Analysis of the Glycoside Hydrolase Family 1 from Wild Jujube Reveals Genes Involved in the Degradation of Jujuboside A
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Acquisition of the Sequences of Potential β-Glucosidases of Wild Jujube
2.3. Phylogenetic Relationship Analysis
2.4. Chromosome Location and Gene Structure Analysis
2.5. Analysis of Physicochemical Properties Concerning the Putative ZsBgl Proteins
2.6. RNA Isolation and Reverse Transcription PCR
2.7. Heterologous Expression of Two ZsBgl Genes in E. coli
2.8. Determination of Enzymatic Activity Using HPLC
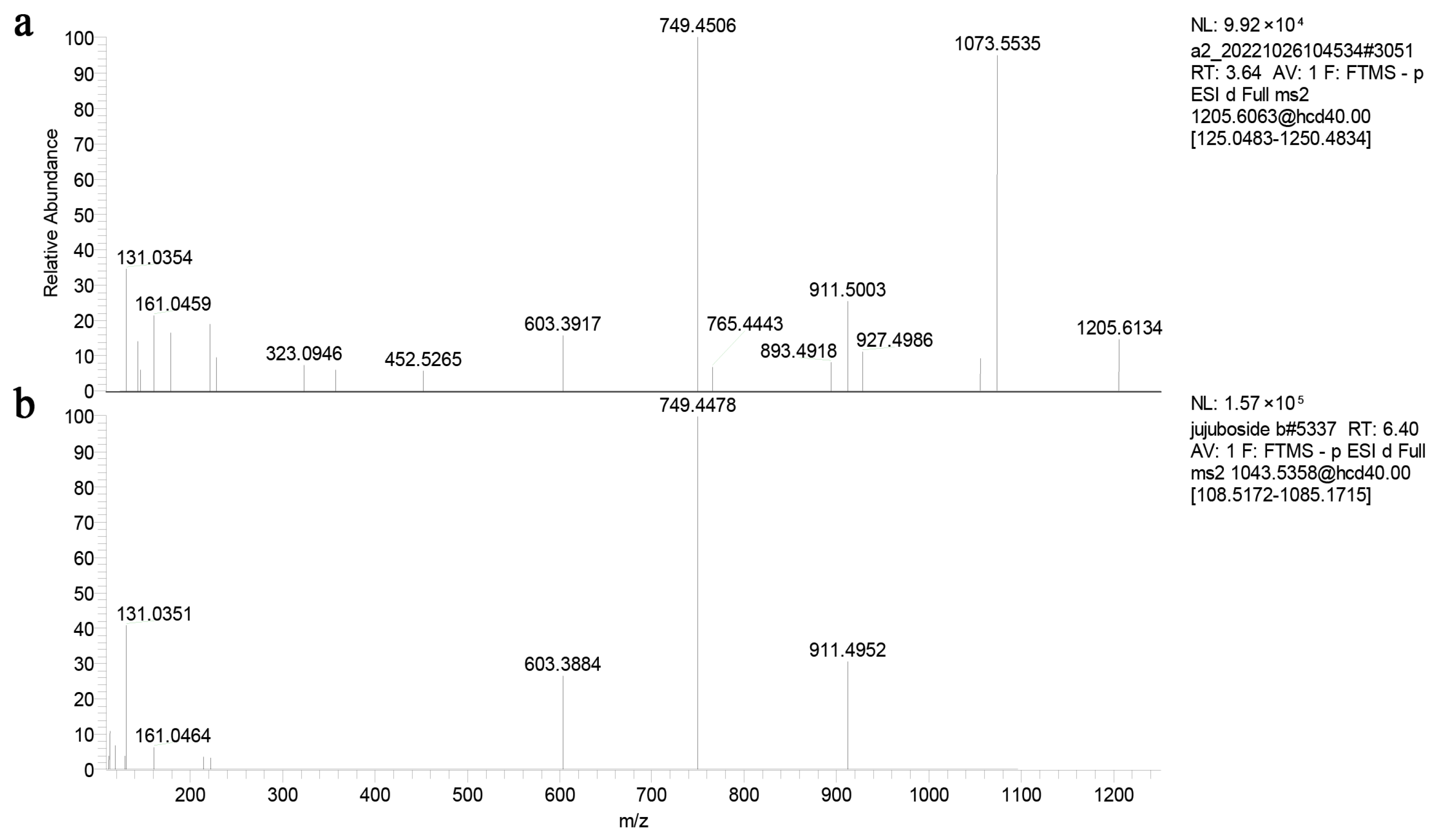
2.9. UPLC-Oribtrap-Exploris-120-MS/MS Analyses
2.10. Bioinformatic Analysis of the Putative Proteins and Molecular Docking
3. Results
3.1. Characteristics of ZsBgl Genes and Their Encoded Proteins
3.2. The Locations and Structures of ZsBgl Genes and Conserved Motif Information for ZsBgl Proteins
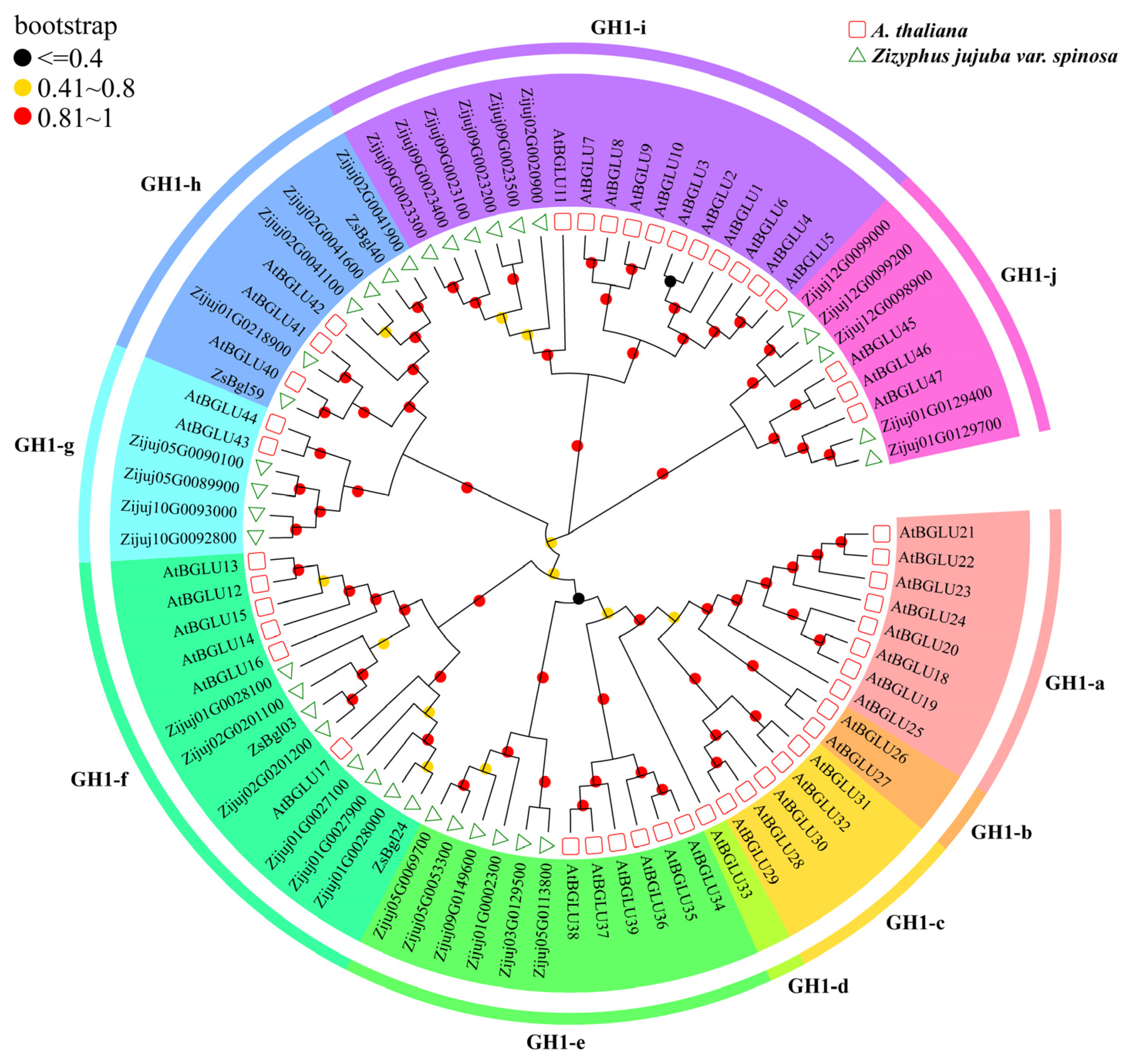
3.3. Phylogenetic Relationships of β-Glucosidases from Wild Jujube and Arabidopsis
3.4. The Prokaryotic Expression of Two ZsBgl Genes
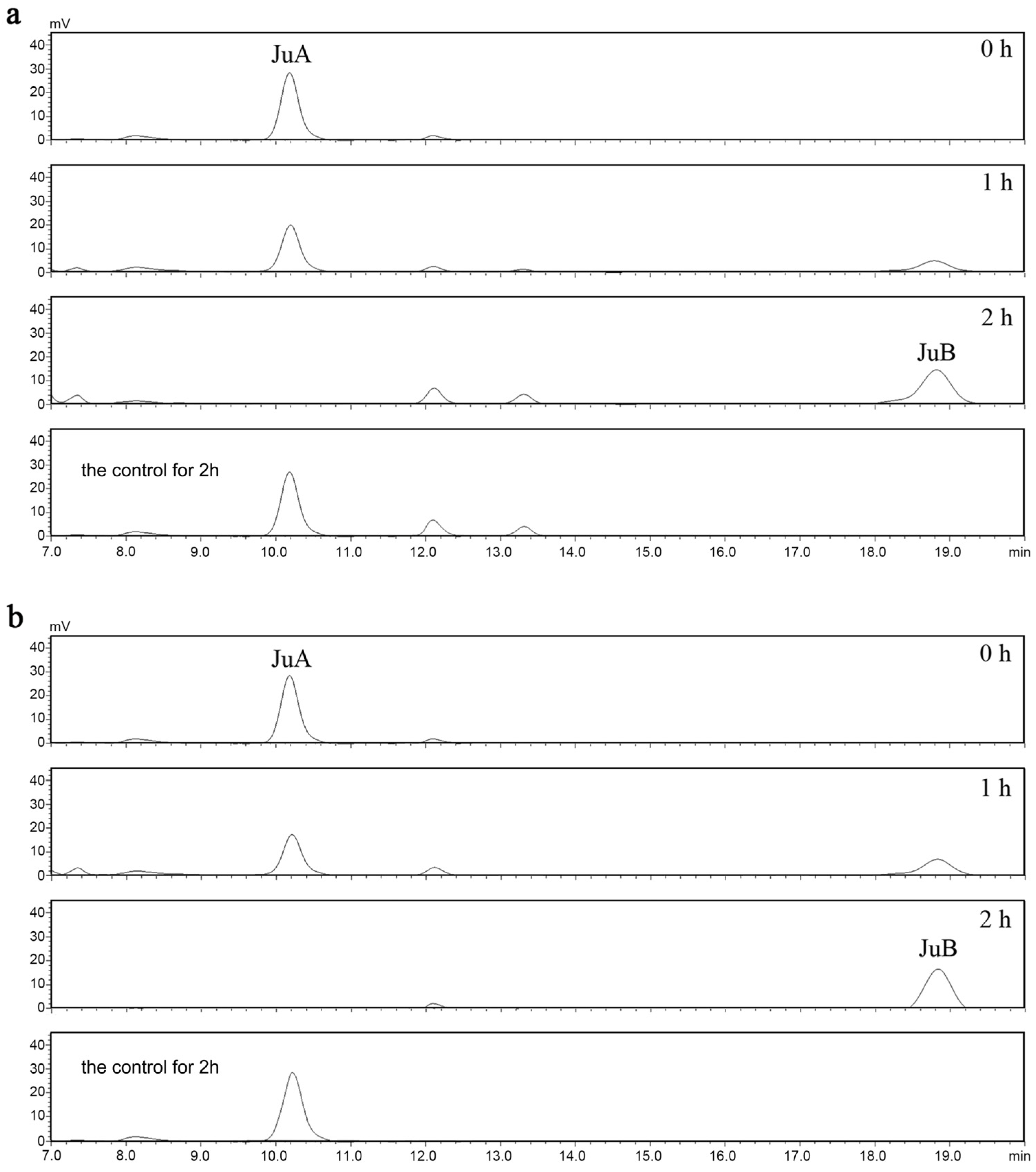
3.5. Determination of the Activity of ZsBgl03 and ZsBgl40
3.6. Protein Structure Prediction and Molecular Modeling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scigelova, M.; Singh, S.; Crout, D.H. Glycosidases—A great synthetic tool. J. Mol. Catal. B Enzym. 1999, 6, 483–494. [Google Scholar] [CrossRef]
- Cairns, J.R.K.; Mahong, B.; Baiya, S.; Jeon, J.S. β-Glucosidases: Multitasking, moonlighting or simply misunderstood? Plant Sci. 2015, 241, 246–259. [Google Scholar] [CrossRef]
- Ketudat Cairns, J.R.; Esen, A. β-Glucosidases. Cell. Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Himeno, N.; Saburi, W.; Wakuta, S.; Takeda, R.; Matsuura, H.; Nabeta, K.; Sansenya, S.; Cairns, J.R.K.; Mori, H.; Imai, R. Identification of rice β-glucosidase with high hydrolytic activity towards salicylic acid β-D-glucoside. Biosci. Biotechnol. Biochem. 2013, 77, 934–939. [Google Scholar] [CrossRef]
- Wakuta, S.; Hamada, S.; Ito, H.; Matsuura, H.; Nabeta, K.; Matsui, H. Identification of a β-glucosidase hydrolyzing tuberonic acid glucoside in rice (Oryza sativa L.). Phytochemistry 2010, 71, 1280–1288. [Google Scholar] [CrossRef]
- Lee, K.H.; Piao, H.L.; Kim, H.Y.; Choi, S.M.; Jiang, F.; Hartung, W.; Hwang, I.; Kwak, J.M.; Lee, I.J.; Hwang, I. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 2006, 126, 1109–1120. [Google Scholar] [CrossRef]
- Hua, Y.L.; Sansenya, S.; Saetang, C.; Wakuta, S.; Cairns, J.R.K. Enzymatic and structural characterization of hydrolysis of gibberellin A4 glucosyl ester by a rice β-D-glucosidase. Arch. Biochem. Biophys. 2013, 537, 39–48. [Google Scholar] [CrossRef]
- Babcock, G.D.; Esen, A. Substrate specificity of maize β-glucosidase. Plant Sci. 1994, 101, 31–39. [Google Scholar] [CrossRef]
- Hösel, W.; Tober, I.; Eklund, S.H.; Conn, E.E. Characterization of β-glucosidases with high specificity for the cyanogenic glucoside dhurrin in Sorghum bicolor (L.) Moench seedlings. Arch. Biochem. Biophys. 1987, 252, 152–162. [Google Scholar] [CrossRef]
- Barleben, L.; Panjikar, S.; Ruppert, M.; Koepke, J.; Stöckigt, J. Molecular architecture of strictosidine glucosidase: The gateway to the biosynthesis of the monoterpenoid indole alkaloid family. Plant Cell 2007, 19, 2886–2897. [Google Scholar] [CrossRef]
- Xia, L.Q.; Ruppert, M.; Wang, M.T.; Panjikar, S.; Lin, H.L.; Rajendran, C.; Barleben, L.; Stöckigt, J. Structures of alkaloid biosynthetic glucosidases decode substrate specificity. ACS Chem. Biol. 2012, 7, 226–234. [Google Scholar] [CrossRef]
- Ahn, Y.O.; Shimizu, B.I.; Sakata, K.; Gantulga, D.; Zhou, Z.H.; Bevan, D.R.; Esen, A. Scopolin-hydrolyzing β-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 2010, 51, 132–143. [Google Scholar] [CrossRef]
- Baba, S.A.; Vishwakarma, R.A.; Ashraf, N. Functional characterization of CsBGlu12, a β-glucosidase from Crocus sativus, provides insights into its role in abiotic stress through accumulation of antioxidant flavonols. J. Biol. Chem. 2017, 292, 4700–4713. [Google Scholar] [CrossRef]
- Roepke, J.; Bozzo, G.G. Arabidopsis thaliana β-glucosidase BGLU15 attacks flavonol 3-O-β-glucoside-7-O-α-rhamnosides. Phytochemistry 2015, 109, 14–24. [Google Scholar] [CrossRef]
- Opassiri, R.; Pomthong, B.; Onkoksoong, T.; Akiyama, T.; Esen, A.; Ketudat Cairns, J.R. Analysis of rice glycosyl hydrolase family 1 and expression of Os4bglu12 β-glucosidase. BMC Plant Biol. 2006, 6, 33. [Google Scholar] [CrossRef]
- Xu, Z.W.; Escamilla Treviño, L.; Zeng, L.H.; Lalgondar, M.; Bevan, D.; Winkel, B.; Mohamed, A.; Cheng, C.L.; Shih, M.C.; Poulton, J. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 2004, 55, 343–367. [Google Scholar] [CrossRef]
- Hino, F.; Okazaki, M.; Miura, Y. Effect of 2, 4-dichlorophenoxyacetic acid on glucosylation of scopoletin to scopolin in tobacco tissue culture. Plant Physiol. 1982, 69, 810–813. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Lee, K.H.; Dong, T.; Jeong, J.C.; Jin, J.B.; Kanno, Y.; Kim, D.H.; Kim, S.Y.; Seo, M.; Bressan, R.A. A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 2012, 24, 2184–2199. [Google Scholar] [CrossRef]
- Miyahara, T.; Sakiyama, R.; Ozeki, Y.; Sasaki, N. Acyl-glucose-dependent glucosyltransferase catalyzes the final step of anthocyanin formation in Arabidopsis. J. Plant Physiol. 2013, 170, 619–624. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.; Feussner, I.; Pieterse, C.M. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef]
- Zamioudis, C.; Hanson, J.; Pieterse, C.M. β-Glucosidase BGLU 42 is a MYB 72 dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. N. Phytol. 2014, 204, 368–379. [Google Scholar] [CrossRef]
- Rouyi, C.; Baiya, S.; Lee, S.K.; Mahong, B.; Jeon, J.S.; Ketudat Cairns, J.R.; Ketudat Cairns, M. Recombinant expression and characterization of the cytoplasmic rice β-glucosidase Os1BGlu4. PLoS ONE 2014, 9, e96712. [Google Scholar] [CrossRef]
- Li, M.; Zhang, C.X.; Hou, L.; Yang, W.C.; Liu, S.S.; Pang, X.M.; Li, Y.Y. Multiple responses contribute to the enhanced drought tolerance of the autotetraploid Ziziphus jujuba Mill. var. spinosa. Cell Biosci. 2021, 11, 119. [Google Scholar] [CrossRef]
- Wu, M.; Gu, X.; Zhang, Z.; Si, M.D.; Zhang, Y.J.; Tian, W.; Ma, D.L. The effects of climate change on the quality of Ziziphus jujuba var. Spinosa in China. Ecol. Indic. 2022, 139, 108934. [Google Scholar] [CrossRef]
- Xia, J.; Zhang, G.; Wang, R.; Zhang, S. Effect of soil water availability on photosynthesis in Ziziphus jujuba var. spinosus in a sand habitat formed from seashells: Comparison of four models. Photosynthetica 2014, 52, 253–261. [Google Scholar] [CrossRef]
- Wu, Y.S.; Wang, Y.C.; Niu, W.T.; Zhang, P.F.; Wu, L.N.; Li, H.; Wang, S.H. Establishment of fitted models for topographical factors and coexisting plants influencing distribution of natural wild jujube. Forests 2023, 14, 439. [Google Scholar] [CrossRef]
- Zhao, X.S.; Xie, L.W.; Wu, H.F.; Kong, W.J.; Yang, M.H. Analysis of six bioactive components in Semen Ziziphi Spinosae by UPLC-ELSD and UPLC-Q/TOF-MS. Anal. Methods 2014, 6, 5856–5864. [Google Scholar] [CrossRef]
- Hua, Y.; Xu, X.X.; Guo, S.; Xie, H.; Yan, H.; Ma, X.F.; Niu, Y.; Duan, J.A. Wild jujube (Ziziphus jujuba var. spinosa): A review of its phytonutrients, health benefits, metabolism, and applications. J. Agric. Food Chem. 2022, 70, 7871–7886. [Google Scholar] [CrossRef]
- He, S.R.; Zhao, C.B.; Zhang, J.X.; Wang, J.; Wu, B.; Wu, C.J. Botanical and traditional uses and phytochemical, pharmacological, pharmacokinetic, and toxicological characteristics of Ziziphi Spinosae Semen: A review. Evid. Based Complement. Alternat Med. 2020, 2020, 5861821. [Google Scholar] [CrossRef]
- Tan, F.; Chen, Y.L.; Tan, X.L.; Ma, Y.Y.; Peng, Y. Chinese materia medica used in medicinal diets. J. Ethnopharmacol. 2017, 206, 40–54. [Google Scholar] [CrossRef]
- Cao, J.X.; Zhang, Q.Y.; Cui, S.Y.; Cui, X.Y.; Zhang, J.; Zhang, Y.H.; Bai, Y.J.; Zhao, Y.Y. Hypnotic effect of jujubosides from Semen Ziziphi Spinosae. J. Ethnopharmacol. 2010, 130, 163–166. [Google Scholar] [CrossRef]
- Hua, Y.; Guo, S.; Xie, H.; Zhu, Y.; Yan, H.; Tao, W.W.; Shang, E.X.; Qian, D.W.; Duan, J.A. Ziziphus jujuba Mill. var. spinosa (Bunge) Hu ex H. F. Chow seed ameliorates insomnia in rats by regulating metabolomics and intestinal flora composition. Front. Pharmacol. 2021, 12, 653767. [Google Scholar] [CrossRef]
- Shergis, J.L.; Ni, X.J.; Sarris, J.; Zhang, A.L.; Guo, X.; Xue, C.C.; Lu, C.J.; Hugel, H. Ziziphus spinosa seeds for insomnia: A review of chemistry and psychopharmacology. Phytomedicine 2017, 34, 38–43. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, J.; Zhang, Y.; Xie, J. Ziziphi Spinosae Semen: A natural herb resource for treating neurological disorders. Curr. Top. Med. Chem. 2022, 22, 1379–1391. [Google Scholar] [CrossRef]
- Chen, C.Y.C.; Chen, Y.F.; Wu, C.H.; Tsai, H.Y. What is the effective component in suanzaoren decoction for curing insomnia? Discovery by virtual screening and molecular dynamic simulation. J. Agric. Food Chem. 2008, 26, 57–64. [Google Scholar] [CrossRef]
- Liu, M.J.; Zhao, J.; Cai, Q.L.; Liu, G.C.; Wang, J.R.; Zhao, Z.H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.J.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef]
- Shen, L.Y.; Luo, H.; Wang, X.L.; Wang, X.M.; Qiu, X.J.; Liu, H.; Zhou, S.S.; Jia, K.H.; Nie, S.; Bao, Y.T. Chromosome-scale genome assembly for Chinese sour jujube and insights into its genome evolution and domestication signature. Front. Plant Sci. 2021, 12, 773090. [Google Scholar] [CrossRef]
- Wen, C.P.; Zhang, Z.; Shi, Q.Q.; Duan, X.S.; Du, J.T.; Wu, C.Y.; Li, X.G. Methyl jasmonate-and salicylic acid-induced transcription factor ZjWRKY18 regulates triterpenoid accumulation and salt stress tolerance in jujube. Int. J. Mol. Sci. 2023, 24, 3899. [Google Scholar] [CrossRef]
- Wen, C.P.; Zhang, Z.; Shi, Q.Q.; Niu, R.Z.; Duan, X.S.; Shen, B.Q.; Li, X.G. Transcription factors ZjMYB39 and ZjMYB4 regulate farnesyl diphosphate synthase-and squalene synthase-mediated triterpenoid biosynthesis in jujube. J. Agric. Food Chem. 2023, 71, 4599–4614. [Google Scholar] [CrossRef]
- Wen, C.P.; Zhang, Z.; Shi, Q.Q.; Yue, R.R.; Li, X.G. Metabolite and gene expression analysis underlying temporal and spatial accumulation of pentacyclic triterpenoids in jujube. Genes 2022, 13, 823. [Google Scholar] [CrossRef]
- Yue, R.R.; Zhang, Z.; Shi, Q.Q.; Duan, X.S.; Wen, C.P.; Shen, B.Q.; Li, X.G. Identification of the key genes contributing to the LOX-HPL volatile aldehyde biosynthesis pathway in jujube fruit. Int. J. Biol. Macromol. 2022, 222, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.H.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhang, X.D.; Li, C.X.; Chen, X.T.; Chio, C.; Shrestha, S.; Qin, W.S. Bacillus velezensis identification and recombinant expression, purification, and characterization of its alpha-amylase. Fermentation 2021, 7, 227. [Google Scholar] [CrossRef]
- Wang, Z.C.; Zhao, M.L.; Zhang, X.J.; Deng, X.M.; Li, J.; Wang, M.N. Genome-wide identification and characterization of active ingredients related β-Glucosidases in Dendrobium catenatum. BMC Genom. 2022, 23, 612. [Google Scholar] [CrossRef]
- Minami, Y.; Takao, H.; Kanafuji, T.; Miura, K.; Kondo, M.; Hara-Nishimura, I.; Nishimura, M.; Matsubara, H. β-Glucosidase in the indigo plant: Intracellular localization and tissue specific expression in leaves. Plant Cell Physiol. 1997, 38, 1069–1074. [Google Scholar] [CrossRef]
- Barth, C.; Jander, G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef]
- Wittstock, U.; Burow, M. Glucosinolate breakdown in Arabidopsis: Mechanism, regulation and biological significance. Arab. Book 2010, 2010, e0134. [Google Scholar] [CrossRef]
- Zhou, C.; Tokuhisa, J.G.; Bevan, D.R.; Esen, A. Properties of β-thioglucoside hydrolases (TGG1 and TGG2) from leaves of Arabidopsis thaliana. Plant Sci. 2012, 191, 82–92. [Google Scholar] [CrossRef]
- Rojas, M.; Ascencio, F.; Tiessen, A.; Arce-Montoya, M.; Gómez-Anduro, G. Infection of maize inbred B73 by Ustilago maydis and Fusarium proliferatum triggers differential expression of the β-glucosidase genes. Physiol. Mol. Plant Pathol. 2018, 104, 127–134. [Google Scholar] [CrossRef]
- Dong, X.S.; Jiang, Y.; Hur, Y. Genome-wide analysis of glycoside hydrolase family 1 β-glucosidase genes in Brassica rapa and their potential role in pollen development. Int. J. Mol. Sci. 2019, 20, 1663. [Google Scholar] [CrossRef]
- Ishihara, H.; Tohge, T.; Viehöver, P.; Fernie, A.R.; Weisshaar, B.; Stracke, R. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6. J. Exp. Bot. 2016, 67, 1505–1517. [Google Scholar] [CrossRef] [PubMed]
- Chapelle, A.; Morreel, K.; Vanholme, R.; Le Bris, P.; Morin, H.; Lapierre, C.; Boerjan, W.; Jouanin, L.; Demont Caulet, N. Impact of the absence of stem-specific β-glucosidases on lignin and monolignols. Plant Physiol. 2012, 160, 1204–1217. [Google Scholar] [CrossRef] [PubMed]
- Escamilla Treviño, L.L.; Chen, W.; Card, M.L.; Shih, M.C.; Cheng, C.L.; Poulton, J.E. Arabidopsis thaliana β-glucosidases BGLU45 and BGLU46 hydrolyse monolignol glucosides. Phytochemistry 2006, 67, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Roepke, J.; Gordon, H.O.; Neil, K.J.; Gidda, S.; Mullen, R.T.; Freixas Coutin, J.A.; Bray-Stone, D.; Bozzo, G.G. An apoplastic β-glucosidase is essential for the degradation of flavonol 3-O-β-glucoside-7-O-α-rhamnosides in Arabidopsis. Plant Cell Physiol. 2017, 58, 1030–1047. [Google Scholar] [CrossRef]
- Yan, C.H.; Yang, N.; Wang, X.Q.; Wang, Y.J. VqBGH40a isolated from Chinese wild Vitis quinquangularis degrades trans-piceid and enhances trans-resveratrol. Plant Sci. 2021, 310, 110989. [Google Scholar] [CrossRef]
- Ma, J.J.; Kang, L.P.; Zhou, W.B.; Yu, H.S.; Liu, P.; Ma, B.P. Identification and characterization of saponins in extract of Ziziphi spinosae Semen (ZSS) by ultra-performance liquid chromatography-electrospray ionization-quadrupole time-offlight tandem mass spectrometry (UPLC-ESI-QTOF-MSE). J. Med. Plants Res. 2011, 5, 6152–6159. [Google Scholar]
- Yang, B.; Yang, H.S.; Chen, F.; Hua, Y.L.; Jiang, Y.M. Phytochemical analyses of Ziziphus jujuba Mill. var. spinosa seed by ultrahigh performance liquid chromatography-tandem mass spectrometry and gas chromatography-mass spectrometry. Analyst 2013, 138, 6881–6888. [Google Scholar] [CrossRef]
- Kang, K.B.; Jang, D.S.; Kim, J.; Sung, S.H. UHPLC-ESI-qTOF-MS analysis of cyclopeptide alkaloids in the seeds of Ziziphus jujuba var. spinosa. Mass. Spectrom. Lett. 2016, 7, 45–49. [Google Scholar] [CrossRef]
- Henrissat, B.; Callebaut, I.; Fabrega, S.; Lehn, P.; Mornon, J.-P.; Davies, G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc. Natl. Acad. Sci. USA 1995, 92, 7090–7094. [Google Scholar] [CrossRef]
- Akiyama, T.; Kaku, H.; Shibuya, N. A cell wall-bound β-glucosidase from germinated rice: Purification and properties. Phytochemistry 1998, 48, 49–54. [Google Scholar] [CrossRef]
- Gänzle, M.G. Food fermentations for improved digestibility of plant foods–an essential ex situ digestion step in agricultural societies? Curr. Opin. Food Sci. 2020, 32, 124–132. [Google Scholar] [CrossRef]
- Morant, A.V.; Jørgensen, K.; Jørgensen, C.; Paquette, S.M.; Sánchez-Pérez, R.; Møller, B.L.; Bak, S. beta-Glucosidases as detonators of plant chemical defense. Phytochemistry 2008, 69, 1795–1813. [Google Scholar] [CrossRef]
- Li, H.T.; Gao, W.L.; Xue, C.L.; Zhang, Y.; Liu, Z.G.; Zhang, Y.; Meng, X.W.; Liu, M.J.; Zhao, J. Genome-wide analysis of the bHLH gene family in Chinese jujube (Ziziphus jujuba Mill.) and wild jujube. BMC Genom. 2019, 20, 568. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, W.L.; Li, H.T.; Wang, Y.K.; Li, D.; Xue, C.L.; Liu, Z.; Liu, M.G.; Zhao, J. Genome-wide analysis of the bZIP gene family in Chinese jujube (Ziziphus jujuba Mill.). BMC Genom. 2020, 21, 483. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Ma, Y.; Si, X.; Liu, X.; Geng, X.; Wen, X.; Li, G.; Zhang, L.; Yang, C.; Zhang, Z. Analysis of the Glycoside Hydrolase Family 1 from Wild Jujube Reveals Genes Involved in the Degradation of Jujuboside A. Genes 2023, 14, 1135. https://doi.org/10.3390/genes14061135
Yang M, Ma Y, Si X, Liu X, Geng X, Wen X, Li G, Zhang L, Yang C, Zhang Z. Analysis of the Glycoside Hydrolase Family 1 from Wild Jujube Reveals Genes Involved in the Degradation of Jujuboside A. Genes. 2023; 14(6):1135. https://doi.org/10.3390/genes14061135
Chicago/Turabian StyleYang, Mingjun, Yimian Ma, Xupeng Si, Xiaofeng Liu, Xin Geng, Xin Wen, Guoqiong Li, Liping Zhang, Chengmin Yang, and Zheng Zhang. 2023. "Analysis of the Glycoside Hydrolase Family 1 from Wild Jujube Reveals Genes Involved in the Degradation of Jujuboside A" Genes 14, no. 6: 1135. https://doi.org/10.3390/genes14061135
APA StyleYang, M., Ma, Y., Si, X., Liu, X., Geng, X., Wen, X., Li, G., Zhang, L., Yang, C., & Zhang, Z. (2023). Analysis of the Glycoside Hydrolase Family 1 from Wild Jujube Reveals Genes Involved in the Degradation of Jujuboside A. Genes, 14(6), 1135. https://doi.org/10.3390/genes14061135





