is a major threat in the poultry industry. Poultry is a major global reservoir of non-typhoidal Salmonellae that cause foodborne diseases. Systemic salmonellosis causes significant losses in the poultry sector in terms of mortality and decreased poultry production [7]. Vaccination, sanitation, and the use of antibiotics are the most common methods used to combat Salmonella infections. As none of the current vaccination programs have been successful in controlling Salmonella infections [8], antibiotics are being preferred by the poultry industry. Owing to widespread antibiotic use, growth of antibiotic-resistant microorganisms and the buildup of antibiotic residues in food intended for human consumption are the two main problems resulting from the emergence of antibiotic-resistant bacteria and the accumulation of antibiotic residues in food. These are the two key difficulties associated with widespread antibiotic use in poultry for human consumption [9,10]. The human immune system has the power to successfully fend off microbial invasion and eradicate microbes. Following bacterial identification, host macrophages induce bactericidal action, which stimulates the maturation and migration of dendritic cells as well as the production of inflammatory chemokines, cytokines, interleukins, and other substances [11]. On the other hand, Salmonella has certain defense mechanisms that help it to combat these host barriers and inhibit host-immune responses via their its virulence genes [11]. After the Salmonella reaches the intestinal macrophages, it senses the phagosomal environment and triggers various virulence mechanisms that help it to survive in the macrophages [12]. Salmonella uses the Salmonella pathogenicity islands during host invasion. Within the Salmonella pathogenicity islands 1 and 2, Salmonella encodes two distinct virulence related T3SSs that act at different times during infection [13]. Once in contact with the host cell, the SPI1-encoded T3SS is activated and translocates bacterial proteins across the plasma membrane, while the SPI2-encoded T3SS that is expressed within phagosomes is involved in the translocation of the effectors across the vacuolar membrane. The SPI1 system plays a key role in invading non-phagocytic cells, induction and activation intestinal inflammatory responses, diarrhea and intestinal colonization. In contrast, the SPI2-encoded T3SS is required for the survival of bacteria in macrophages and the onset of systemic disease [12]. After the Salmonella has entered the cell, it resides within a vacuolar compartment known as a spacious phagosome (SP) [14]. This spacious phagosome shrinks within minutes to hours and forms an adherent membrane that wraps one or more bacteria and is referred as the Salmonella-containing vacuole (SCV). Intracellular persistence of SCV can range from hours to days, which makes it a unique phagosome in terms of normal maturation and recycling of phagolysosomes. Despite controversies, some studies report that Salmonella can live in macrophages that have lysosomal compartments fused with the SCV [15,16].
In the context of the above statements, genetic resistance is a long-term approach for a disease-control strategy [17]. Selecting more resistant hens may be an alternative option to reduce illness occurrence. Genetic disease resistance is often more relevant in underdeveloped nations since indigenous breeds are more resistant to local diseases [18]. K. favorella is a well-known indigenous chicken breed from the north Indian state Jammu and Kashmir. It is regarded as the most significant source of animal protein and is raised largely for meat and egg production [19]. The local climate conditions, feed, and stress management are well matched to this native breed, which has a good disease resistance [20].
Recent developments in molecular science have opened up new possibilities for improving quantitative traits genetically, especially those that are resistant to disease. The use of gene introgression or marker-assisted selection would be made easier with the discovery of direct or indirect molecular markers. Molecular markers are essential tools for marker-assisted breeding. The simple sequence repeats (SSRs) (SNP) markers are two attractive and widely used markers because of many merits including locus-specificity, reproducibility, co-dominance, and random genome-wide distribution in many organisms [21,22]. These features, however, are not only regulated at the DNA level, but also at the mRNA level before and after transcription, and this level of regulation is more extensive, systematic, and accurate. SNP detection by RNA-seq is of great interest to researchers as whole genome sequencing is expensive as well as exome sequencing tools are uncommon. The detection of SNPs in coding regions is used to understand variants affecting protein functions and analyze allele-specific expressions. Gene expression can be highly variable and which makes SNP detection and genotype calling by RNA-seq a challenging endeavor [23]. The study aims to identify SNPs, which are potentially involved in disease resistance against Salmonella infection in poultry and thus may lay a foundation step for future in-depth studies of disease resistance mechanisms.
2. Materials and Methods
2.1. Experimental Birds and Sample Collection
Experimentation and animal tissue collection was carried out with the proper consent of the Institutional Animal Ethics Committee on Ethical Standards in Animal Experiments (AU/FVSc/PS-57/16021). During all of the experimental studies, the Institutional Animal Ethics Committee’s rules were rigorously followed. The experimental chicks from two breeds, i.e., K. faverolla and broiler (Cobb), were procured from the Division of Livestock Production and Management, Sher-e-Kashmir University of Agricultural Sciences and Technology of Kashmir (SKUAST-K)-India. From day one of hatching, the experimental birds were kept in the animal house facilities center at SKUAST-K, under standard sanitary, temperature, and pressure conditions. The birds were monitored on a daily basis and provided unlimited access to food and water.
To ensure that all the birds under experimental study from both the breeds were free from Salmonella infection, fecal swabs from all the birds were taken and inoculated in tetrathionate broth and further streaked on BGA and MacConkey plates. Only Salmonella negative birds were taken for the study. After 12 h post infection, fecal swabs were taken from all the birds and incubated overnight in tetrathionate broth at 42 °C. The overnight culture was streaked on BGA and MacConkey plates. The plates were kept overnight at 37 °C. Following overnight incubation, the colonies on the plates were then examined for Salmonella species.
After giving an acclimatization time of 3 days to the chicks from both the breeds, they were orally infected with S. typhimurium (2 × 108 CFU/mL) at 4 days of age and were initially assessed for disease resistance up to 10 days post infection. The two chicken breeds were classified as Salmonella-resistant and -susceptible breeds by taking into consideration the clinical symptoms and bacterial loads. The clinical scores were recorded twice daily following points based a scoring system. Chicks with severe clinical signs (progressive weakening, anorexia, diarrhea, and head lowering) as well as significant liver disease and greater bacterial loads in fecal swabs were identified as the challenged susceptible group. The challenged-resistant birds were recognized as chicks with little clinical and pathological symptoms and low bacterial burdens. The K. favorella was determined to be resistant based on the aforementioned clinical signs and bacterial levels, and broilers were determined to be susceptible to Salmonella infection. Samples (liver and spleen) were collected from Salmonella-resistant (K. favorella) and susceptible (broiler) chicken breeds [1] (Table S1).
Carcass of sacrificed birds were subjected to a systemic necropsy technique for the examination and documentation of the Salmonella specific lesions. Lesions primarily included bronze colored discoloration of liver, typhilitis and splenomegaly. For comparative histopathological analysis, representative liver and spleen samples from commercial broilers and K. favorella were collected in 10% buffered formalin and processed using the standard paraffin embedding technique using alcohol and acetone as dehydrating agents, benzene as a clearing agent, and paraffin wax with a melting point of 60 °C. For routine investigation, sections of 5 m thickness were cut and stained with Harris Hematoxylin and Eosin.
In this study, 10 samples from broiler chicken (5 liver and 5 spleen) and 9 samples from favorella chicken (5 liver and 4 spleen) were utilized to identify SNPs that could potentially mediate the Salmonella disease resistance in chicken. To reduce the effect of confounding factors, and increase sensitivity and specificity of SNPs, both groups have control and infected samples. In the broiler liver group, 3 are infected and 2 are control samples. Similarly, in the broiler spleen group, 3 are infected and 2 are control samples. In the K. favorella liver group, 2 are infected and 2 are control samples. In the K. favorella spleen group, 3 are infected and 2 are control samples. The reason for including infected samples with control for SNP identification was to reduce any biological, technical or genomic factors that could possibly affect the key SNP identification with the phenotype of interest, i.e., Salmonella resistance.
2.2. Total RNA Isolation, cDNA Library Construction, and Sequencing
The sequencing data were downloaded from our previously published dataset NCBI (Accession ID: GSE168060). All sample processing and sequencing steps are described previously [24]. Briefly, total RNA was extracted using Trizol method (Ambion, Naugatuck, CT, USA) following the manufacturers guidelines. The RNA quality and integrity were examined using a spectrophotometer (ThermoFisher, Waltham, MA, USA) and a bioanalyzer (Agilent, Santa Clara, CA, USA). Libraries were prepared using RNA samples with RIN values ≥ 8. The Illumina TruSeq stranded mRNA sample preparation kit was used to construct cDNA libraries, and the manufacturer’s protocol was followed. The 4 μg/sample total RNA was utilized to prepare libraries. Poly-T attached magnetic beads were used to purify poly-A containing mRNA molecules. Following purification, divalent cations were used in a high-temperature process to break the RNA down into smaller bits. Using the enzyme reverse transcriptase and random primers, the RNA fragments were utilized to create first strand cDNA (Illumina, San Diego, CA, USA). After DNA fragments were adenylated (at their 3 ends), the hybridization process was started by ligating the Illumina paired-end adaptor and index. Using an Illumina PCR primer cocktail, the cDNA fragments (150 bp) were produced and used to create the sequencing paired end cDNA library. Libraries were pooled in equimolar levels using a High Throughput Model flow cell on an Illumina HiSeq 2500 platform and paired end sequenced by SciGenom, Cochin, Kerala-India.
2.3. Quality Control, Aligning and Mapping Reads to the Genome
The FASTQC program v0.11.1 was used to examine read quality control [25]. Following preprocessing, low-quality sequence filtering and adaptor trimming with Cutadapt v3.40 [26], high-quality sequencing reads that exceeded thresholds (Phred Score > 30) were combined for SNP identification analysis. For each sample, more than 40 million clean, high-quality readings were gathered. HISAT2 was used to map the cleaned reads to the reference genome assembly ARS-UCD1.2.99 [27]. Before variant identification, the data pretreatment stages suggested in the Genome Analysis Toolkit (GATK) best practices workflow were carried out [28]. MarkDuplicates from Picard tools were used to identify PCR duplicates [29]. Additionally, we used GATK to recalibrate the base quality scores, examine intron–exon junctions, and perform local realignment around InDels. SNP and INDELs discovery across 10 broiler and 10 K. favorella transcriptome samples independently was carried out using two distinct variant callers: (i) mpileup from SAMtools v1.4 [30] in multi-sample calling mode with default parameters; (ii) GATK utilizing the HaplotypeCaller tool in multi-sample calling mode (modality “GATK”). The final set for analysis contains SNPs and InDels, which are common in both datasets. Chicken genetic variants from dbSNP 2.0 build 153 dated: 8 August 2019 were incorporated in SNP calling to populate the RS_ID column of the known SNPs. Filtering [base quality score (Q Score) > 30, mapping quality > 30, and minimum depth > 10] of generated variants and annotation were performed using VCFtools version 0.1.8 and SnpEff program v4.1. Biological significance was further evaluated for the genes that had high-impact variations. The KEGG pathway enrichment analysis was performed using KOBAS server version 3 [31,32].
3. Results
3.1. Quality Control, Mapping, and Post Treatment
A total of 46.95 million reads (range 30.51–68.56 million reads/library) and 41.74 million reads (range 26.92–63.61 million reads/library) were generated by the liver and spleen transcriptome libraries, respectively. Overall, 43.76 million reads (92.82%) of the 46.95 million hepatic transcriptome reads passed quality control and were mapped to the Gallus gallus genome GRCg6a. The remaining reads were deleted, and 41.46 million uniquely mapped reads in total were processed further. In the spleen, 38.65 million reads (92.24%) of the total 41.74 million reads passed the quality check and were aligned to the G. gallus CRCg6a genome. Additionally, 35.45 million uniquely mapped reads were processed further, while the remaining reads were discarded. K. favorella and broiler yielded a total of 1,141,122 and 1,151,874 variations, respectively. The chromosomal distribution of SNPs and INDELs is shown in Table S2 and the variant types are shown in Tables S3 and S4. Kashmir favorella had a total of 26.036% missense, 0.138% nonsense, and 73.785% silent alterations, while broiler chicken had a total of 26.845% missense, 0.167% nonsense, and 72.988% silent mutations. The transitions to transversions ratio (Ts/Tv) for K. favorella and broiler was determined to be 2.7 (6080998/2236247) and 2.7 (6993925/2563136), respectively, in line with earlier studies. SNP distribution on different chromosomes in both K. favorella and commercial broiler are shown (Figure 1). It was found that there were 758 common genes with SNPs (Figure 2). The common SNPs were filtered out, and the high-impact SNPs and INDELs specific to broiler 1778 (1070 SNPs and 708 INDELs) and K. favorella 1459 (859 SNPs and 600 INDELs) (Supplementary File S1 and S2) were further studied. List of genes with high impact SNPs involved in Salmonella disease resistance in chickens are shown in Table 1.
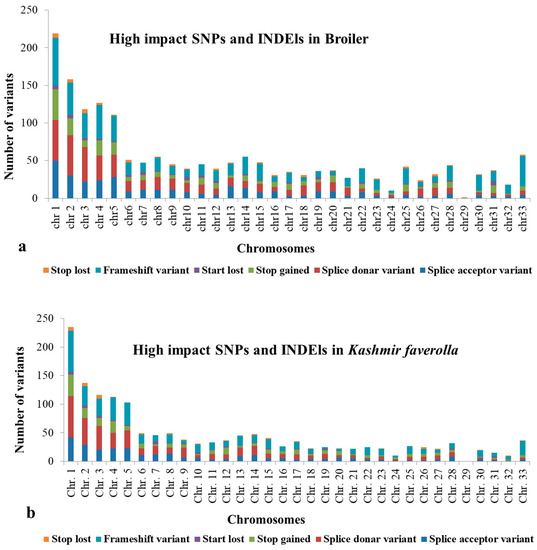
Figure 1.
Chromosomal distribution of high impact SNPs and Indels in (a) Broiler chicken and (b) K. favorella.
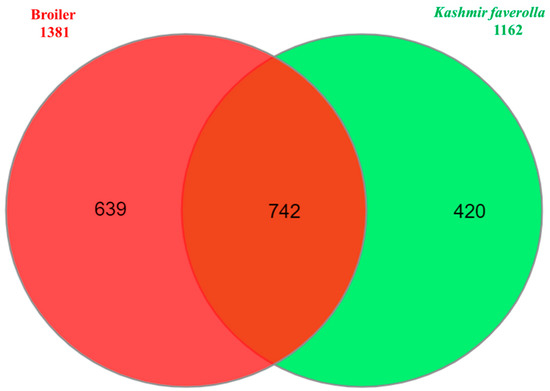
Figure 2.
Venn diagram showing common genes with SNPs between K. favorella and broiler chicken breeds.

Table 1.
List of genes with high impact SNPs involved in Salmonella disease resistance in chicken.
3.2. Analysis of Genes with SNPs and INDELs
Functional annotation indicates that signaling pathways, such as those involved in metabolism, herpes simplex virus type 1 infection, Influenza A, fatty acid biosynthesis, fatty acid metabolism, carbon metabolism and citrate cycle in broiler chicken are enriched (p < 0.05) (Table S5, Figure 3a). The enhanced pathways in K. favorella chicken comprised the Wnt signaling route, the FoxO signaling pathway, the cellular senescence pathway, and the NOD-like receptor signaling pathway (Table S6, Figure 3b). In K. favorella and broiler chicken, gene ontology (GO) research shows genes (p < 0.05) with SNPs were primarily involved in the binding process (enzyme, ribonucleotide, and histone binding), as well as antigen processing (Figure 4a,b, Tables S7 and S8).
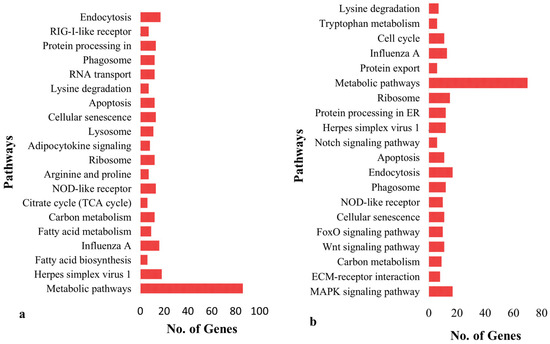
Figure 3.
KEGG enrichment of genes with high impact variations. (a) Broiler. (b) K. favorella.
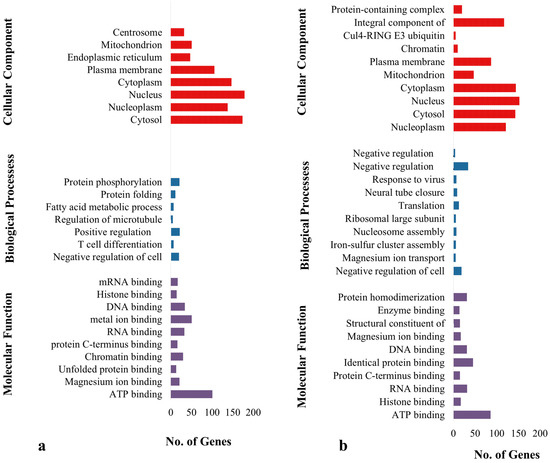
Figure 4.
Gene ontology analysis of genes with high impact SNPs and INDELs. (a) Broiler. (b) K. favorella.
3.3. Protein-Protein Interaction
Salmonella infection affects every chicken breed; however, every breed has its own defense mechanism to counter the infection. In both the chicken breeds, we found similar hub genes; however, in the resistant breed (K. favorella) PLK1, MK1671P gene mutations were hyperactive, suggesting a possible role in this particular breed. Plk-1 is the member of the serine/threonine polio-like kinase family has a vital role in immune signaling [33]. Further PLK-1 interacts with BRCA-2 and MLF1P, which regulate autophagy, antigen presentation, immune response, angiogenesis and apoptosis [34,35] (Figure 5).
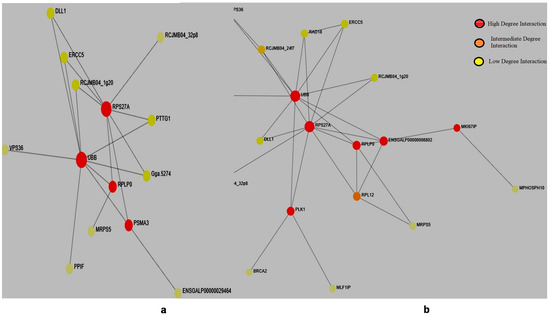
Figure 5.
Protein–protein interaction network of genes with high impact variations. (a) Broiler. (b) K. favorella.
4. Discussion
Salmonella, one of the major infectious diseases in poultry, causes considerable economic losses in terms of mortality and morbidity, especially in countries that lack effective vaccination programs. Besides being resistant to diseases, indigenous chicken breeds are also a potential source of animal protein in developing countries. For understanding the disease resistance, an indigenous chicken line K. favorella, and commercial broilers were selected [1]. The severity of clinical symptoms, pathological manifestations and bacterial load post infection were used to assess the disease susceptibility and resistance.
An effective immune response against the invading pathogen requires a balance between the pathogen clearance and self-damage. However, this balance likely to change dynamically as the infection will progress. The host will destroy the invading pathogen at the initial stage of infection and towards the end, the host repairs the damage so that it can return to its original state. An effective host response is referred to as a balancing resistance and infection tolerance mechanism [36]. Indigenous chickens are genetically more diversified than commercial breeds due to their extensive history of breeding and improvement through methods remarkably distinct from those employed for commercial varieties. Therefore, it is essential to preserve regional chicken breeds as genetic resources in order to be prepared for unforeseen breeding demands in the future [37,38].
In our pilot study, we found the bacterial load in the resistant chicken breed (K. favorella) was lower than the bacterial load in the susceptible chicken breed (broiler). Our studies were in accordance with the previous studies, which showed the bacterial load in the local chicken was lower than other chicken breeds [39,40]. K. favorella showed minor lesions in the liver and spleen while the broiler chicken showed major necrotic lesions. This was in consensus with previous studies, which showed that the higher the bacterial count, the greater the pathological score [41].
The K. favorella, a well-known indigenous chicken breed from the north Indian state of Jammu and Kashmir, is regarded as the most important source of animal protein [13]. This native breed is disease-resistant and highly adapted to local climate circumstances, feed, and stress management [14]. For understanding the disease resistance mechanism, we have analyzed RNA sequencing data and performed a comparative study between K. favorella and broiler chicken breeds [1]. To evaluate putative polymorphisms in disease resistance genes, SNP detection analysis was performed using RNA seq data from 10 K. favorella and 10 broiler chicken.
A total of 1,141,122 and 1,151,874 variants were identified from K. favorella and broiler, respectively. A total of 1778 (1070 SNPs and 708 INDELs) and 1459 (859 SNPs and 600 INDELs) were identified in broiler and K. favorella, respectively. The KEGG and gene ontology analyses revealed that the genes were engaged in a variety of significant immune-related pathways. The MAPK signaling pathway, ECM-receptor interaction, Wnt signaling route, FoxO signaling pathway, and cellular senescence were shown to be substantially enriched in Ka. favorella. These pathways stimulate the immune response against Salmonella infection [42,43]. WNT signaling is essential for maintaining tissue homeostasis, epithelial barrier functioning, inflammatory cytokine production and modulation, host cell innate defense mechanisms, and the integration of innate and adaptive immunity [44]. In K. favorella, amplification of the WNT signaling pathway in response to Salmonella infection could increase B cell survival or proliferation [45]. Recent studies suggest that Wnt signaling performs an essential function in immune cell modulation and counteracts various disorders [46]. We found variations in different genes that regulate Wnt signaling (Figure S1) (TCF7, LRP5, CaMKII, WNT5A, NLK, etc.). TCF7 plays a vital role in tissue repair, remodeling and disease pathogenesis [47]. LRP5 has been found to possess a novel role in IL-10 signaling, thereby exerting a protective role during inflammation [48]. CaM-dependent proteins (CaMKII) have a critical role in infectious diseases through involvement in inflammatory processes, apoptosis and necroptosis [49]. Wnt5A promotes the death of numerous bacterial pathogens by altering actin assembly in macrophages, and thus resulting in bacterial phagocytosis [50]. Nemo-like kinase (NLK) has a role in modulating immune responses through regulation of NF-κB signaling by interfering with different signaling molecules [7]. In broilers, the enrichment analysis revealed that genes with high impact SNPs were involved mainly in metabolic pathways, fatty acid metabolism, carbon metabolism and amino acid metabolism (Arginine and proline metabolism). Salmonella utilizes these metabolites as an energy source for its intracellular survival and proliferation [51].
The phylogenomic analysis revealed the exhaustive similarity between commercially available chicken breeds and possible similar mechanism of weak resistance against Salmonella infection (Figure 6). Resistance to salmonellosis in chicken greatly varies among the chicken line [52]. Due to breed differences, there is significant genetic heterogeneity in chicken for resistance to S. typhimurium [39].
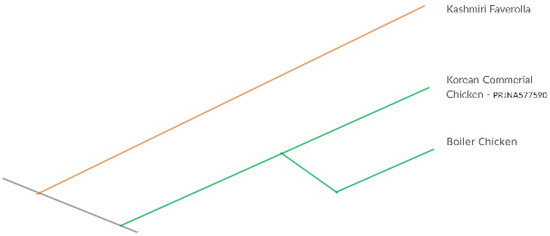
Figure 6.
Comparative phylogenetic analysis between commercial and native chicken breeds.
MADPRT1, PPARD, IL18, IL18R1, TNFRSF10B, IL1R1, TNFAIP1, MMP28, SLC9A9, SLC5A10, SLC13A2 genes were identified with high impact SNPs involved in Salmonella disease resistance in chicken. This correlates with our previous study, which highlights their role in innate and adaptive immune responses [1]. Genetic variation in IL-18 has been associated with increased risk of atopy and asthma [53,54]. Further, IL-18 polymorphism has been linked to an increased or decreased progression of hepatocellular carcinoma [55]. Polymorphisms and haplotypes in TNFRSF10B are associated with an increased risk of death in non-small cell lung cancer [56].
5. Conclusions
SNP analysis demonstrated a significant difference between the K. favorella and broiler chickens in disease resistance. The high impact SNP variations in K. favorella and broilers were mostly engaged in metabolic pathways followed by the differential expression of some immune-related genes. Phylogenomic analysis based on the SNP studies showed clear genetic variations of the two breeds, based on the resistant mechanisms of indigenous poultry breeds (resistant) and susceptibility of broiler to Salmonella infection. These findings will offer fresh perspectives on the genetic diversity in chicken breeds and will aid in genomic selection of poultry birds.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14061283/s1, Figure S1: Genes with high impact (in Red) in Wnt signaling pathway; Table S1: Assessment of disease resistance and susceptibility by different parameters; Table S2: The chromosomal distribution of SNPs and INDELs; Table S3: High-impact SNPs and INDELs specific to K. favorella; Table S4: High-impact SNPs and INDELs specific to broiler chicken; Table S5: Pathways affected by high-impact SNPs in K. favorella; Table S6: Pathways affected by high-impact SNPs in broiler; Table S7: Gene Ontology analysis of genes with high impact SNPs and INDELs in K. favorella; Table S8: Gene Ontology analysis of genes with high impact SNPs and INDELs in broiler.
Author Contributions
M.A.D. and B.B. contributed equally to this work. They performed the experiment and wrote the manuscript. J.N., A.S. and T.M. performed the data analysis. M.K. and Z.H. summarized results and helped in proof reading of manuscript. S.S.B. and S.M.A. designed and supervised the experiments. All authors have read and agreed to the published version of the manuscript.
Funding
The Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India supported the project under grant number EMR/2017/000580.
Institutional Review Board Statement
The tissue collection was carried out only after receiving approval from institutional animal ethics committee on ethical standards in animal experimentation (AU/FVSc/PS-57/16021).
Informed Consent Statement
Not applicable.
Data Availability Statement
The sequencing data used in the current study were downloaded from NCBI GEO database (Accession ID GSE168060).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| DNA | Deoxyribonucleic acid |
| mRNA | Messenger RNA |
| TCF | Transcription Factor |
| CaMK | CaM-dependent proteins |
| SSRs | Simple sequence repeats |
| INDELs | insertions and deletions |
| BRCA | Breast cancer gene |
| SNP | Single nucleotide polymorphism |
| KEGG | Kyoto encyclopaedia of genes and genomes |
| IL | Interleukin |
| NLK | Nemo-like kinase |
| LRP | Low-density lipoprotein receptor-related protein |
| GO | Gene ontology |
| MLF1P | Myeloid leukemia factor |
References
- Dar, M.A.; Ahmad, S.M.; Bhat, B.A.; Dar, T.A.; ul Haq, Z.; Wani, B.A.; Shabir, N.; Kashoo, Z.A.; Shah, R.A.; Ganai, N.A.; et al. Comparative RNA-Seq analysis reveals insights in Salmonella disease resistance of chicken; and database development as resource for gene expression in poultry. Genomics 2022, 114, 110475. [Google Scholar] [CrossRef]
- Dar, M.A.; Ahmed, R.; Urwat, U.; Ahmad, S.M.; Dar, P.A.; Kushoo, Z.A.; Dar, T.A.; Mumtaz, P.T.; Bhat, S.A.; Amin, U.; et al. Expression kinetics of natural resistance associated macrophage protein (NRAMP) genes in Salmonella typhimurium-infected chicken. BMC Vet. Res. 2018, 14, 180. [Google Scholar] [CrossRef]
- Dar, M.A.; Mumtaz, P.T.; Bhat, S.A.; Taban, Q.; Khan, S.A.; Banday, T.; Ahmad, S.M. Immunopathogenesis of Salmonellosis. In New Insight into Brucella Infection and Foodborne Diseases; IntechOpen: London, UK, 2019. [Google Scholar]
- Jain, P.; Chowdhury, G.; Samajpati, S.; Basak, S.; Ganai, A.; Samanta, S.; Okamoto, K.; Mukhopadhyay, A.K.; Dutta, S. Characterization of non-typhoidal Salmonella isolates from children with acute gastroenteritis, Kolkata, India, during 2000–2016. Braz. J. Microbiol. 2020, 51, 613–627. [Google Scholar] [CrossRef]
- Scharff, R.L. Food Attribution and Economic Cost Estimates for Meat- and Poultry-Related Illnesses. J. Food Prot. 2020, 83, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Dar, M.A.; Urwat, U.; Ahmad, S.M.; Ahmad, R.; Kashoo, Z.A.; Dar, T.A.; Bhat, S.A.; Mumtaz, P.T.; Shabir, N.; Shah, R.A.; et al. Gene expression and antibody response in chicken against Salmonella typhimurium challenge. Poult. Sci. 2019, 98, 2008–2013. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Nie, C.; Liu, Y.; Chen, Y.; Lv, X.; Wang, L.; Zhang, J.; Li, K.; Jia, Y.; Ban, L.; et al. A genome-wide association study explores the genetic determinism of host resistance to Salmonella pullorum infection in chickens. Genet. Sel. Evol. 2019, 51, 51. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Renu, S.; Patil, V.; Schrock, J.; Feliciano-Ruiz, N.; Selvaraj, R.; Renukaradhya, G.J. Immune response to Salmonella enteritidis infection in broilers immunized orally with chitosan-based Salmonella subunit nanoparticle vaccine. Front. Immunol. 2020, 19, 935. [Google Scholar] [CrossRef]
- Rodrigues, G.L.; Panzenhagen, P.; Ferrari, R.G.; Dos Santos, A.; Paschoalin, V.M.; Conte-Junior, C.A. Frequency of antimicrobial resistance genes in Salmonella from Brazil by in silico whole-genome sequencing analysis: An overview of the last four decades. Front. Microbiol. 2020, 11, 1864. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 30, 795. [Google Scholar] [CrossRef]
- Wang, M.; Qazi, I.H.; Wang, L.; Zhou, G.; Han, H. Salmonella virulence and immune escape. Microorganisms 2020, 8, 407. [Google Scholar] [CrossRef]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008, 6, 53–66. [Google Scholar] [CrossRef]
- Hansen-Wester, I.; Hensel, M. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 2001, 3, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Alpuche-Aranda, C.M.; Racoosin, E.L.; Swanson, J.A.; Miller, S.I. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J. Exp. Med. 1994, 179, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Carrol, M.E.W.; Jackett, P.S.; Aber, V.R.; Lowrie, D.B. Phagolysosome formation, cyclic adenosine 3′: 5′-monophosphate and the fate of Salmonella typhimurium within mouse peritoneal macrophages. Microbiology 1979, 110, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Buchmeier, N.A.; Heffron, F.R.E.D. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect. Immun. 1991, 59, 2232–2238. [Google Scholar] [CrossRef] [PubMed]
- Rautenschlein, S.; Cheng, H.H.; Lamont, S.J. Host factors for disease resistance. Dis. Poult. 2020, 13, 79–108. [Google Scholar]
- Pal, A.; Chakravarty, A.K. Disease resistance for different livestock species. Genet. Breed. Dis. Resist. Livest. 2020, 2020, 271–296. [Google Scholar]
- Wani, H.; Darzi, M.M.; Kamil, S.A.; Wani, S.A.; Munshi, Z.H.; Shakoor, A.; Raja, T.A.; Shoukat, S.; Kashani, B.; Shah, A. Histological and histochemical studies on the reproductive tract of Kashmir faverolla chicken. J. Etnomology Zool. Stud. 2017, 5, 2256–2262. [Google Scholar]
- Iqbal, S.; Pampori, Z.A. Production potential and qualitative traits of indigenous chicken of Kashmir. Livest. Res. Rural. Dev. 2008, 20, 14. [Google Scholar]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 30, 484. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Nawaz, M.A.; Shahid, M.Q.; Doğan, Y.; Comertpay, G.; Yıldız, M.; Hatipoğlu, R.; Ahmad, F.; Alsaleh, A.; Labhane, N.; et al. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnol. Biotechnol. Equip. 2018, 32, 26185. [Google Scholar] [CrossRef]
- Jehl, F.; Degalez, F.; Bernard, M.; Lecerf, F.; Lagoutte, L.; Désert, C.; Coulée, M.; Bouchez, O.; Leroux, S.; Abasht, B.; et al. RNA-Seq Data for Reliable SNP Detection and Genotype Calling: Interest for Coding Variant Characterization and Cis-Regulation Analysis by Allele-Specific Expression in Livestock Species. Front. Genet. 2021, 12, 655707. [Google Scholar] [CrossRef]
- Ahmad, S.M.; Bhat, S.S.; Shafi, S.; Dar, M.A.; Saleem, A.; Haq, Z.; Farooq, N.; Nazir, J.; Bhat, B. Identification of key transcription factors and their functional role involved in Salmonella typhimurium infection in chicken using integrated transcriptome analysis and bioinformatics approach. BMC Genom. 2023, 24, 214. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data 2010; BibSonomy, Babraham Bioinformatics: Cambridgeshire, UK, 2018; Volume 17. [Google Scholar]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Poplin, R.; Chang, P.C.; Alexander, D.; Schwartz, S.; Colthurst, T.; Ku, A.; Newburger, D.; Dijamco, J.; Nguyen, N.; Afshar PTGross, S.S. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018, 36, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Toolkit, P. Broad Institute, GitHub Repository; Broad Institute: Cambridge, MA, USA, 2019. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Biswas, A.; Zhou, D.; Fiches, G.N.; Wu, Z.; Liu, X.; Ma, Q.; Zhao, W.; Zhu, J.; Santoso, N.G. Inhibition of polo-like kinase 1 (PLK1) facilitates reactivation of γ-herpesviruses and their elimination. PLoS Pathog. 2021, 23, e1009764. [Google Scholar] [CrossRef]
- Morand, S.; Stanbery, L.; Walter, A.; Rocconi, R.P.; Nemunaitis, J. BRCA1/2 Mutation Status Impact on Autophagy and Immune Response: Unheralded Target. JNCI Cancer Spectr. 2020, 4, pkaa077. [Google Scholar] [CrossRef]
- Irawan, C.; Atmakusumah, D.; Siregar, N.C.; Tean, T.B.; Kong, L.W.; Kiat, O.C.; Mansyur, M. Expression of biomarkers CXCR4, IL11-RA, TFF1, MLF1P in advanced breast cancer patients with bone metastatic: A diagnostic study. Acta Med. Indones 2016, 48, 261–268. [Google Scholar]
- Chambers, M.C.; Schneider, D.S. Balancing resistance and infection tolerance through metabolic means. Proc. Natl. Acad. Sci. USA 2012, 109, 13886–13887. [Google Scholar] [CrossRef]
- Chang, C.S.; Chen, C.F.; Berthouly-Salazar, C.; Chazara, O.; Lee, Y.P.; Chang, C.M.; Chang, K.H.; Bed’Hom, B.; Tixier-Boichard, M. A global analysis of molecular markers and phenotypic traits in local chicken breeds in Taiwan. Anim. Genet. 2012, 43, 172–182. [Google Scholar] [CrossRef]
- Khanyile, K.S.; Dzomba, E.F.; Muchadeyi, F.C. Population genetic structure, linkage disequilibrium and effective population size of conserved and extensively raised village chicken populations of Southern Africa. Front. Genet. 2015, 3, 13. [Google Scholar] [CrossRef]
- Girmay, G.; Pal, M.; Dessie, T.; Sissay, T.; Wubete, A. Evaluating the relative resistance of different poultry breeds to Salmonella typhimurium. Afr. J. Agric. Res. 2015, 10, 2928–2939. [Google Scholar]
- Wang, F.; Zhang, J.; Zhu, B.; Wang, J.; Wang, Q.; Zheng, M.; Wen, J.; Li, Q.; Zhao, G. Transcriptome analysis of the cecal tonsil of Jingxing yellow chickens revealed the mechanism of differential resistance to Salmonella. Genes 2019, 10, 979. [Google Scholar] [CrossRef]
- Lacharme-Lora, L.; Owen, S.V.; Blundell, R.; Canals, R.; Wenner, N.; Perez-Sepulveda, B.; Fong, W.Y.; Gilory, R.; Wigley, P.; Hinton, J.C. The use of chicken and insect infection models to assess the virulence of African Salmonella typhimurium ST313. PLoS Negl. Trop. Dis. 2019, 13, e0007540. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Tong, C.; Ma, S.; Zhou, L.; Zhao, L.; Zhao, X. Involvement of microRNAs in probiotics-induced reduction of the caecal inflammation by Salmonella typhimurium. Front. Immunol. 2017, 8, 704. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Z.; Shu, Q.P.; Song, Y.; Zhang, H.H.; Liu, Y.; Jin, B.X.; Liuyu, T.Z.; Li, C.; Huang, X.C.; Du, R.L.; et al. Phosphorylation of MAVS/VISA by Nemo-like kinase (NLK) for degradation regulates the antiviral innate immune response. Nat. Commun. 2019, 10, 3233. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, J.K.; Kling, J.C.; Tran, T.T.; Blumenthal, A. Functions of the WNT signaling network in shaping host responses to infection. Front. Immunol. 2019, 10, 2521. [Google Scholar] [CrossRef] [PubMed]
- Monson, M.S.; Van Goor, A.G.; Ashwell, C.M.; Persia, M.E.; Rothschild, M.F.; Schmidt, C.J. Immunomodulatory effects of heat stress and lipopolysaccharide on the bursal transcriptome in two distinct chicken lines. BMC Genom. 2018, 19, 643. [Google Scholar] [CrossRef] [PubMed]
- Haseeb, M.; Pirzada, R.H.; Ain, Q.U.; Choi, S. Wnt Signaling in the Regulation of Immune Cell and Cancer Therapeutics. Cells 2019, 8, 1380. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, W.; Wang, X. Roles of transcriptional factor 7 in production of inflammatory factors for lung diseases. J. Transl. Med. 2015, 13, 273. [Google Scholar] [CrossRef]
- Swafford, D.; Shanmugam, A.; Ranganathan, P.; Manoharan, I.; Hussein, M.S.; Patel, N.; Sifuentes, H.; Koni, P.A.; Prasad, P.D.; Thangaraju, M.; et al. The Wnt–β-Catenin–IL-10 Signaling Axis in Intestinal APCs Protects Mice from Colitis-Associated Colon Cancer in Response to Gut Microbiota. J. Immunol. 2020, 205, 2265–2275. [Google Scholar] [CrossRef]
- Beckendorf, J.; van den Hoogenhof, M.M.G.; Backs, J. Physiological and unappreciated roles of CaMKII in the heart. Basic Res. Cardiol. 2018, 113, 29. [Google Scholar] [CrossRef]
- Jati, S.; Sengupta, S.; Sen, M. Wnt5A-Mediated Actin Organization Regulates Host Response to Bacterial Pathogens and Non-Pathogens. Front. Immunol. 2021, 11, 628191. [Google Scholar] [CrossRef]
- Thompson, A.; Fulde, M.; Tedin, K. The metabolic pathways utilized by Salmonella typhimurium during infection of host cells. Environ. Microbiol. Rep. 2018, 10, 140–154. [Google Scholar] [CrossRef]
- Calenge, F.; Kaiser, P.; Vignal, A. Genetic control of resistance to salmonellosis and to Salmonella carrier-state in fowl: A review. Genet. Sel. Evol. 2010, 42, 11. [Google Scholar] [CrossRef]
- Izakovicova, H.L. Interleukin-18 in asthma and other allergies. Clin. Exp. Allergy 2003, 33, 1023–1025. [Google Scholar] [CrossRef]
- Imboden, M.; Nicod, L.; Nieters, A.; Glaus, E.; Matyas, G.; Bircher, A.J.; Ackermann-Liebrich, U.; Berger, W.; Probst-Hensch, N.M.; APALDIA Team. The common G-allele of interleukin-18 single-nucleotide polymorphism is a genetic risk factor for atopic asthma. The SAPALDIA Cohort Study. Clin. Exp. Allergy 2006, 36, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Bakr, N.M.; Awad, A.; Moustafa, E. Association of genetic variants in the interleukin-18 gene promoter with risk of hepatocellular carcinoma and metastasis in patients with hepatitis C virus infection. IUBMB Life 2018, 70, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Schabath, M.B.; Giuliano, A.R.; Thompson, Z.J.; Amankwah, E.K.; Gray, J.E.; Fenstermacher, D.A.; Jonathan, K.A.; Beg, A.A.; Haura, E.B. TNFRSF10B polymorphisms and haplotypes associated with increased risk of death in non-small cell lung cancer. Carcinogenesis 2013, 34, 2525–2530. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).