Abstract
HSPA8 is involved in many stroke-associated cellular processes, playing a pivotal role in the protein quality control system. Here we report the results of the pilot study aimed at determining whether HSPA8 SNPs are linked to the risk of ischemic stroke (IS). DNA samples from 2139 Russians (888 IS patients and 1251 healthy controls) were genotyped for tagSNPs (rs1461496, rs10892958, and rs1136141) in the HSPA8 gene using probe-based PCR. SNP rs10892958 of HSPA8 was associated with an increased risk (risk allele G) of IS in smokers (OR = 1.37; 95% CI = 1.07–1.77; p = 0.01) and patients with low fruit and vegetable consumption (OR = 1.36; 95% CI = 1.14–1.63; p = 0.002). SNP rs1136141 of HSPA8 was also associated with an increased risk of IS (risk allele A) exclusively in smokers (OR = 1.68; 95% CI = 1.23–2.28; p = 0.0007) and in patients with a low fruit and vegetable intake (OR = 1.29; 95% CI = 1.05–1.60; p = 0.04). Sex-stratified analysis revealed an association of rs10892958 HSPA8 with an increased risk of IS in males (risk allele G; OR = 1.30; 95% CI = 1.05–1.61; p = 0.01). Thus, SNPs rs10892958 and rs1136141 in the HSPA8 gene represent novel genetic markers of IS.
1. Introduction
Ischemic stroke is the most frequent type of brain attack, the leading cause of death worldwide [1]. IS is considered a multifactorial disease due to the involvement of genetic and environmental factors [2]. A lot of studies have been done to evaluate the genetic nature of IS [3,4,5,6,7] and IS-related modifiable risk factors, like hypertension [8,9], atherosclerosis [10,11], thrombosis [12]. However, disclosure of genetic correlates of IS remains an important task on the way to predicting and fighting the disease.
In ischemic conditions, a cell relies on multiple interplaying ensembles preserving macromolecules from degradation caused by oxygen deprivation, acidosis and oxidation. Heat-shock proteins (HSPs) are a type of chaperones that are considered to be the most conservative molecular machines providing homeostasis in ischemic conditions. HSPs encourage the refolding of misfolded or immature proteins and prevent aggregation of misfolded proteins, which are involved in apoptosis, necrosis, and inflammation, which determines their significant role under pathological conditions, in particular ischemia-reperfusion [13]. Accordingly, a large body of evidence has shown HSPs to be among the most impactful players in response to ischemia as well as in recovering after ischemia-reperfusion injury [14].
The HSPA8 gene encodes a member of the heat shock protein 70 (HSP70) family known as the heat shock cognate 71 kDa protein (HSC70). HSP70 is known for its neuroprotective functions [15,16,17,18], and as well as other members, HSP70 maintains cellular homeostasis [19]. Precise roles of HSC70 include folding and transport of newly synthesised polypeptides and assembly of protein complexes [20,21,22], regulation of mitochondrial import [23], and the ER-associated degradation quality control system [24]. According to our knowledge, no research has looked into the relationship between HSPA8 genetic variants and IS. Therefore, the objective of our study was to challenge whether polymorphisms of HSPA8 gene are related to IS risk.
2. Materials and Methods
Figure 1 depicts the research design’s general layout.
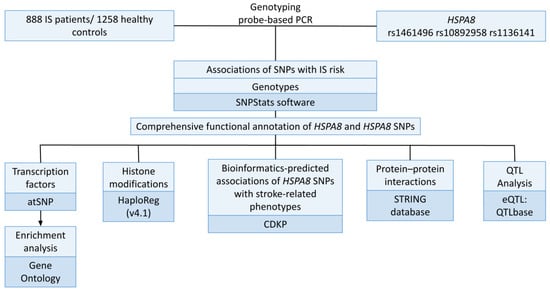
Figure 1.
Study design.
For the study, a total of 2139 unrelated Russians (888 IS patients and 1251 healthy individuals) from Central Russia were enrolled. The study protocol was approved by the Kursk State Medical University’s Ethical Review Committee. All participants provided written informed consent prior to being accepted into the study, with the following inclusion requirements: self-declared Russian ancestry and a birthplace in Central Russia. Table 1 provides the baseline and clinical characteristics of the study cohort.

Table 1.
Baseline and clinical characteristics of the studied groups.
The participants were enrolled in the study in two time periods: between 2010 and 2012 at the Kursk Emergency Medicine Hospital’s neurology clinics [25] and between 2015 and 2017 at the Regional Vascular Centre of Kursk Regional Clinical Hospital [26]. A team of qualificated neurologists assessed each case. The diagnosis of IS during the acute phase of stroke was confirmed using the findings of brain computed tomography and/or magnetic resonance imaging. The patients were recruited consecutively. Intracerebral hemorrhage, hemodynamic or dissection-related stroke, traumatic brain injury, hepatic or renal failure, autoimmune, oncological, or other disorders that can produce an acute cerebrovascular event were considered exclusion criteria. All of the IS patients had a history of hypertension and were receiving antihypertensive medications.
According to the WHO recommendations, low fruit and vegetable consumption was defined as an intake of less than 400 g per day. Normal consumption of fresh vegetables and fruits was defined as an intake of 400 g or more in 3–4 servings per day (excluding potatoes and other starchy tubers). [27]. The control group was made up of healthy volunteers with normal blood pressure who did not take antihypertensive therapy and did not exhibit any clinical symptoms of cardiovascular, cerebrovascular, or other major diseases. Healthy persons were included in the control group if their systolic blood pressure was less than 130 mm Hg and their diastolic blood pressure was less than 85 mm Hg on at least three different measurements. In the Kursk region, controls were chosen from the hospitals in Kursk during routine medical exams at public institutions and industrial businesses [28,29]. This group was recruited from the same population and during the same period.
The following criteria were used to choose the SNPs: they had to be tagging, have a minor allele frequency of at least 0.05 in the European population, and be distinguished by a high regulatory potential. The bioinformatic tools LD TAG SNP Selection (TagSNP) and SNPinfo Web Server (https://snpinfo.niehs.nih.gov/(accessed on 15 March 2022)), which were utilised to choose SNPs based on the reference haplotypic structure of the Caucasian population (CEU) of the project HapMap, showed that the HSPA8 (heat shock protein family A (Hsp70) member 8, ID: 3312) gene contains three tag SNPs (rs1461496, rs10892958, and rs1136141). SNPs rs1461496 and rs1136141 are located in the 3 prime UTRs; rs10892958 is located in the intron.
Several bioinformatic approaches were used to examine the HSPA8 tag SNPs’ regulatory potential. SNP FuncPred reports that the rs10892958 has a Regulatory Potential Score of 0.154 and the rs1136141 has a Regulatory Potential Score of 0.132 (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html (accessed on 15 February 2023)) [30].
The RegulomeDB instrument revealed that rs1461496 is characterized by a regulatory coefficient of 3a (TF binding + any motif + DNase peak); rs10892958 and rs1136141 are characterized by a regulatory coefficient of 4 (TF binding + DNase peak) (https://regulomedb.org/regulome-search/ (accessed on 15 February 2023)) [31].
According to the HaploReg (v4.1) database, these SNPs have properties such as enhancer histone tags in various tissues (rs1461496), regions of hypersensitivity for DNAse 1 (rs1461496, rs10892958, rs1136141), binding sites for regulatory proteins (rs10892958, rs1136141), and DNA regulatory motifs (rs10892958, rs1136141) (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php (accessed on 15 February 2023)) [32].
According to information from the NCBI source (https://www.ncbi.nlm.nih.gov/snp/ (accessed on 15 February 2023)), these genetic variants are identified by an average frequency of the minor alleles in European populations of >0.05. As a result, all three SNPs were chosen for our investigation, which satisfied the requirements for study inclusion.
2.1. Genetic Analysis
The Laboratory of Genomic Research at the Research Institute for Genetic and Molecular Epidemiology of Kursk State Medical University (Kursk, Russia) performed genotyping. Up to 5 mL of venous blood from each participant was collected from a cubital vein, put into EDTA-coated tubes, and kept at −20 °C until it was processed. Defrosted blood samples were used to extract genomic DNA using the standard methods of phenol/chloroform extraction and ethanol precipitation. The purity, quality, and concentration of the extracted DNA solution were assessed using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Genotyping of the SNPs was done using allele-specific probe-based polymerase chain reaction (PCR) according to the protocols designed in the Laboratory of Genomic Research. The Primer3 software was used for primer design [33]. Table S1 lists the primers and probes used for genotyping. A real-time PCR procedure was performed in a 25 mL reaction solution containing 1.5 units of Hot Start Taq DNA polymerase (Biolabmix, Novosibirsk, Russia), approximately 1 μg of DNA, 0.25 μM each of the primers 0.25 μM each primer, 250 μM dNTPs, 3.0 mM MgCl2 (for rs1136141 and rs1461496), 4.5 mM MgCl2 (for rs10892958); 1×PCR buffer (67 mM Tris-HCl, pH 8.8, 16.6 mM (NH4)2SO4, 0.01% Tween-20). The PCR procedure comprised an initial denaturation for 10 minutes at 95°C, followed by 39 cycles of 92 °C for 30 s and 65 °C, 61.5 °C, 51 °C for 1 min (for rs10892958, rs1136141 and rs1461496 respectively). Figures S1–S3 show allelic discrimination plots for HSPA8 assays designed for this study. 10% of the DNA samples were genotyped twice, blinded to the case-control status, in order to assure quality control. Over 99% of the data were concordant.
2.2. Statistical and Bioinformatic Analysis
The statistical power for the study was determined using the genetic association study power calculator, a tool available online at http://csg.sph.umich.edu/abecasis/gas_power_calculator/ (accessed on 16 February 2023). Given a sample size of 888 cases and 1251 controls, an association study between the HSPA8 gene polymorphisms and IS risk might identify a genotype relative risk of 1.20–1.32 assuming 0.80 power and a 5% type I error (α = 0.05).
The STATISTICA software (v13.3, Santa Clara, California, USA) was used to conduct all statistical analyses. The Shapiro-Wilk test was used to determine the normality of the distribution of quantitative data. The median (Me) and first and third quartiles [Q1 and Q3] were used to express the biochemical parameters and body mass index because they deviated from the normal distribution. The Pearson’s chi-squared test with Yates’s continuity correction was used to determine the statistical significance of differences for categorical variables.
Using Fisher’s exact test, the genotype distributions were evaluated for Hardy-Weinberg equilibrium compliance. Using the SNPStats software (https://www.snpstats.net/start.htm (accessed on 18 January 2023)), the genotype frequencies in the study groups and their relationships to disease risk were analyzed [34]. For the analysis of associations of genotypes, additive models were used. The associations of genotypes in the whole group of IS patients and controls were adjusted for age, gender, and smoking status. When the environmental risk factor in the control group was unknown, relationships were examined based on whether the risk factor was present or absent in the IS group relative to the entire control group. The Bonferroni correction was also used in this instance.
The functional effects of HSPA8 SNPs were examined using the bioinformatics resources listed below:
- The expression quantitative trait loci (eQTLs) in the brain, whole blood, and blood vessels have been evaluated using the bioinformatic tool QTLbase (http://www.mulinlab.org/qtlbase/index.html (accessed on 21 February 2023)) [35].
- The STRING database’s bioinformatic tools were utilised to analyse the main functional partners of HSPA8 (https://string-db.org/ (accessed on 21 February 2023)) [36]. Additionally, the STRING database was used to assess biological processes and molecular functions data describing interactions between HSPA8 and its functionally significant partner proteins. For the interpretation of interactions only experimentally confirmed data was used.
- The effect of HSPA8 SNPs on the binding of transcription factors (TFs) to DNA was assessed using the atSNP Function Prediction online tool (http://atsnp.biostat.wisc.edu/search (accessed on 21 February 2023)) [37]. Based on a positional weight matrix-based calculation of the impact of SNPs on how well TFs interact with DNA, certain TFs were added.
- The online Gene Ontology tool was used to conduct the subsequent study of the potential joint involvement of TFs linked with the reference and SNP alleles in overrepresented biological processes that are related to the mechanisms of IS (http://geneontology.org/ (accessed on 21 February 2023)) [38]. As functional groups, we employed biological processes governed by transcription factors connected to HSPA8 SNPs.
- HaploReg (v4.1), a bioinformatics tool (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php (accessed on 20 February 2023)) was used to evaluate the relationships between HSPA8 SNPs and the following histone modifications that mark promoters and enhancers: acetylation of the lysine residues at positions 27 and 9 of the histone H3 protein, as well as mono-methylation at position 4 of the histone H3 protein (H3K4me1) and tri-methylation at position 4 of the histone H3 protein (H3K4me3). Additionally, this tool has been employed to examine the localization of SNPs in DNase hypersensitive areas, regulatory motif regions, and locations that bind to regulatory proteins [32].
- The interpretation of environment-associated correlates of HSPA8 polymorphism was done using the Comparative Toxicogenomics Database (CTD) resource at http://ctdbase.org (accessed on 24 February 2023) [39]. Based on data gathered from internationally published scientific studies, CTD offers the capability to investigate particular interactions between genes and chemicals in vertebrates and invertebrates. Using this method, bidirectional interactions comprising a single chemical and a single gene or protein were examined.
- The Cerebrovascular Disease Knowledge Portal (CDKP) is available at https://cd.hugeamp.org/ (accessed on 24 February 2023) was employed for a bioinformatic investigation of the relationships between HSPA8 SNPs and stroke-related traits, intermediate phenotypes, and risk factors for IS (such as blood pressure, heart rate, etc.) [40].
3. Results
3.1. Bioinformatic Analysis of the HSPA8 Gene
Brain tissues, blood vessels, and whole blood have high levels of HSPA8 gene expression. HSPA8 gene expression levels (MeTPM) range from 197.4 to 662.8 in brain tissues, from 327.8 to 509.0 in blood vessels, and are 177.0 in whole blood (Figure 2).
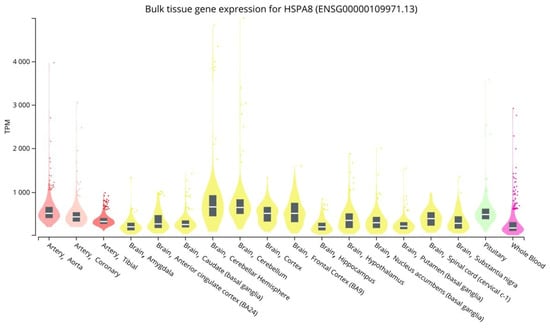
Figure 2.
HSPA8 expression levels in vessels, brain, and peripheral blood (https://gtexportal.org/home/ (accessed on 15 February 2023)).
Protein–Protein Interactions
Ten proteins having the most prominent interactions with HSPA8 were found after searching for the primary functional partners of HSPA8 using the STRING database data: STIP1, BAG1, BAG2, STUB1, HSP90AA1, BAG3, HSPA4, HSPBP1, LRRK2, DNAJA1 (Figure 3, Table S2).
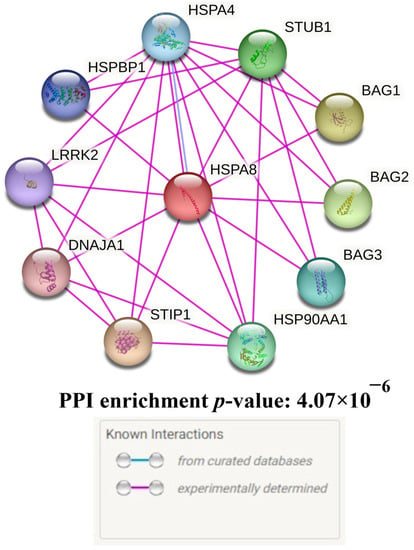
Figure 3.
Predicted functional partners of HSPA8.
HSPA8 and its key functional partners participate in 40 GO, predominantly reflecting proteostasis (for example, “protein folding” (GO = 0006457, FDR = 8.40 × 10−8), “response to unfolded protein” (GO = 0006986, FDR = 8.04 × 10−7), “regulation of protein ubiquitination” (GO = 0031396, FDR = 1.68 × 10−6), “regulation of protein stability” (GO = 0031647, FDR = 8.40 × 10−6), and “regulation of protein ubiquitination” (GO = 0031396, FDR = 0.004)), heat shock (for example, “regulation of cellular response to heat” (GO = 1900034, FDR = 1.50 × 10−6) and “response to heat” (GO = 0009408, FDR = 1.70 × 10−4)) and autophagy (“chaperone-mediated autophagy” (GO = 0061684, FDR = 1.71 × 10−7) and “autophagy” (GO: 0006914, FDR = 1.70 × 10−4)) (the full list of biological processes of HSPA8 and its main functional partners is presented in Table S3).
The presented data provide additional confirmation of the significant role of HSPA8 in protein homeostasis, heat shock, and autophagy. Additionally, the data confirm the potentially high pathogenetic significance of this gene in relation to the risk of developing IS.
3.2. HSPA8 SNPs and the Ischemic Stroke Risk: An Analysis of Associations
Table S4 displays the genotype frequencies of the HSPA8 variants (rs1461496, rs10892958, and rs1136141) in the study groups. In the control group, the distribution of genotype frequencies for each of the investigated SNPs matched the Hardy-Weinberg equilibrium (p > 0.05). The observed heterozygosity for rs1136141 (0.2280) in IS patients, nonetheless, was lower than expected (0.2495); p < 0.05 (Table S4).
There were no links between IS risk and HSPA8 SNPs in the analysis of the entire cohort (Table 2). Following sub-group analysis, it was revealed that rs10892958 was linked to a higher risk of IS only in males (risk allele G; OR = 1.30; 95% CI = 1.05–1.61; p = 0.01). The genetic variants rs10892958 (risk allele G; OR = 1.37; 95% CI = 1.07–1.77; p = 0.01) and rs1136141 (risk allele A; OR = 1.68; 95% CI = 1.23–2.28; p = 7.0 × 10−4) were associated with the development of IS only in smokers. It is noteworthy that the associations between rs10892958 (risk allele G; OR = 1.36; 95% CI = 1.14–1.63; p = 9.0 × 10−4; Pbonf = 0.002) and rs1136141 (risk allele A; OR = 1.29; 95% CI = 1.05–1.60; p = 0.02; Pbonf = 0.04) with IS risk were observed only under the condition of low fruit and vegetable intake (Table 2; Table S5).

Table 2.
Analysis of relationships between HSPA8 SNPs IS risk.
3.3. Functional Annotation of HSPA8 SNPs
3.3.1. QTL-Effects
Table 3 displays the results of the eQTL analysis for the HSPA8 SNPs. The QTLbase browser data show that the risk allele G rs10892958 is associated with decreased expression of HSPA8 in Brain-Hippocampus; risk allele A rs1136141 is related to reduced expression of HSPA8 and increased expression of CLMP in Brain-Hippocampus (Table 3).

Table 3.
Cis-eQTL-mediated effects of HSPA8 SNPs (http://www.mulinlab.org/qtlbase (accessed on 21 February 2023)).
3.3.2. Histone Modifications
A further examination revealed the substantial effect of IS-related HSPA8 SNPs on histone tags. SNPs rs10892958 and rs1136141 are located in the region of DNA binding to H3K4me1 in brain tissues (rs10892958), blood (rs10892958, rs1136141), as well as H3K4me3 in the brain tissues and blood (rs10892958, rs1136141). The effect of these histone tags is increased by the H3K27ac, marking enhancers in blood cells and all brain tissues (rs10892958, rs1136141), as well as the H3K9ac, marking promoters in blood cells and all brain tissues, except the Brain Hippocampus Middle (rs10892958, rs1136141). It is noteworthy that the SNPs rs10892958 and rs1136141 are also localized in DNA regions hypersensitive to DNase-1 in the blood (Table 4).

Table 4.
The impact of HSPA8 SNPs on histone tags in various tissues.
Moreover, using the resource HaploReg (v4.1), it was found that these SNPs are highly regulated by regulatory proteins: rs1136141 is located in the binding site DNA with 23 regulatory proteins: CMYC, ELF1, ELK4, GTF2B, GTF2F1, HEY1, MXI1, NELFE, NFKB, OCT2, P300, POL2, POL24H8, POL2B, POL2S2, POU2F2, SIN3AK20, TAF1, TAF7, TBP, TCF12, TCF4, YY1. SNP rs10892958 is localized at the DNA binding site with 6 regulatory proteins: CMYC, OCT2, POL2, POL24H8, POU2F2, TAF7.
3.3.3. Analysis of Transcription Factors
The risk allele G rs10892958 of HSPA8 generates DNA binding sites for 31 TFs that are simultaneously implicated in 6 overrepresented GO regulating oxidative stress and neurogenesis: “regulation of transcription from RNA polymerase II promoter in response to oxidative stress” (GO = 0043619; FDR = 0.02), “glial cell fate commitment” (GO = 0021781; FDR = 0.04), “regulation of oligodendrocyte differentiation” (GO = 0048713; FDR = 0.01), “neuron fate commitment” (GO = 0048663; FDR = 0.0009), “negative regulation of neuron differentiation” (GO = 0045665; FDR = 0.04), and “oligodendrocyte differentiation” (GO = 0048709; FDR = 0.04) (Table S6). Meanwhile, protective allele C rs10892958 HSPA8 provides DNA binding regions for 53 TFs jointly involved in the regulation of proatherosclerotic mechanisms, inflammation, cell signaling, neurogenesis, and apoptosis: “macrophage derived foam cell differentiation” (GO = 0010742; FDR = 0.0149), “cellular response to transforming growth factor beta stimulus” (GO = 0071560; FDR = 0.02), “cellular response to cytokine stimulus” (GO = 0071345; FDR = 0.008), “negative regulation of interferon-beta production” (GO = 0032688; FDR = 0.035), “glial cell fate commitment” (GO = 0021781; FDR = 0.03), and “regulation of apoptotic process” (GO = 0042981; FDR = 0.01) (Table S6).
Risk allele A, rs1136141, of HSPA8 creates DNA binding sites for 43 TFs, which together participate in 5 overrepresented GOs co-controlling “positive regulation of CD8-positive, alpha-beta T cell differentiation” (GO = 0043378; FDR = 0.007), “regulation of blood vessel endothelial cell migration” (GO = 0043535; FDR = 0.04), “positive regulation of angiogenesis” (GO = 0045766; FDR = 0.02), “response to growth factor” (GO = 0070848; FDR = 0.015), and “negative regulation of apoptotic process” (GO = 0043066; FDR = 0.04). Meanwhile, no common GO was defined for 19 TFs, binding with the protective allele G (Table S7).
3.3.4. Bioinformatic Analysis of the Associations of HSPA8 SNPs with IS-Related Phenotypes
According to the bioinformatic resource CDKP, which combines and analyzes the results of genetic associations from the largest consortiums for the study of cerebrovascular diseases, risk allele A rs1136141 of HSPA8 is associated with increases in systolic blood pressure, heart rate, peripheral artery disease in ever-smokers and stroke (TOAST, other-determined) (Table 5).

Table 5.
Results of aggregated analyses of associations between HSPA8 SNPs and cerebrovascular diseases/ their intermediate phenotypes (CDKP: Cerebrovascular Disease Knowledge Portal data).
4. Discussion
Our study is the first to show that HSPA8 SNPs are associated with the risk and clinical features of IS, and this relationship is significantly modified by gender and environmental risk factors. SNP rs10892958 (risk allele G) was found to be associated with an increased risk of IS exclusively in males, smokers, and individuals with low fruit and vegetable consumption. SNP rs1136141 was associated with an increased risk of IS only in smokers and on the condition of insufficient fruit and vegetable consumption.
HSPA8 is a molecular chaperone highly expressed in vessels, brain tissues, and blood [19,41,42,43]. Bioinformatic analysis revealed that HSPA8 also interacts with other proteins that control the regulation of protein constancy, protein folding, refolding, response to unfolded proteins, regulation of protein ubiquitination, and cellular response to unfolded proteins. The possible significance of HSPA8 (HSC70) in the molecular mechanisms of ischemic stroke has already been noted in previous studies. For example, according to Bustamente et al., elevated serum HSC70 levels are typical of ischemic stroke compared to hemorrhagic stroke [44]. Stankowski et al. noted that the heat shock C-terminus related to 70 interacting proteins increases after stroke and impairs survival in acute oxidative stress [45]. Previous studies also demonstrated that vascular expression of HSPA8 had been up-regulated in individuals with atherosclerotic diseases, perhaps as a compensatory response [46,47]. HSPA8 was shown to interact with NLRC5 and suppress the NF-kB pathway [48] in macrophages, suggesting its role in vascular wall inflammation [49]. Noteworthy is that apparently HSPA8 may also display prothrombotic activities [50].
The functional impacts of genetic variants were interpreted using a bioinformatic approach because there has not been any research investigating the association of HSPA8 SNPs with IS risk up to this point. This approach made it possible to study the molecular mechanisms behind the involvement of HSPA8 polymorphic loci in IS pathogenesis.
Firstly, IS-linked SNPs are characterized by high regulatory potential: they significantly affect histone modifications in brain and blood tissues, mainly trimethylation in the 4th lysine residue of histone H3 and acetylation of lysine residues in the N-terminal position 27 of histone; they are largely regulated by regulatory proteins.
Secondly, according to bioinformatic resources, the risk alleles G rs10892958 and A rs1136141 have a relationship to cis-eQTL-mediated down-regulation of HSPA8 in brain tissues. Allele A rs1136141 also affects an increase in CLMP expression in the brain. Probably, CLMP plays a substantial role in the formation of cerebral atherosclerosis since it encodes a type I transmembrane protein that is found in junctional complexes between endothelial and epithelial cells. This protein may also play a function in cell-cell adhesion [51].
Thirdly, risk allele G rs10892958 and risk allele A rs1136141 HSPA8 produce DNA binding sites for TFs involved in oxidative stress, neurogenesis, angiogenesis, apoptosis, and migration of blood vessel endothelial cells; this adds more support to the notion that HSPA8 plays a significant role in the molecular mechanisms underlying IS.
Fourth, information obtained with the bioinformatic tool CDKP revealed that the risk allele A rs1136141 is linked to a higher risk of cerebral stroke (TOAST other determined), increased level of systolic blood pressure, and an increased heart rate. As a result, this genetic variation may have a significant impact on the development of hypertension, the main risk factor for IS. Atrial fibrillation, another key risk factor for the cardioembolic type of IS, is also made more likely by a higher heart rate. Notably, CDKP data also indicates that the A allele rs1136141 has a role in risking peripheral artery disease in ever-smokers —a pathogenetically related cardiovascular pathology. This finding correlates with the data obtained in our study, which provide proof of the involvement of this genetic variant in the formation of IS risk in smokers.
On the other hand, HSPA8 may play a protective role in IS the process of ischemic stroke by protecting nerve cells and inhibiting neuronal apoptosis [52,53,54]. As a chaperone, HSPA8 orchestrates the folding and compartmentalization of the client proteins, contributing to the cellular stress response [55,56]. HSPA8 also down-regulates apoptotic cell death, both via direct suppression of several members of the apoptotic pathway and via activation of the anti-death protein bcl-2 [56]. Additionally, HSP70s have been shown to be critical for the biogenesis of extracellular vesicles, participating in neurological recovery after stroke [57]. Moreover, HSPA8 is considered a negative regulator of oxidative stress. Some authors revealed that constitutively expressed HSPA8 plays a role in protecting cardiomyocytes from oxidative damage, increasing cell survival [58,59]. Finally, Hspa1a/ Hspa1b knock-out mice subjected to different thrombotic challenges developed thrombosis significantly earlier than their wild-type counterparts, suggesting the important role of members of HSP70 family in regulation of haemostasis [60]. More recent studies have reported that HSPA8 is released from cardiomyocytes under oxidative stress [61]. Altogether, these data indicate that HSPA8 may take an important place during the cellular response to ischemia, and future research should address its role in the course of IS.
In our study, we also revealed sex-specific correlates of HSPA8 genetic variants. Numerous proofs are presented in the literature that candidate genes for cardiovascular diseases are characterized by pronounced gender-specific effects in the manifestation of associations [62,63,64,65,66]. The association of rs10892958 with the development of IS in men found in our study is explained by the regulation of HSPA8 expression by sex hormones. There is a lot of evidence that estradiol increases the expression of HSPA8 [67,68,69,70,71]. Considering that the carriage of the G rs10892958 allele is associated with HSPA8 down-regulation, the absence of a compensating effect of estrogens on the HSPA8 mRNA level increases the risk effects of this genetic variant. In addition, it has been found that in women, HSPA8 levels are inversely correlated with the level of Toll-like receptor 4, which triggers oxidative stress and inflammation, which are key elements of vascular diseases [72]. At the same time, no such correlation has been found in men.
In the present research, we also report environmentally-dependent effects of HSPA8 polymorphisms. It is known that environmental risk factors can modify the contribution of genetic variants on the risk of diseases [28,73,74]. Our study provided significant evidence of the role of smoking and low consumption of fresh vegetables and fruits in the manifestation of associations rs10892958 and rs1136141 HSPA8 and the development of IS. It is noteworthy that both of these risk factors are associated with high levels of reactive oxygen species [75,76] and, accordingly, oxidative stress, a major link contributing to the development of cardiovascular disorders [25,77,78,79]. Smoking-specific correlates of candidate genes with risk of IS and stroke-related phenotypes have been demonstrated in many of our prior investigations [65,80], suggesting a significant role for smoking-induced endothelial dysfunction, vascular tone disorders, and atherosclerosis in the development of IS. The Comparative Toxigenomics Database offered proof of smoking’s effects on the reduction of HSPA8 expression [81] as well as accelerating the metabolism of HSPA8 protein [82]. Thus, most likely, smoking exacerbates the effect of IS-related alleles on the risk of disease by down-regulating HSPA8 expression.
In addition to smoking, low fruit and vegetable intake is linked to high levels of reactive oxygen species, in particular hydroperoxides [83,84], and increased consumption of antioxidant-rich foods may have prompted a decrease in the total amount of lipid peroxide [85]. In vitro research demonstrates that polyphenols, which are rich in fresh vegetables and fruits, shield arteries and neural cells from the toxicity of oxidative glutamate and H2O2 [86,87]. Many polyphenols have strong anti-inflammatory activities in addition to their anti-oxidant characteristics [88]. The conducted studies have shown that hydrogen peroxide may affect the level of HSPA8 mRNA [89], resulting in decreased expression of the HSPA8 protein [90]. Thus, low fresh fruit and vegetable consumption related to oxidative stress and high levels of reactive oxygen species may affect the decrease in HSPA8 expression, thereby showing more pronounced effects of the risk alleles rs10892958 and rs1136141 HSPA8.
In conclusion, the results of our case-control study indicate that susceptibility to IS may be determined by genetic variants in HSPA8. However, we examined only tagSNPs and excluded from the analysis SNPs that are in linkage disequilibrium with tagSNPs, which can lead to false-positive results since the relationships we identified may reflect the effects of genetic variants linked to analysed SNPs. This is a limitation of our study. Further research is needed to confirm our findings and explore potential molecular mechanisms underlying the associations between IS risk and polymorphisms in the HSPA8 gene.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/genes14061171/s1. Figure S1: Plot showing a clear separation between the signals derived from allele 1 (rs1136141-G, FAM fluorescent dye) or allele 2 (rs1136141-A, ROX fluorescent dye). Genotypes GG, GA, and AA are shown as circles, triangles, and squares, respectively; Figure S2: Plot showing a clear separation between the signals derived from allele 1 (rs1461496-A, FAM fluorescent dye) or allele 2 (rs1461496-G, ROX fluorescent dye). Genotypes GG, GA, and AA are shown as squares, triangles, and circles, respectively; Figure S3: Plot showing a clear separation between the signals derived from allele 1 (rs1136141-C, FAM fluorescent dye) or allele 2 (rs1136141-G, ROX fluorescent dye). Genotypes CC, CG, and GG are shown as circles, triangles, and squares, respectively; Table S1: Primers and probes designed for this study; Table S2: Main functional characteristics of predicted functional partners of HSPA8; Table S3: Functional enrichments of HSPA8 network; Table S4: Distribution of HSPA8 genotypes in ischemic stroke patients/healthy controls and their correspondence to the Hardy–Weinberg equilibrium; Table S5: Distribution of HSPA8 genotypes in sub-group analysis of patients/healthy controls; Table S6: Analysis of the effect of rs10892958 of HSPA8 on binding of DNA to transcription factors; Table S7: Analysis of the effect of rs1136141 of HSPA8 on binding of DNA to transcription factors.
Author Contributions
Conceptualization, O.Y.B.; methodology, O.Y.B. and K.A.K.; validation, A.V.P. and M.I.C.; formal analysis, M.A.B. and M.B.F.; investigation, M.O.S., K.A.K., T.A.S. and M.A.B.; resources, M.O.S. and T.A.S.; data curation, O.Y.B.; writing—original draft preparation, K.A.K. and A.V.D.; writing—review and editing, O.Y.B.; visualization, V.O.S.; supervision, O.Y.B.; project administration, A.V.P. and M.I.C.; funding acquisition, O.Y.B., V.O.S., K.A.K., A.V.D. and M.B.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Russian Science Foundation (№ 22-15-00288, https://rscf.ru/en/project/22-15-00288/ (assessed on 30 March 2022).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and was approved by the Ethical Review Committee of Kursk State Medical University, Russia (protocol no. 12, 7 December 2015). All the participants gave written informed consent before enrollment in this study.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 30 March 2023).
- Orlacchio, A.; Bernardi, G. Research actuality in the genetics of stroke. Clin. Exp. Hypertens. 2006, 28, 191–197. [Google Scholar] [CrossRef]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.-K.; van der Laan, S.W.; Gretarsdottir, S.; et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef]
- Debette, S.; Markus, H.S. Stroke Genetics: Discovery, Insight Into Mechanisms, and Clinical Perspectives. Circ. Res. 2022, 130, 1095–1111. [Google Scholar] [CrossRef] [PubMed]
- Bushueva, O.; Barysheva, E.; Markov, A.; Belykh, A.; Koroleva, I.; Churkin, E.; Polonikov, A.; Ivanov, V.; Nazarenko, M. DNA Hypomethylation of the MPO Gene in Peripheral Blood Leukocytes Is Associated with Cerebral Stroke in the Acute Phase. J. Mol. Neurosci. 2021, 71, 1914–1932. [Google Scholar] [CrossRef] [PubMed]
- Bushueva, O.Y.; Stetskaya, T.A.; Polonikov, A.V.; Ivanov, V.P. The relationship between polymorphism 640A > G of the CYBA gene with the risk of ischemic stroke in the population of the Central Russia. Zh. Nevrol. Psikhiatr. Im. S. S. Korsakova 2015, 115 Pt 2, 38. [Google Scholar] [CrossRef]
- Polonikov, A.; Bocharova, I.; Azarova, I.; Klyosova, E.; Bykanova, M.; Bushueva, O.; Polonikova, A.; Churnosov, M.; Solodilova, M. The Impact of Genetic Polymorphisms in Glutamate-Cysteine Ligase, a Key Enzyme of Glutathione Biosynthesis, on Ischemic Stroke Risk and Brain Infarct Size. Life 2022, 12, 602. [Google Scholar] [CrossRef]
- Polonikov, A.V.; Ushachev, D.V.; Ivanov, V.P.; Churnosov, M.I.; Freidin, M.B.; Ataman, A.V.; Harbuzova, V.Y.; Bykanova, M.A.; Bushueva, O.Y.; Solodilova, M.A. Altered erythrocyte membrane protein composition mirrors pleiotropic effects of hypertension susceptibility genes and disease pathogenesis. J. Hypertens. 2015, 33, 2265–2277. [Google Scholar] [CrossRef]
- Wang, M.; Brage, S.; Sharp, S.J.; Luo, S.; Yeung, S.L.A.; Kim, Y. Associations of genetic susceptibility and healthy lifestyle with incidence of coronary heart disease and stroke in individuals with hypertension. Eur. J. Prev. Cardiol. 2022, 29, 2101–2110. [Google Scholar] [CrossRef]
- Borén, J.; Chapman, M.J.; Krauss, R.M.; Packard, C.J.; Bentzon, J.F.; Binder, C.J.; Daemen, M.J.; Demer, L.L.; Hegele, R.A.; Nicholls, S.J.; et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: Pathophysiological, genetic, and therapeutic insights: A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 2020, 41, 2313–2330. [Google Scholar] [CrossRef]
- Sorokin, A.; Kotani, K.; Bushueva, O.; Taniguchi, N.; Lazarenko, V. The Cardio-Ankle Vascular Index and Ankle-Brachial Index in Young Russians. J. Atheroscler. Thromb. 2015, 22, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Akhter, A.S.; Zakeri, A.; Herson, P.S. Sex differences in thrombosis as it affects acute ischemic stroke. Neurobiol. Dis. 2022, 165, 105647. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhao, G.; Hua, C.; Ju, W.-N.; Jin, H. Molecular chaperones and hypoxic-ischemic encephalopathy. Neural Regen. Res. 2017, 12, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, J.W.; Yenari, M.A. Heat shock protein signaling in brain ischemia and injury. Neurosci. Lett. 2020, 715, 134642. [Google Scholar] [CrossRef]
- Doeppner, T.R.; Doehring, M.; Kaltwasser, B.; Majid, A.; Lin, F.; Bähr, M.; Kilic, E.; Hermann, D.M. Ischemic Post-Conditioning Induces Post-Stroke Neuroprotection via Hsp70-Mediated Proteasome Inhibition and Facilitates Neural Progenitor Cell Transplantation. Mol. Neurobiol. 2017, 54, 6061–6073. [Google Scholar] [CrossRef] [PubMed]
- Dukay, B.; Csoboz, B.; Tóth, M.E. Heat-Shock Proteins in Neuroinflammation. Front. Pharmacol. 2019, 10, 920. [Google Scholar] [CrossRef]
- Zhan, X.; Ander, B.P.; Liao, I.H.; Hansen, J.E.; Kim, C.; Clements, D.; Weisbart, R.H.; Nishimura, R.N.; Sharp, F.R. Recombinant Fv-Hsp70 Protein Mediates Neuroprotection After Focal Cerebral Ischemia in Rats. Stroke 2010, 41, 538–543. [Google Scholar] [CrossRef]
- Weiss, Y.G.; Maloyan, A.; Tazelaar, J.; Raj, N.; Deutschman, C.S. Adenoviral transfer of HSP-70 into pulmonary epithelium ameliorates experimental acute respiratory distress syndrome. J. Clin. Investig. 2002, 110, 801–806. [Google Scholar] [CrossRef]
- Stricher, F.; Macri, C.; Ruff, M.; Muller, S. HSPA8/HSC70 chaperone protein: Structure, Function, and Chemical Targeting. Autophagy 2013, 9, 1937–1954. [Google Scholar] [CrossRef]
- Yamamoto, Y.-H.; Kimura, T.; Momohara, S.; Takeuchi, M.; Tani, T.; Kimata, Y.; Kadokura, H.; Kohno, K. A Novel ER J-protein DNAJB12 Accelerates ER-associated Degradation of Membrane Proteins Including CFTR. Cell Struct. Funct. 2010, 35, 107–116. [Google Scholar] [CrossRef]
- Grove, D.E.; Fan, C.-Y.; Ren, H.Y.; Cyr, D.M. The endoplasmic reticulum–associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3–dependent degradation of nascent CFTRΔF508. Mol. Biol. Cell 2011, 22, 301–314. [Google Scholar] [CrossRef]
- Sopha, P.; Kadokura, H.; Yamamoto, Y.-H.; Takeuchi, M.; Saito, M.; Tsuru, A.; Kohno, K. A Novel Mammalian ER-located J-protein, DNAJB14, Can Accelerate ERAD of Misfolded Membrane Proteins. Cell Struct. Funct. 2012, 37, 177–187. [Google Scholar] [CrossRef]
- Young, J.C.; Hoogenraad, N.J.; Hartl, F. Molecular Chaperones Hsp90 and Hsp70 Deliver Preproteins to the Mitochondrial Import Receptor Tom70. Cell 2003, 112, 41–50. [Google Scholar] [CrossRef]
- Matsumura, Y.; Sakai, J.; Skach, W.R. Endoplasmic Reticulum Protein Quality Control Is Determined by Cooperative Interactions between Hsp/c70 Protein and the CHIP E3 Ligase. J. Biol. Chem. 2013, 288, 31069–31079. [Google Scholar] [CrossRef] [PubMed]
- Vialykh, E.K.; A Solidolova, M.; Bushueva, O.; Bulgakova, I.V.; Polonikov, A. Catalase gene polymorphism is associated with increased risk of cerebral stroke in hypertensive patients. Zh. Nevrol. Psikhiatr. im. S. S. Korsakova 2012, 112, 3–7. [Google Scholar] [PubMed]
- Kobzeva, K.A.; Shilenok, I.V.; Belykh, A.E.; Gurtovoy, D.E.; Bobyleva, L.A.; Krapiva, A.B.; Stetskaya, T.A.; Bykanova, M.A.; Mezhenskaya, A.A.; Lysikova, E.A.; et al. C9orf16 (BBLN) gene, encoding a member of Hero proteins, is a novel marker in ischemic stroke risk. Res. RESULTS Biomed. 2022, 8, 278. [Google Scholar] [CrossRef]
- Who, J.; Consultation, F.E. Diet, nutrition and the prevention of chronic diseases. World Health Organ Tech. Rep. Ser. 2003, 916, 1–149. [Google Scholar]
- Bushueva, O.; Solodilova, M.; Ivanov, V.; Polonikov, A. Gender-specific protective effect of the −463G > A polymorphism of myeloperoxidase gene against the risk of essential hypertension in Russians. J. Am. Soc. Hypertens. 2015, 9, 902–906. [Google Scholar] [CrossRef]
- Bushueva, O.Y. Single nucleotide polymorphisms in genes encoding xenobiotic metabolizing enzymes are associated with predisposition to arterial hypertension. Res. Results Biomed. 2020, 6, 447–456. [Google Scholar] [CrossRef]
- Xu, Z.; Taylor, J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009, 37, W600–W605. [Google Scholar] [CrossRef]
- Dong, S.; Boyle, A.P. Predicting functional variants in enhancer and promoter elements using RegulomeDB. Hum. Mutat. 2019, 40, 1292–1298. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef] [PubMed]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–1929. [Google Scholar] [CrossRef]
- Zheng, Z.; Huang, D.; Wang, J.; Zhao, K.; Zhou, Y.; Guo, Z.; Zhai, S.; Xu, H.; Cui, H.; Yao, H.; et al. QTLbase: An integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 2020, 48, D983–D991. [Google Scholar] [CrossRef]
- Von Mering, C.; Jensen, L.J.; Snel, B.; Hooper, S.D.; Krupp, M.; Foglierini, M.; Jouffre, N.; Huynen, M.A.; Bork, P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005, 33, D433–D437. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Hudson, R.; Harrison, C.; Craven, M.; Keleş, S. atSNP Search: A web resource for statistically evaluating influence of human genetic variation on transcription factor binding. Bioinformatics 2019, 35, 2657–2659. [Google Scholar] [CrossRef]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Grondin, C.J.; Johnson, R.J.; Sciaky, D.; Wiegers, J.; Wiegers, T.C.; Mattingly, C.J. Comparative Toxicogenomics Database (CTD): Update 2021. Nucleic Acids Res. 2021, 49, D1138–D1143. [Google Scholar] [CrossRef]
- Crawford, K.M.; Gallego-Fabrega, C.; Kourkoulis, C.; Miyares, L.; Marini, S.; Flannick, J.; Burtt, N.P.; Von Grotthuss, M.; Alexander, B.; Costanzo, M.C.; et al. Cerebrovascular Disease Knowledge Portal. Stroke 2018, 49, 470–475. [Google Scholar] [CrossRef]
- Rashedan, M. Characterization of the Localization of Heat-Shock Proteins, HSPA8 and HSPA5, in Mammalian Cells; California State University: Fullerton, CA, USA, 2013. [Google Scholar]
- Sirtori, R.; Riva, C.; Ferrarese, C.; Sala, G. HSPA8 knock-down induces the accumulation of neurodegenerative disorder-associated proteins. Neurosci. Lett. 2020, 736, 135272. [Google Scholar] [CrossRef]
- Bonam, S.R.; Ruff, M.; Muller, S. HSPA8/HSC70 in Immune Disorders: A Molecular Rheostat that Adjusts Chaperone-Mediated Autophagy Substrates. Cells 2019, 8, 849. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, A.; López-Cancio, E.; Pich, S.; Penalba, A.; Giralt, D.; García-Berrocoso, T.; Ferrer-Costa, C.; Gasull, T.; Hernández-Pérez, M.; Millan, M.; et al. Blood Biomarkers for the Early Diagnosis of Stroke: The Stroke-Chip Study. Stroke 2017, 48, 2419–2425. [Google Scholar] [CrossRef]
- Stankowski, J.N.; Zeiger, S.L.; Cohen, E.L.; DeFranco, D.B.; Cai, J.; McLaughlin, B. C-Terminus of Heat Shock Cognate 70 Interacting Protein Increases Following Stroke and Impairs Survival Against Acute Oxidative Stress. Antioxid. Redox Signal. 2011, 14, 1787–1801. [Google Scholar] [CrossRef] [PubMed]
- Dybdahl, B.; A Slørdahl, S.; Waage, A.; Kierulf, P.; Espevik, T.; Sundan, A. Myocardial ischaemia and the inflammatory response: Release of heat shock protein 70 after myocardial infarction. Heart 2005, 91, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Timofeeva, A.; Goryunova, L.; Khaspekov, G.; Kovalevskii, D.; Scamrov, A.; Bulkina, O.; Karpov, Y.; Talitskii, K.; Buza, V.; Britareva, V.; et al. Altered Gene Expression Pattern in Peripheral Blood Leukocytes from Patients with Arterial Hypertension. Ann. N. Y. Acad. Sci. 2006, 1091, 319–335. [Google Scholar] [CrossRef]
- Yu, Q.; Ju, P.; Kou, W.; Zhai, M.; Zeng, Y.; Maimaitiaili, N.; Shi, Y.; Xu, X.; Zhao, Y.; Jian, W.; et al. Macrophage-Specific NLRC5 Protects from Cardiac Remodeling Through Interaction With HSPA8. JACC Basic Transl. Sci. 2023, 8, 479–496. [Google Scholar] [CrossRef]
- Xu, H.; Jiang, J.; Chen, W.; Li, W.; Chen, Z. Vascular Macrophages in Atherosclerosis. J. Immunol. Res. 2019, 2019, 4354786. [Google Scholar] [CrossRef]
- Gerges, M.; Gerges, C.; Publig, M.; Skoro-Sajer, N.; Bonderman, D.; Frey, M.; Schwarzinger, I.; Lechner, K.; Seidl, V.; Alimohammadi, A.; et al. Chronic inflammation after splenectomy is a risk factor for increased thrombotic cardiovascular events. Eur. Heart J. 2017, 38 (Suppl. 1), A5706. [Google Scholar] [CrossRef]
- Guo, Y.-L.; Bai, R.; Chen, C.X.-J.; Liu, D.-Q.; Liu, Y.; Zhang, C.-Y.; Zen, K. Role of Junctional Adhesion Molecule-Like Protein in Mediating Monocyte Transendothelial Migration. Arter. Thromb. Vasc. Biol. 2009, 29, 75–83. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, S.; Yang, P.-Y.; Zhang, Y.-F.; Li, T.-J.; Rui, Y.-C. EF1A1/HSC70 Cooperatively Suppress Brain Endothelial Cell Apoptosis via Regulating JNK Activity. CNS Neurosci. Ther. 2016, 22, 836–844. [Google Scholar] [CrossRef]
- Chen, S.; Brown, I.R. Translocation of constitutively expressed heat shock protein Hsc70 to synapse-enriched areas of the cerebral cortex after hyperthermic stress. J. Neurosci. Res. 2007, 85, 402–409. [Google Scholar] [CrossRef]
- McLaughlin, B.; Hartnett, K.A.; Erhardt, J.A.; Legos, J.J.; White, R.F.; Barone, F.C.; Aizenman, E. Caspase 3 activation is essential for neuroprotection in preconditioning. Proc. Natl. Acad. Sci. USA 2003, 100, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Schmitt, E.; Candé, C.; Vahsen, N.; Parcellier, A.; Kroemer, G. HSP27 and HSP70: Potentially oncogenic apoptosis inhibitors. Cell Cycle 2003, 2, 578–583. [Google Scholar] [CrossRef]
- Giffard, R.G.; Yenari, M.A. Many Mechanisms for Hsp70 Protection from Cerebral Ischemia. J. Neurosurg. Anesthesiol. 2004, 16, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhang, L.; Kuang, Y.; Venkataramani, V.; Jin, F.; Hein, K.; Zafeiriou, M.P.; Lenz, C.; Moebius, W.; Kilic, E.; et al. Extracellular Vesicles Derived from Neural Progenitor Cells––A Preclinical Evaluation for Stroke Treatment in Mice. Transl. Stroke Res. 2021, 12, 185–203. [Google Scholar] [CrossRef]
- Chong, K.-Y.; Lai, C.-C.; Lille, S.; Chang, C.; Su, C.-Y. Stable Overexpression of the Constitutive Form of Heat Shock Protein 70 Confers Oxidative Protection. J. Mol. Cell. Cardiol. 1998, 30, 599–608. [Google Scholar] [CrossRef]
- Su, C.-Y.; Chong, K.-Y.; E Owen, O.; Dillmann, W.H.; Chang, C.; Lai, C.-C. Constitutive and Inducible hsp70s are Involved in Oxidative Resistance Evoked by Heat Shock or Ethanol. J. Mol. Cell. Cardiol. 1998, 30, 587–598. [Google Scholar] [CrossRef]
- Canton, M.; Menabo, R.; Carpi, A.; Di Lisa, F. P253Oxidative stress causes the release of a specific subset of proteins from viable cardiac myocytes. Cardiovasc. Res. 2014, 103 (Suppl. 1), S45. [Google Scholar] [CrossRef]
- Allende, M.; Molina, E.; Guruceaga, E.; Tamayo, I.; González-Porras, J.R.; Gonzalez-López, T.J.; Toledo, E.; Rabal, O.; Ugarte, A.; Roldán, V.; et al. Hsp70 protects from stroke in atrial fibrillation patients by preventing thrombosis without increased bleeding risk. Cardiovasc. Res. 2016, 110, 309–318. [Google Scholar] [CrossRef]
- Silander, K.; Alanne, M.; Kristiansson, K.; Saarela, O.; Ripatti, S.; Auro, K.; Karvanen, J.; Kulathinal, S.; Niemelä, M.; Ellonen, P.; et al. Gender Differences in Genetic Risk Profiles for Cardiovascular Disease. PLoS ONE 2008, 3, e3615. [Google Scholar] [CrossRef]
- Bushueva, O.; Solodilova, M.; Churnosov, M.; Ivanov, V.; Polonikov, A. The Flavin-Containing Monooxygenase 3 Gene and Essential Hypertension: The Joint Effect of Polymorphism E158K and Cigarette Smoking on Disease Susceptibility. Int. J. Hypertens. 2014, 2014, 712169. [Google Scholar] [CrossRef] [PubMed]
- Bushueva, O.Y.; Bulgakova, I.V.; Ivanov, V.P.; Polonikov, A.V. Association of Flavin Monooxygenase Gene E158K Polymorphism with Chronic Heart Disease Risk. Bull. Exp. Biol. Med. 2015, 159, 776–778. [Google Scholar] [CrossRef]
- Bushueva, O.; A Stetskaya, T.; Korogodina, G.V.; Ivanov, V.P.; Polonikov, A. Gender-specific differences of endothelial nitric oxide synthase e298d polymorphism and the risk of stroke. Clin. Med. (Russ. J.) 2015, 93, 34–40. [Google Scholar]
- Stetskaia, T.A.; Bushueva, O.I.; Bulgakova, I.V.; Vialykh, E.K.; Shuteeva, T.V.; Biriukov, A.E.; Ivanov, V.P.; Polonikov, A.V. Association of T174M polymorphism of the angiotensinogen gene with the higher risk of cerebral stroke in women. Ter. Arkhiv 2014, 86, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Jennen, D.G.J.; Magkoufopoulou, C.; Ketelslegers, H.B.; van Herwijnen, M.H.M.; Kleinjans, J.C.S.; van Delft, J.H.M. Comparison of HepG2 and HepaRG by Whole-Genome Gene Expression Analysis for the Purpose of Chemical Hazard Identification. Toxicol. Sci. 2010, 115, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Scafoglio, C.; Ambrosino, C.; Cicatiello, L.; Altucci, L.; Ardovino, M.; Bontempo, P.; Medici, N.; Molinari, A.M.; Nebbioso, A.; Facchiano, A.; et al. Comparative gene expression profiling reveals partially overlapping but distinct genomic actions of different antiestrogens in human breast cancer cells. J. Cell. Biochem. 2006, 98, 1163–1184. [Google Scholar] [CrossRef]
- Wang, D.-Y.; Fulthorpe, R.; Liss, S.N.; Edwards, E.A. Identification of Estrogen-Responsive Genes by Complementary Deoxyribonucleic Acid Microarray and Characterization of a Novel Early Estrogen-Induced Gene: EEIG1. Mol. Endocrinol. 2004, 18, 402–411. [Google Scholar] [CrossRef]
- Krebs, C.J.; Jarvis, E.D.; Pfaff, D.W. The 70-kDa heat shock cognate protein (Hsc73) gene is enhanced by ovarian hormones in the ventromedial hypothalamus. Proc. Natl. Acad. Sci. USA 1999, 96, 1686–1691. [Google Scholar] [CrossRef]
- Kwekel, J.C.; Burgoon, L.D.; Burt, J.W.; Harkema, J.R.; Zacharewski, T.R. A cross-species analysis of the rodent uterotrophic program: Elucidation of conserved responses and targets of estrogen signaling. Physiol. Genom. 2005, 23, 327–342. [Google Scholar] [CrossRef]
- De Oliveira, A.A.; Faustino, J.; Nunes, K.P. Transcriptomic Data Analysis Reveals Sex-Related Differences in the Interplay Between Toll-Like Receptor 4 and Heat-Shock Protein 70 in the Aorta of Type 2 Diabetic Donors. Hypertension 2019, 74 (Suppl. 1), AP190. [Google Scholar] [CrossRef]
- Polonikov, A.V.; Samgina, T.A.; Nazarenko, P.M.; Bushueva, O.Y.; Ivanov, V.P. Alcohol Consumption and Cigarette Smoking are Important Modifiers of the Association Between Acute Pancreatitis and the PRSS1-PRSS2 Locus in Men. Pancreas 2017, 46, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Bushueva, O.Y.; Ivanov, V.P.; Ryzhaeva, V.N.; Ponomarenko, I.V.; I Churnosov, M.; Polonikov, A.V. Association of the -844G>A polymorphism in the catalase gene with the increased risk of essential hypertension in smokers. Ter. Arkhiv 2016, 88, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Scicchitano, P.; Cortese, F.; Gesualdo, M.; De Palo, M.; Massari, F.; Giordano, P.; Ciccone, M.M. The role of endothelial dysfunction and oxidative stress in cerebrovascular diseases. Free. Radic. Res. 2019, 53, 579–595. [Google Scholar] [CrossRef]
- Bacchetti, T.; Turco, I.; Urbano, A.; Morresi, C.; Ferretti, G. Relationship of fruit and vegetable intake to dietary antioxidant capacity and markers of oxidative stress: A sex-related study. Nutrition 2019, 61, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Sorokin, A.V.; Kotani, K.; Bushueva, O.Y.; Polonikov, A.V. Antioxidant-related gene polymorphisms associated with the cardio-ankle vascular index in young Russians. Cardiol. Young 2016, 26, 677–682. [Google Scholar] [CrossRef]
- Sorokin, A.V.; Kotani, K.; Bushueva, O.Y. Association of matrix metalloproteinase 3 and γ-glutamyltransferase 1 gene polymorphisms with the cardio-ankle vascular index in young Russians. Cardiol. Young 2016, 26, 1238–1240. [Google Scholar] [CrossRef]
- Polonikov, A.; Vialykh, E.; Vasil’eva, O.; Bulgakova, I.; Bushueva, O.; Illig, T.; Solodilova, M. Genetic Variation in Glutathione S-Transferase Genes and Risk of Nonfatal Cerebral Stroke in Patients Suffering from Essential Hypertension. J. Mol. Neurosci. 2012, 47, 511–513. [Google Scholar] [CrossRef]
- Vasil’eva Yu, I.; Bushueva, O.; Zhabin, S.; Ivanov, S.V.; Polonikov, A. Smoking as a trigger factor in the development of diabetic angiopathy of lower extremities in men with methylenetetrahydrofolatereductase 677tt genotype. Clin. Med. (Russ. J.) 2015, 93, 45–49. [Google Scholar]
- Iskandar, A.R.; Mathis, C.; Schlage, W.K.; Frentzel, S.; Leroy, P.; Xiang, Y.; Sewer, A.; Majeed, S.; Ortega-Torres, L.; Johne, S.; et al. A systems toxicology approach for comparative assessment: Biological impact of an aerosol from a candidate modified-risk tobacco product and cigarette smoke on human organotypic bronchial epithelial cultures. Toxicol. Vitr. 2017, 39, 29–51. [Google Scholar] [CrossRef]
- Sheridan, J. Partnership between the Aryl Hydrocarbon Receptor (AHR) and RELB Regulates Cigarette Smoke-Induced Cyclooxygenase-2 (COX-2) Expression; McGill University: Montréal, QC, Canada, 2014. [Google Scholar]
- Bin-Jumah, M.N.; Nadeem, M.S.; Gilani, S.J.; Mubeen, B.; Ullah, I.; Alzarea, S.I.; Ghoneim, M.M.; Alshehri, S.; Al-Abbasi, F.A.; Kazmi, I. Lycopene: A Natural Arsenal in the War against Oxidative Stress and Cardiovascular Diseases. Antioxidants 2022, 11, 232. [Google Scholar] [CrossRef]
- Ciumărnean, L.; Milaciu, M.V.; Runcan, O.; Vesa, S.C.; Răchișan, A.L.; Negrean, V.; Perné, M.-G.; Donca, V.I.; Alexescu, T.-G.; Para, I.; et al. The effects of favonoids in cardiovascular diseases. Molecules 2020, 25, 4320. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.B.; A Niaz, M.; Agarwal, P.; Begom, R.; Rastogi, S.S. Effect of Antioxidant-Rich Foods on Plasma Ascorbic Acid, Cardiac Enzyme, and Lipid Peroxide Levels in Patients Hospitalized with Acute Myocardial Infarction. J. Am. Diet. Assoc. 1995, 95, 775–780. [Google Scholar] [CrossRef] [PubMed]
- Ishige, K.; Schubert, D.; Sagara, Y. Flavonoids protect neuronal cells from oxidative stress by three distinct mechanisms. Free. Radic. Biol. Med. 2001, 30, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Spada, P.D.; Dani, C.; Bortolini, G.V.; Funchal, C.; Henriques, J.A.; Salvador, M. Frozen Fruit Pulp of Euterpe oleraceae Mart. (Acai) Prevents Hydrogen Peroxide-Induced Damage in the Cerebral Cortex, Cerebellum, and Hippocampus of Rats. J. Med. Food 2009, 12, 1084–1088. [Google Scholar] [CrossRef] [PubMed]
- Scalbert, A.; Williamson, G. Dietary Intake and Bioavailability of Polyphenols. J. Nutr. 2000, 130 (Suppl. 8), 2073S–2085S. [Google Scholar] [CrossRef]
- Chadwick, W.; Zhou, Y.; Park, S.-S.; Wang, L.; Mitchell, N.; Stone, M.D.; Becker, K.G.; Martin, B.; Maudsley, S. Minimal Peroxide Exposure of Neuronal Cells Induces Multifaceted Adaptive Responses. PLoS ONE 2010, 5, e14352. [Google Scholar] [CrossRef] [PubMed]
- Law, C.-H.; Li, J.-M.; Chou, H.-C.; Chen, Y.-H.; Chan, H.-L. Hyaluronic acid-dependent protection in H9C2 cardiomyocytes: A cell model of heart ischemia–reperfusion injury and treatment. Toxicology 2013, 303, 54–71. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).