Comparative Proteomic Analysis of Roots from a Wild Eggplant Species Solanum sisymbriifolium in Defense Response to Verticillium dahliae Inoculation
Abstract
1. Introduction
2. Materials and Methods
2.1. Eggplant V. dahliae
2.2. Plant Materials and Inoculation
2.3. Measurement of Physiological and Biochemical Indexes
2.4. Protein Extraction
2.5. iTRAQ Labeling
2.6. LC-MS/MS Analysis
2.7. iTRAQ Protein Identification and Quantification
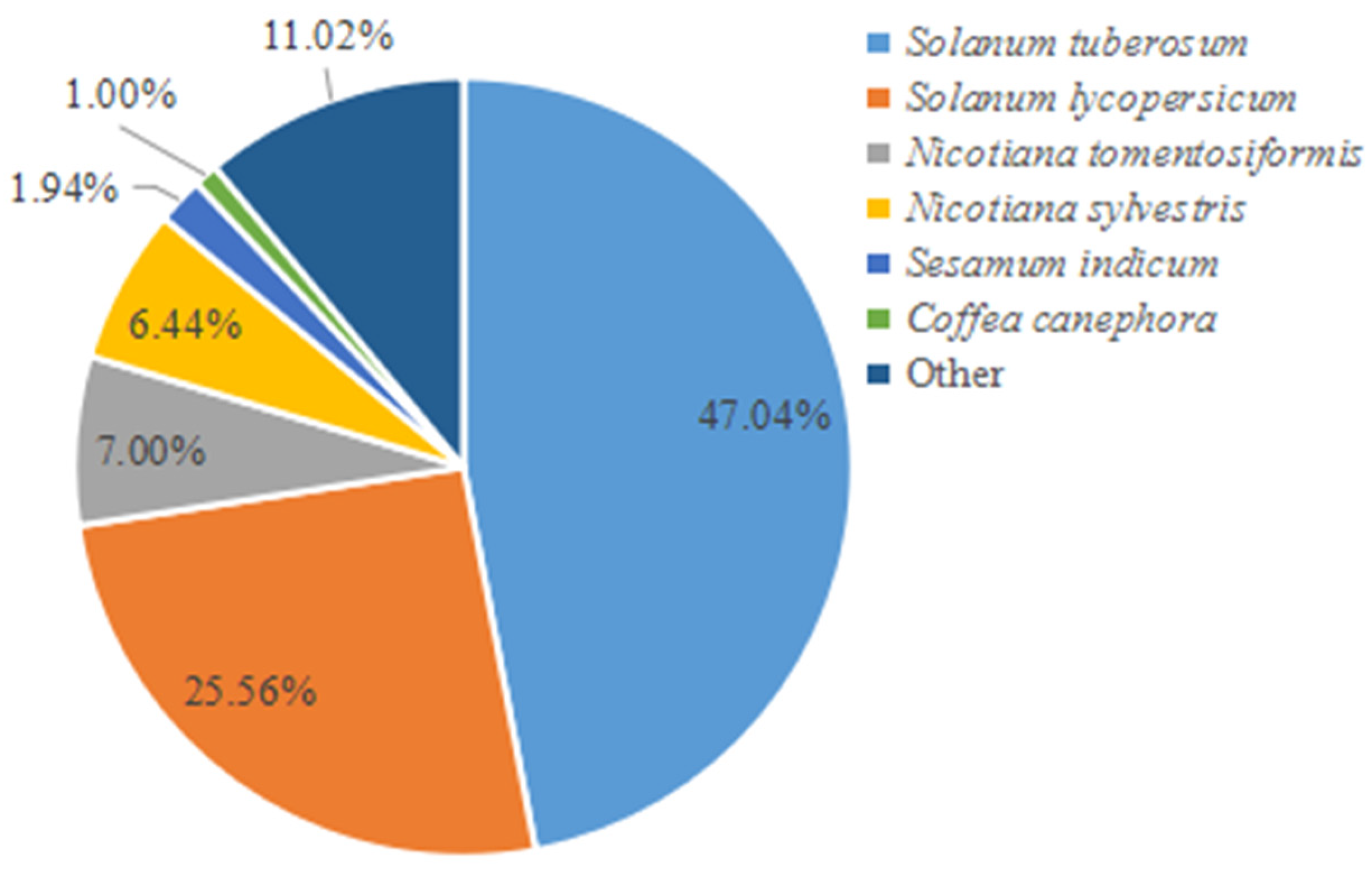
2.8. Bioinformatics Analysis
2.9. Targeted Protein Verified by Parallel Reaction Monitoring (PRM) Analysis
3. Results
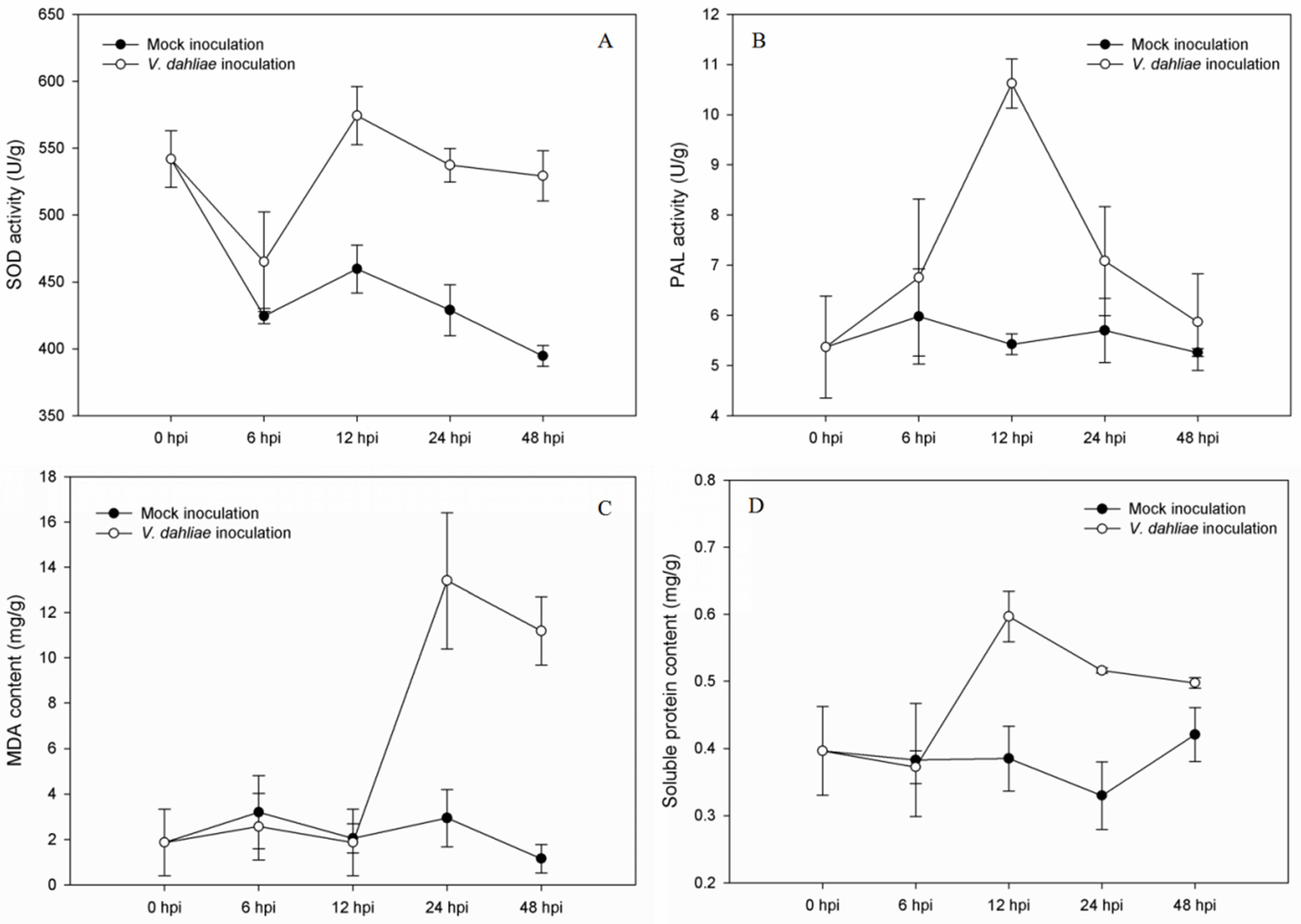
3.1. Analysis of Physiological Indexes in Roots of S. sisymbriifolium
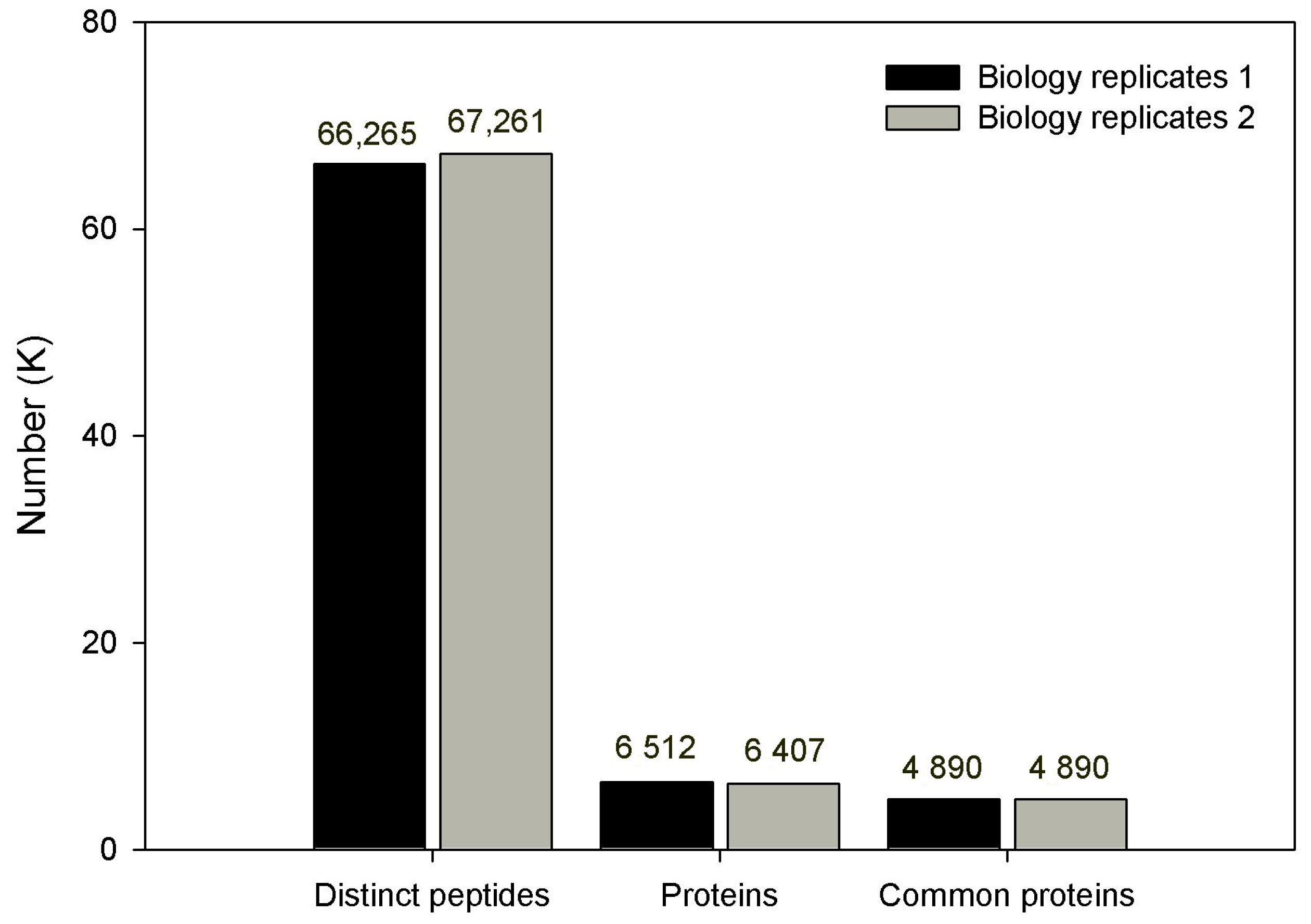
3.2. Proteins Identified by iTRAQ Analysis
3.3. Differentially Expressed Proteins (DEPs)
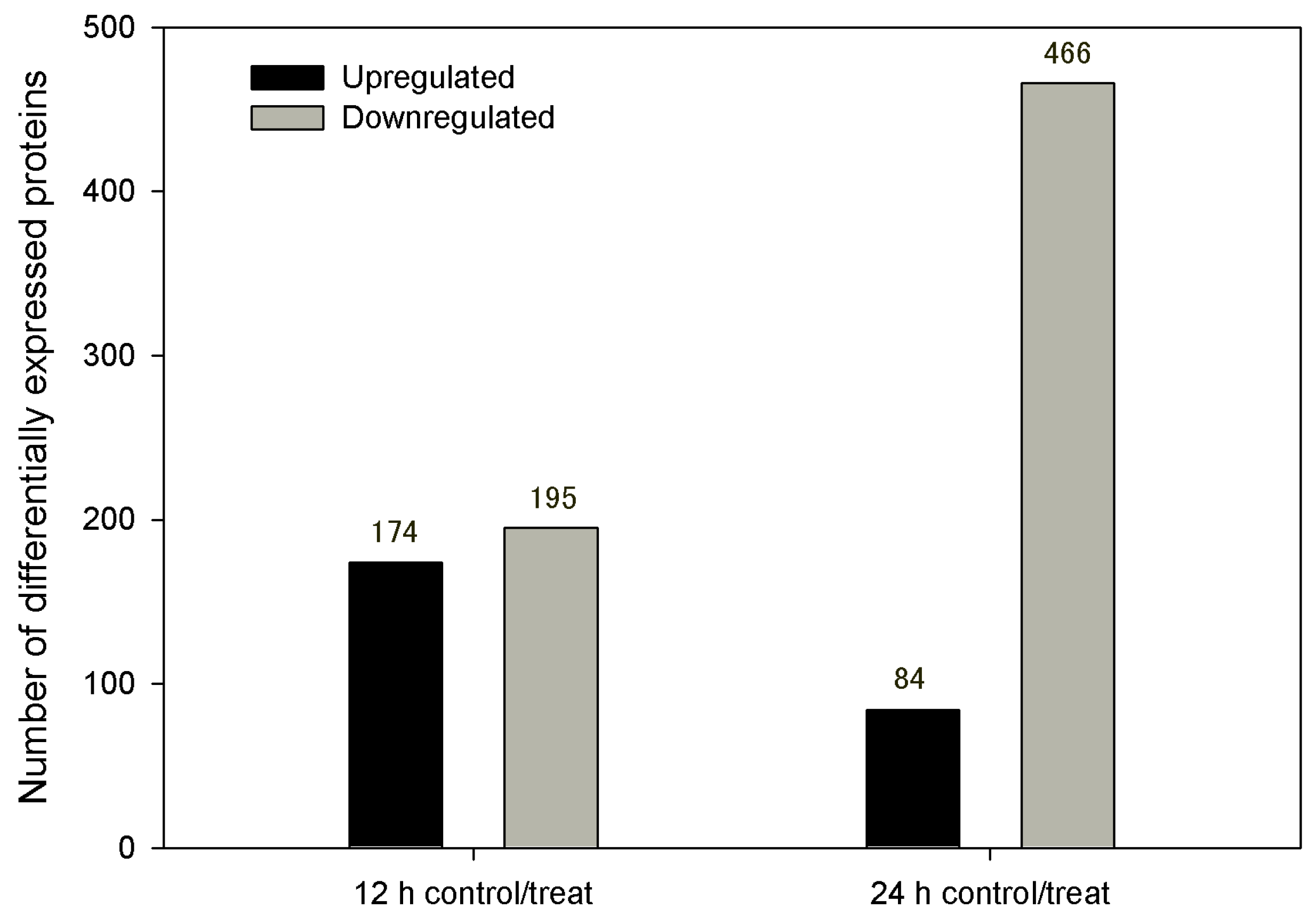
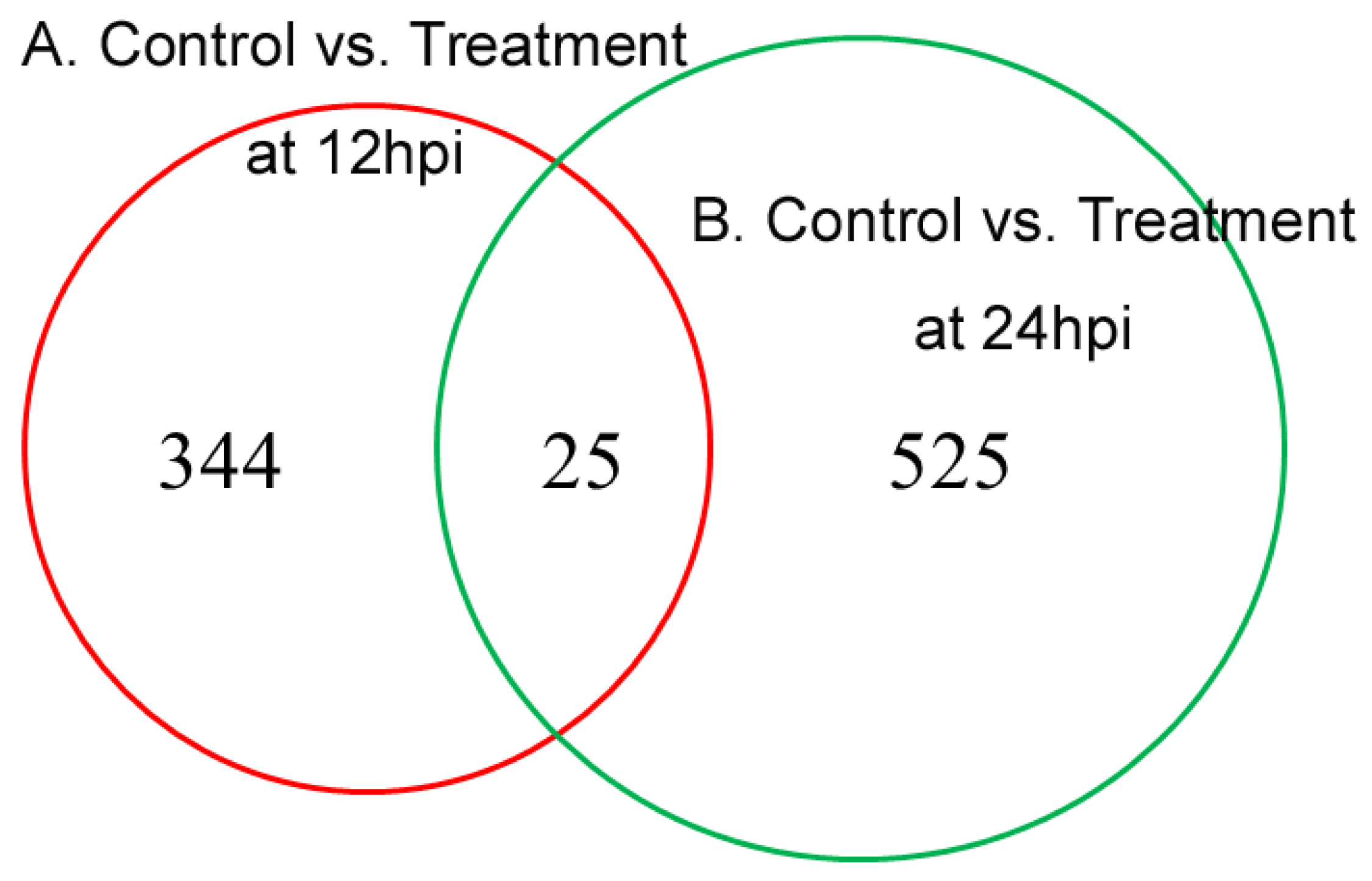
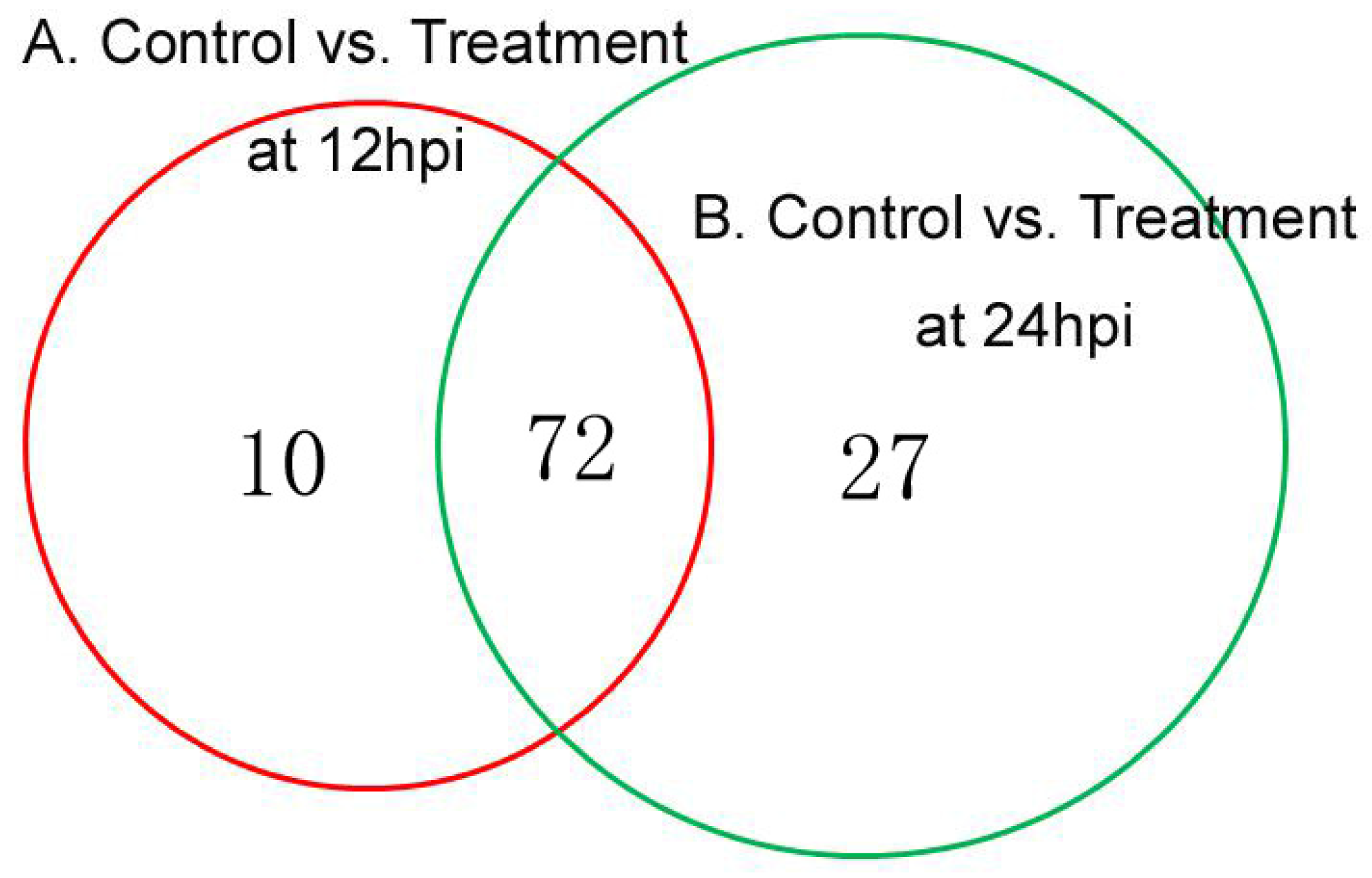
3.3.1. DEPs between Control/Treatment at 12 hpi and 24 hpi
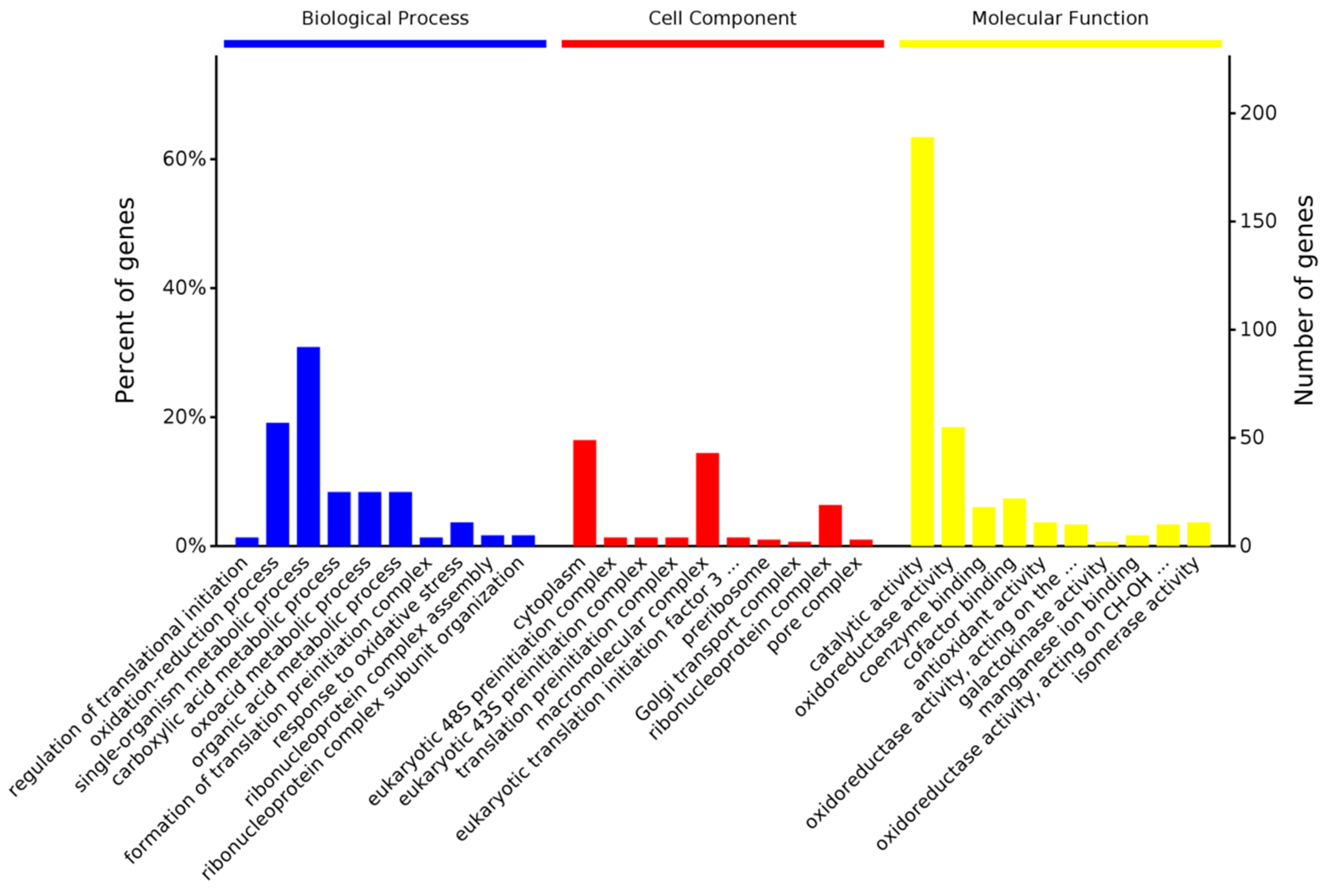
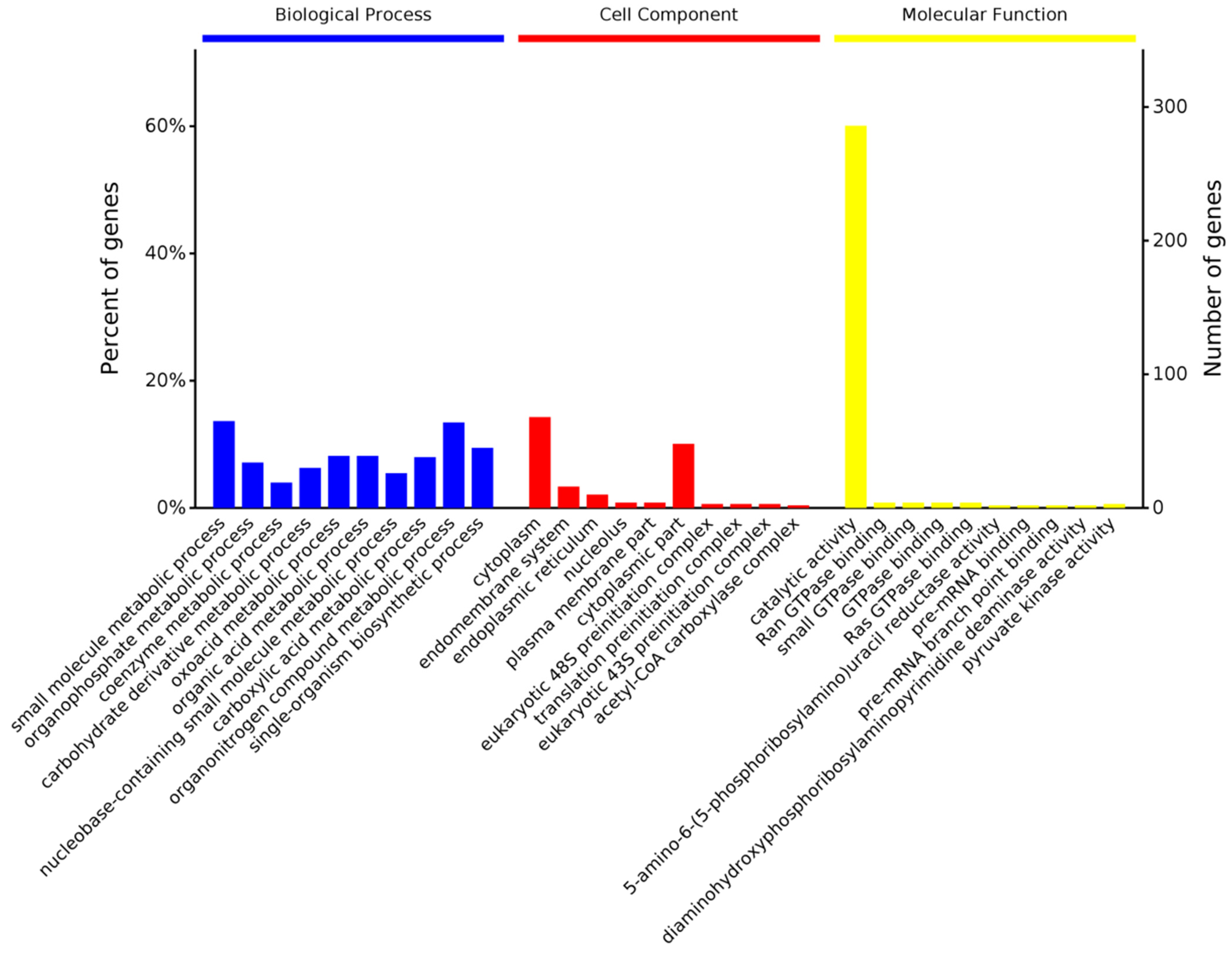
3.3.2. Gene Ontology Enrichment Analysis of DEPs
3.3.3. KEGG Pathway Enrichment Analysis of DEPs
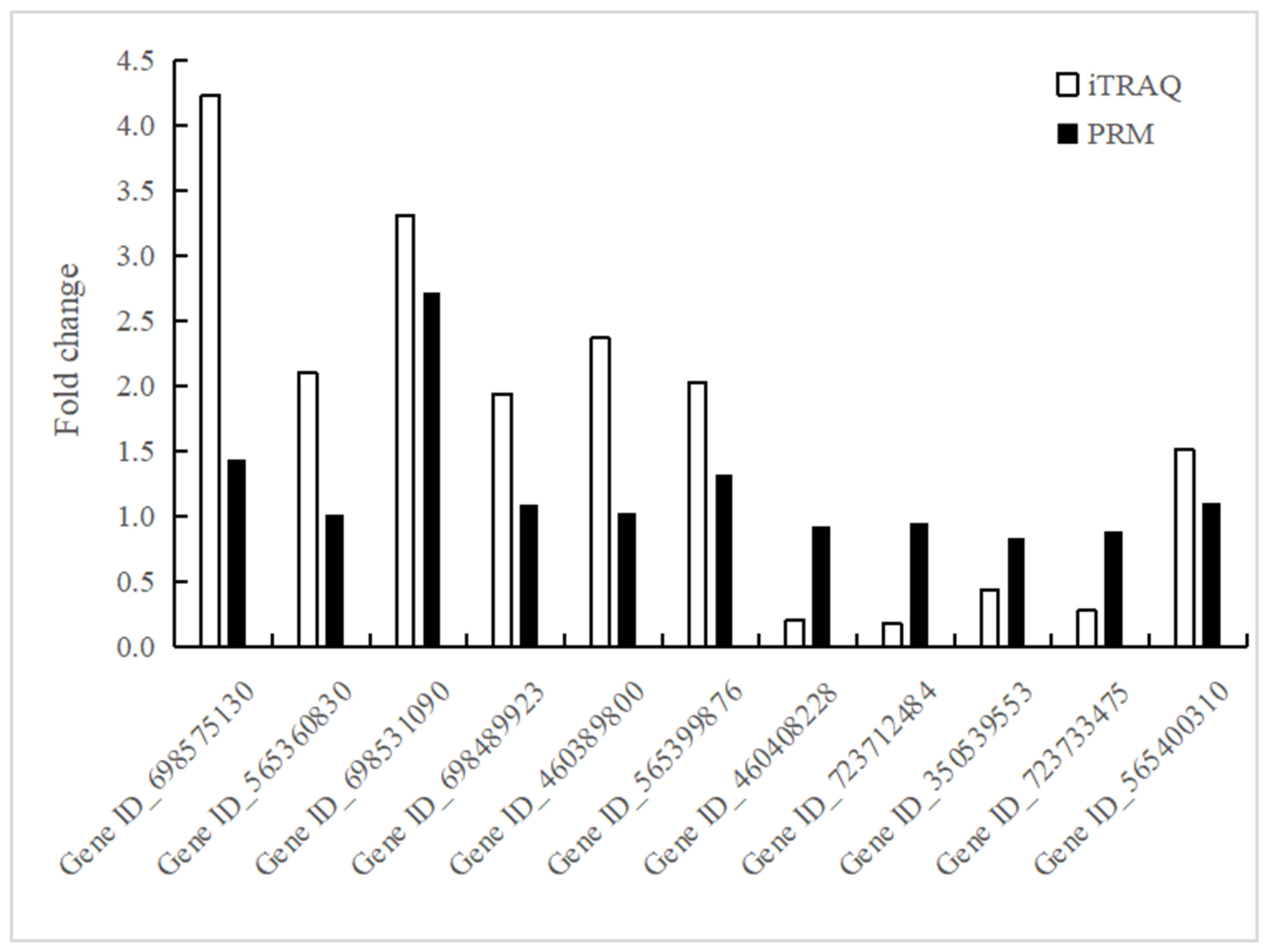
3.4. Validation of iTRAQ Data for Selected Proteins by PRM Analysis
4. Discussion
4.1. Wild Eggplant Species S. sisymbriifolium
4.2. Inoculation and Sampling
4.3. DEPs Analysis at 12 hpi and 24 hpi
4.4. Proteins Involved in the Defense Responses
4.4.1. Phenylpropanoid-Pathway-Related Proteins
4.4.2. Stress and Defense Response Proteins
4.4.3. Plant–Pathogen Interaction Pathway and Pathogenesis-Related (PR) Proteins
4.4.4. Proteins Involved in Cell Wall Organization and Reinforcement
4.4.5. Phytohormones-Pathway-Related Proteins
4.4.6. Other Defense-Related Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dubery, I.A.; Meyer, R. Specific binding of a Verticillium dahliae phytotoxin to protoplasts of cotton, Gossypium hirsutum. Plant Cell Rep. 1996, 15, 777–780. [Google Scholar] [CrossRef]
- Daunay, M.C. Eggplant. In Vegetables II; Prohens, J., Nuez, F., Eds.; Springer: New York, NY, USA, 2008; pp. 163–220. [Google Scholar]
- Wang, F.; Ma, Y.; Yang, C.; Zhao, P.; Yao, Y.; Jian, G.; Luo, Y.; Xia, G. Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 2011, 11, 4296–4309. [Google Scholar] [CrossRef]
- Fradin, E.F.; Thomma, B.P.H.J. Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 2006, 7, 71–86. [Google Scholar] [CrossRef]
- Yang, L.; Jue, D.; Li, W.; Zhang, R.; Chen, M.; Yang, Q. Identification of MiRNA from eggplant (Solanum melongena L.) by small RNA deep sequencing and their response to Verticillium dahliae infection. PLoS ONE 2013, 8, e72840. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cheng, Y.F.; Deng, C.; Ma, Y.; Wang, Z.; Chen, X.; Xue, L. Comparative transcriptome analysis of eggplant (Solanum melongena L.) and turkey berry (Solanum torvum Sw.): Phylogenomics and disease resistance analysis. BMC Genom. 2014, 15, 412. [Google Scholar] [CrossRef]
- Zhou, X.; Bao, S.; Liu, J.; Zhuang, Y. De novo sequencing and analysis of the transcriptome of the wild eggplant species Solanum aculeatissimum in response to Verticillium dahliae. Plant Mol. Biol. Rep. 2016, 34, 1193–1203. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Cheng, Y.; Chen, X. Transcriptome analysis reveals multiple signal network contributing to the Verticillium wilt resistance in eggplant. Sci. Hortic. 2019, 256, 108576. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Xue, J.; Liu, F.; Cheng, Y. Analysis of small RNAs from Solanum torvum Swartz by deep sequencing. Trop. Plant Biol. 2019, 12, 44–54. [Google Scholar] [CrossRef]
- Wu, L.Y.; Guo, Z.X.; Zeng, L.; Bao, R.; Li, Z.B.; Gong, Y.J. Resistance identification of Yunnan wild eggplant resources to Verticillium wilt. J. Plant Genet. Res. 2017, 18, 1046–1054. [Google Scholar]
- Wu, L.; Du, G.; Bao, R.; Li, Z.; Liu, F.; Gong, Y. De novo assembly and discovery of genes involved in the response of Solanum sisymbriifolium to Verticillium dahlia. Physiol. Mol. Biol. Plant. 2019, 25, 1009–1027. [Google Scholar] [CrossRef] [PubMed]
- Coumans, J.V.F.; Poljak, A.; Raftery, M.J.; Backhouse, D.; Pereg-Gerk, L. Analysis of cotton (Gossypium hirsutum) root proteomes during a compatible interaction with the black root rot fungus Thielaviopsis basicola. Proteomics 2009, 9, 335–349. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Zhang, J.; Cao, J.; Geng, A.; Wang, H.; Liu, H. Comprehensive proteomic characterization of the pectoralis major at three chronological ages in beijing-you chicken. Front. Physiol. 2021, 12, 658711. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Chai, Y.; Wang, J.; Lin, J.; Sun, X.; Sun, C.; Zuo, K.; Tang, K. cDNA cloning and characterization of the Ve homologue gene StVe from Solanum torvum Swartz. DNA Seq. 2004, 15, 88–95. [Google Scholar] [CrossRef]
- Liu, J.; Zheng, Z.; Zhou, X.; Feng, C.; Zhuang, Y. Improving the resistance of eggplant (Solanum melongena) to Verticillium wilt using wild species Solanum linnaeanum. Euphytica 2015, 201, 463–469. [Google Scholar] [CrossRef]
- Zhao, F.; Fang, W.; Xie, D.; Zhao, Y.; Tang, Z.; Li, W.; Nie, L.; Lv, S. Proteomic identification of differentially expressed proteins in Gossypium thurberi inoculated with cotton Verticillium dahliae. Plant Sci. 2012, 185, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.; Magalhães, B.S.; Souza, D.S.L.; Vasconcelos, E.; Silva, L.; Grossi-de-Sa, M.; Franco, O.; da Costa, P.; Rocha, T. Rooteomics: The challenge of discovering plant defense-related proteins in roots. Curr. Protein Pept. Sci. 2008, 9, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.J.; Reddy, M.S.S.; Wang, L. The phenylpropanoid pathway and plant defence—A genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- Passardi, F.; Cosio, C.; Penel, C.; Dunand, C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005, 24, 255–265. [Google Scholar] [CrossRef]
- Cheeseman, J.M. Hydrogen peroxide and plant stress: A challenging relationship. Plant Stress 2007, 1, 4–15. [Google Scholar]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.M.; Senthil-Kumar, M.; Tzin, V.; Mysore, K.S. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212, 29–37. [Google Scholar] [CrossRef]
- Smit, F.; Dubery, I.A. Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry 1997, 44, 811–815. [Google Scholar] [CrossRef]
- Swain, S.; Singh, N.; Nandi, A.K. Identification of plant defence regulators through transcriptional profiling of Arabidopsis thaliana cdd1 mutant. J. Biosci. 2015, 40, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.H.; Bauw, G.; Welinder, K.G.; Montagu, M.V.; Boerjan, W. Purification and characterization of peroxidases correlated with lignification in poplar xylem. Plant Physiol. 1998, 118, 125–135. [Google Scholar] [CrossRef]
- Zenoni, S.; Reale, L.; Tornielli, G.B.; Lanfaloni, A.L.; Porceddu, C.A. Downregulation of the Petunia hybrida a-expansin gene PhEXP1 reduces the amount of crystalline cellulose in cell walls and leads to phenotypic changes in petal limbs. Plant Cell 2004, 16, 295–308. [Google Scholar] [CrossRef]
- Philippe, F.; Pelloux, J.; Rayon, C. Plant pectin acetylesterase structure and function: New insights from bioinformatic analysis. BMC Genom. 2017, 18, 456. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Oskar, M.; Buchwald, W.; Nawrot, R. Plant defense responses against viral and bacterial pathogen infections. Focus on RNA-binding proteins (RBPs). Herba Polonica 2014, 60, 60–73. [Google Scholar] [CrossRef]
| No. | Enriched KEGG Pathways (ID) | DEPs | p-Value | |
|---|---|---|---|---|
| Up | Down | |||
| 1 | Selenocompound metabolism (sly00450) | 2 | 2 | 1.36 × 10−3 |
| 2 | Ubiquinone and other terpenoid-quinone biosynthesis (sly00130) | 3 | 3 | 2.03 × 10−3 |
| 3 | Fatty acid biosynthesis (sly00061) | 4 | 2 | 3.16 × 10−3 |
| 4 | Lysine biosynthesis (sly00300) | 1 | 2 | 4.75 × 10−3 |
| 5 | Citrate cycle (TCA cycle) (sly00020) | 1 | 5 | 6.74 × 10−3 |
| 6 | Biosynthesis of secondary metabolites (sly01110) | 22 | 25 | 1.25 × 10−2 |
| 7 | Fatty acid metabolism (sly01212) | 4 | 3 | 1.58 × 10−2 |
| 8 | Phenylpropanoid biosynthesis (sly00940) | 6 | 6 | 1.62 × 10−2 |
| 9 | Ascorbate and aldarate metabolism (sly00053) | 1 | 4 | 1.89 × 10−2 |
| 10 | Nucleotide excision repair (sly03420) | 1 | 4 | 2.71 × 10−2 |
| 11 | Phenylalanine metabolism (sly00360) | 2 | 3 | 2.90 × 10−2 |
| 12 | Monobactam biosynthesis (sly00261) | 1 | 1 | 3.43 × 10−2 |
| 13 | Glycolysis/Gluconeogenesis (sly00010) | 7 | 1 | 4.04 × 10−2 |
| 14 | Ribosome biogenesis in eukaryotes (sly03008) | 2 | 4 | 4.75 × 10−2 |
| 15 | Biosynthesis of amino acids (sly01230) | 7 | 5 | 4.96 × 10−2 |
| No. | Enriched KEGG Pathways (ID) | DEPs | p-Value | |
|---|---|---|---|---|
| Up | Down | |||
| 1 | Glycolysis/Gluconeogenesis (sly00010) | 1 | 16 | 8.93 × 10−5 |
| 2 | Biosynthesis of secondary metabolites (sly01110) | 13 | 63 | 4.46 × 10−4 |
| 3 | Linoleic acid metabolism (sly00591) | 2 | 3 | 1.40 × 10−3 |
| 4 | Pyruvate metabolism (sly00620) | 2 | 9 | 4.57 × 10−3 |
| 5 | Cyanoamino acid metabolism (sly00460) | 2 | 5 | 4.82 × 10−3 |
| 6 | Fructose and mannose metabolism (sly00051) | 2 | 7 | 5.15 × 10−3 |
| 7 | Carbon metabolism (sly01200) | 4 | 18 | 7.78 × 10−3 |
| 8 | Phenylpropanoid biosynthesis (sly00940) | 3 | 14 | 1.09 × 10−2 |
| 9 | Amino sugar and nucleotide sugar metabolism (sly00520) | 3 | 10 | 1.12 × 10−2 |
| 10 | Metabolic pathways (sly01100) | 15 | 97 | 1.70 × 10−2 |
| 11 | Purine metabolism (sly00230) | 1 | 12 | 2.12 × 10−2 |
| 12 | Protein processing in endoplasmic reticulum (sly04141) | 1 | 17 | 2.39 × 10−2 |
| 13 | Alanine, aspartate, and glutamate metabolism (sly00250) | 2 | 4 | 3.20 × 10−2 |
| 14 | Selenocompound metabolism (sly00450) | 0 | 3 | 4.14 × 10−2 |
| 15 | Biosynthesis of amino acids (sly01230) | 2 | 15 | 4.59 × 10−2 |
| 16 | Base excision repair (sly03410) | 0 | 5 | 4.72 × 10−2 |
| 17 | Nonhomologous end-joining (sly03450) | 0 | 2 | 4.99 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, L.; Gui, M.; Liu, J.; Cheng, J.; Li, Z.; Bao, R.; Chen, X.; Gong, Y.; Du, G. Comparative Proteomic Analysis of Roots from a Wild Eggplant Species Solanum sisymbriifolium in Defense Response to Verticillium dahliae Inoculation. Genes 2023, 14, 1247. https://doi.org/10.3390/genes14061247
Wu L, Gui M, Liu J, Cheng J, Li Z, Bao R, Chen X, Gong Y, Du G. Comparative Proteomic Analysis of Roots from a Wild Eggplant Species Solanum sisymbriifolium in Defense Response to Verticillium dahliae Inoculation. Genes. 2023; 14(6):1247. https://doi.org/10.3390/genes14061247
Chicago/Turabian StyleWu, Liyan, Min Gui, Jiaxun Liu, Jie Cheng, Zhibin Li, Rui Bao, Xia Chen, Yaju Gong, and Guanghui Du. 2023. "Comparative Proteomic Analysis of Roots from a Wild Eggplant Species Solanum sisymbriifolium in Defense Response to Verticillium dahliae Inoculation" Genes 14, no. 6: 1247. https://doi.org/10.3390/genes14061247
APA StyleWu, L., Gui, M., Liu, J., Cheng, J., Li, Z., Bao, R., Chen, X., Gong, Y., & Du, G. (2023). Comparative Proteomic Analysis of Roots from a Wild Eggplant Species Solanum sisymbriifolium in Defense Response to Verticillium dahliae Inoculation. Genes, 14(6), 1247. https://doi.org/10.3390/genes14061247





