Abstract
Background: The PCSK3 gene encodes for the protease enzyme Furin, which promotes proteolytic maturation of important regulators of the immune response, and also enhances the secretion of interferon-γ (IFN). Several studies have suggested its possible involvement in the pathogenesis of chronic inflammatory diseases. Methods: We investigated the PCSK3 gene expression level in peripheral blood mononuclear cells isolated from Sjögren’s Syndrome (SS) patients and healthy controls and we evaluated a possible correlation with IFN-γ gene expression. Moreover, we also explored the variability of two PCSK3 genetic polymorphisms (rs4932178 and rs4702) to evaluate a possible association between these polymorphisms and the expression levels of this gene. Results: We observed, by RT-qPCR, that the PCSK3 expression level was significantly higher in SS patients compared to the controls (p = 0.028), and we confirmed a positive correlation between PCSK3 and IFN-γ expression levels (p < 0.001). Moreover, we reported that the variant homozygous genotype of rs4932178 SNP is associated with a higher expression of the PCSK3 gene (p = 0.038) and with the SS susceptibility (p = 0.016). Conclusions: Our data suggest that Furin could play a role in SS development, also promoting IFN-γ secretion.
1. Introduction
Sjögren’s Syndrome (SS; OMIM 270150) is a chronic inflammatory autoimmune disease. It is characterized by chronic lymphocytic infiltrates in the exocrine glands, resulting in dry eye and dry mouth. The spectrum of this disease may also include systemic manifestations such as arthritis, interstitial lung involvement, and neurological involvement [1]. Interactions between environmental stimuli and genetic susceptibility factors are involved in disease development [2], but SS etiology is still partially unknown. The identification of new genes associated with this disease could therefore help to gain a more complete view on the mechanisms involved in SS development.
Several studies have suggested the involvement of the PCSK3 gene in the pathogenesis of chronic inflammatory diseases, such as atherosclerosis, rheumatoid arthritis (RA), and systemic lupus erythematous (SLE) [3,4,5]. The PCSK3 gene, consisting of sixteen exons, is located on chromosome 15q26.1 and encodes for a convertase subtilisin/kexin enzyme (Furin) that promotes proteolytic maturation of pro-proteins, thanks to its cleavage properties [6]. Furin is an important regulator of the immune response, and it is known to exert a pro-inflammatory function [3]. Indeed, among its substrates, some have immunoregulatory functions such as cytokines, integrins, and proteins of the viral envelope [7]. However, Cordova et al. have shown that Furin expression could also reduce the production of pro-inflammatory cytokines in myeloid cells [8]. In addition, it is demonstrated that the expression of Furin in T cells is critical for the maintenance of peripheral immune tolerance [9]; in fact, its deletion in mouse T cells results in a loss of tolerance [10]. As in all autoimmune inflammatory conditions, tolerance defects also contribute to SS pathogenesis [11]. An upregulation of Furin has been observed in SS, particularly in salivary gland biopsies and peripheral circulation [12,13]. Moreover, Furin is a ubiquitously expressed protein, preferentially in Th1 cells, and promotes the secretion of interferon-γ (IFN) by activated T cells [10,14]. IFN-γ is known to mediate Th1 cell functions, macrophage activation, and immunoglobulin class switching, and it has been widely associated with systemic autoimmunity [15]. Recent studies in both animal models and patients have demonstrated that IFN type II (IFN-γ) is involved in the pathogenesis of SS. Indeed, over half of SS patients exhibit an IFN signature and high levels of IFN-γ were found in serum, saliva, and Th cells from salivary gland biopsies of these patients [16,17,18]. Moreover, in SS patients, IFN-γ levels correlate with the disease activity score, and the presence of inflammatory infiltration is closely related to IFN-γ activity [18].
Genetic variants located in regulatory regions of the PCSK3 gene could influence its transcript levels. Indeed, it is known that the PCSK3 expression is regulated by three promoters that activate the transcription of three mRNA isoforms with a different 5′-untranslated region (UTR) [19]. The rs4932178 SNP (Single Nucleotide Polymorphism) in the most active promoter of this gene has been described as associated with a higher mRNA level in vitro. PCSK3 expression levels are regulated also by 3′UTR, where several miRNAs binding sites are located. Several studies suggest that a SNP (rs4702) in this region causes a differential binding of miR-338-3p and could influence the gene expression [20,21].
Here, we aimed to investigate the PCSK3 gene expression level in peripheral blood mononuclear cells (PBMCs) isolated from SS patients and healthy controls (CTRLs) and to evaluate a possible correlation with IFN-γ gene expression. Moreover, we explored the variability of two PCSK3 genetic polymorphisms (rs4932178 and rs4702), located in regulatory regions, to evaluate a possible association between these polymorphisms and their expression levels.
2. Materials and Methods
SS patients were enrolled from the Sjögren’s Clinic of Sapienza University of Rome and diagnosed according to the 2016 ACR-EULAR Classification Criteria [22]. Study protocol included a complete physical examination and blood drawing. The clinical and laboratory data were collected in a standardized filled form including demographics, past medical history, date of diagnosis, comorbidities, and previous/concomitant treatments. Regarding laboratory data, the following has been performed for each patient: a complete cell blood count, including the leucocyte, erythrocyte, and platelet counts; serum protein electrophoresis to determine levels of C3 and C4 complements, hypergammaglobulinemia, and monoclonal components; immunofluorescence (IIF) on Hep-2 to reveal antinuclear antibodies (ANA) (considered present at a titer 1:160); Enzyme-Linked Immunosorbent Assay (ELISA) to detect anti-SSA and anti-SSB antibodies; Waaler–Rose test and/or Ra test to detect the rheumatoid factor presence; cryoprecipitate detection to determine the cryoglobulins (samples kept at 37 °C, warm centrifugation, warm cell precipitation, serum conservation at 4 °C, and cryoprecipitate detection after 7 days).
Healthy controls were enrolled at the University of Rome “Tor Vergata”. Written informed consent was obtained from each participant and the ethical committee of Sapienza University of Rome approved the study design. Peripheral blood samples from all patients and controls have been collected and stored at −20 °C until usage. The expression study was performed in a group of 27 SS patients and 18 healthy controls. Demographic and clinical features of both groups were reported in Table 1.

Table 1.
Demographic and clinical features of SS patients and healthy controls.
Total RNA was isolated from PBMCs using the TRIzol reagent (Ambion, CA, USA) protocol, followed by reverse transcription using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA, USA). Expression analysis was performed by quantitative RT-polymerase chain reaction (RT-qPCR) (SYBR Green Assay, Applied Biosystems), using the 7500 Real-Time PCR System (Applied Biosystems). PCSK3 expression analysis was performed on SS patients and healthy controls, while IFN-γ mRNA levels was evaluated only on the SS group. The primers used to detect gene expression are: 5′-AAGATGACCCAGATCATGTTTGAGACC and 3′-AGCCAGGTCCAGACGCAGGAT for β-Actin; 5′-CGGAAAGTGAGCCACTCATA and 3′-TGTCTTTGGGCTCGGTGAG for PCSK3; 5′-GCATCCAAAAGAGTGTGGAG and 3′-GACAGTTCAGCCATCACTTGG for IFN-γ. Each sample was analysed in triplicate and, to standardize the results, each assay was run with an endogenous control (β-Actin). Relative expression levels were calculated using the 2−ΔΔCt method and data were reported as mean values ± standard deviation.
Genomic DNA was isolated from PBMCs of 195 SS patients using a Qiagen blood DNA mini-kit. A demographic and clinical description of patients was previously reported [23]. Patients were analysed for two polymorphisms in the PCSK3 gene (rs4932178 and rs4702), and genotyping was performed by direct sequencing (ABI 3130xl Automated Sequencer (Applied Biosystems, Foster City, CA, USA)) using the following primers: 5′-CATAATTGTGGCAGCACTGG and 3′-AGCACCTGGGATTCATCCTG for PCSK3 rs4932178; 5′-TTCCTGGTACCCAGCCATCT and 3′-CAGGCAGGCCACTGTGTAG for PCSK3 rs4702.
DNA and RNA quality and concentration were evaluated by the NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and RNA integrity was assessed by standard denaturing agarose gel electrophoresis.
Pearson’s χ2 test has been used to verify the Hardy–Weinberg equilibrium for SNPs and to compare genotype frequencies of patients with respect to those listed in the 1000 Genomes Project database for the European non-Finnish population. Odds ratios with a 95% CI were calculated. The ANOVA test was used to compare expression values among the different phenotypic and genotypic groups. The linear regression analysis was used to evaluate the correlation between PCSK3 and IFN-γ mRNA levels. A p-value < 0.05 was considered as significant.
All graphs were performed by GraphPad Prism 9 (GraphPad Software, Boston, MA, USA). All statistical analyses were performed by the SPSS program, version 25 (IBM Corp, Armonk, NY, USA).
3. Results
For this study, we included 27 patients with SS (88.5% women), with a mean age of 58.79 ± 10.33 years and a disease duration of 4.64 ± 5.01 years. Among the patients, 82.1% developed ANA, 64.3% anti-SSA, and 53.6% anti-SSB antibodies. Additionally, 14.3% of cases presented arthritis, 7.1% lymphoproliferative complications (non-Hodgkin lymphoma), and 3.6 salivary gland swelling. For the control group, we enrolled 18 sex- and age-matched healthy subjects (83.3% women; mean age of 59.3 ± 10.21 years) (Table 1).
Firstly, we analysed the PCSK3 gene expression in PBMCs of 27 SS patients and 18 healthy controls. We observed, by RT-qPCR, that PCSK3 expression level was significantly higher in SS patients, compared to the controls (p = 0.028; Figure 1).
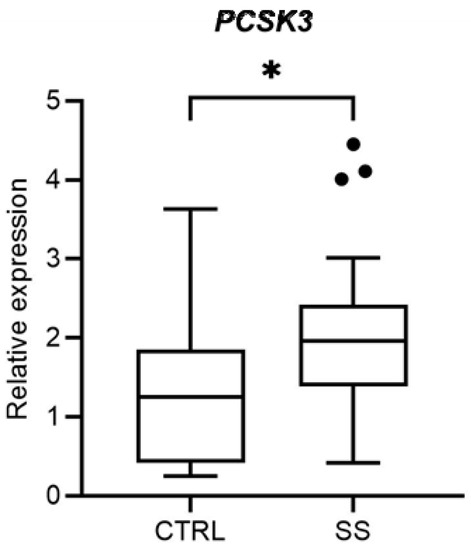
Figure 1.
Comparison of PCSK3 expression levels between healthy controls (CTRL) and Sjögren Syndrome patients (SS). * p = 0.028.
We also examined the possible correlation between PCSK3 expression levels and specific clinical features of SS patients reported in Table 1, but we did not find any significant association.
We then evaluated whether PCSK3 expression affects IFN-γ regulation in SS patients. We observed that SS patients with high PCSK3 mRNA levels showed also elevated mRNA levels of IFN-γ; indeed, we confirmed a positive correlation between PCSK3 and IFN-γ expression levels (R2 = 0.41; p < 0.001; Figure 2).
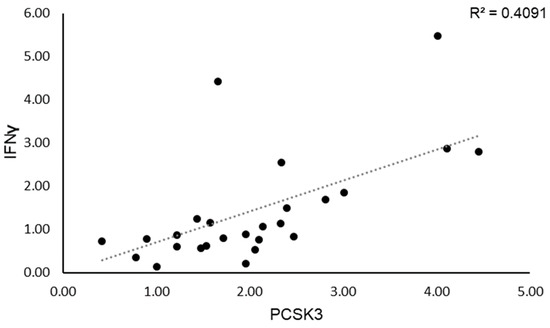
Figure 2.
Correlation between IFN-γ and PCSK3 expression levels.
To verify the possible association between common polymorphisms localized in the regulatory regions of the PCSK3 gene and the different expression levels of the same gene, we compared the distribution of the mean values of PCSK3 expression in the different genotypic classes for two selected SNPs in the SS patients. The analysed SNPs were rs4932178 and rs4702. As shown in Figure 3, the rs4932178 polymorphism variant allele in the PCSK3 promoter region was associated with a higher expression of this gene. In particular, the patients with homozygous variant genotype (TT) showed an increase of PCSK3 expression compared with the patients with homozygous wild-type (CC) and heterozygous (CT) genotypes (p = 0.038). No significant association was observed between the rs4702 polymorphism located in 3′-UTR region and the expression levels of PCSK3 gene.
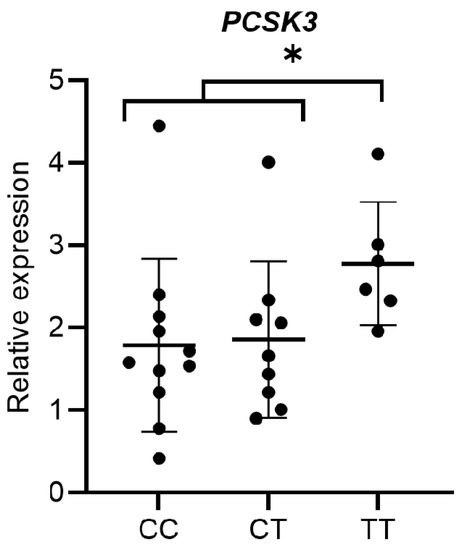
Figure 3.
Distribution of mean expression levels of PCSK3 among the genotypic classes for the polymorphism rs4932178. * p = 0.038.
Lastly, we investigated the genotypes distribution of the two polymorphisms previously evaluated, in a cohort of 195 SS patients. We compared the distributions of genotypic frequencies of the two PCSK3 polymorphisms (rs4932178 and rs4702) in the SS patients to the frequencies listed in the 1000 Genomes Project database for the European non-Finnish population. In the light of the higher expression level seen in the homozygous TT genotype, the associations were evaluated under a recessive genetic model. The PCSK3 genotype distribution and the case/control association analysis are reported in Table 2.

Table 2.
Association analysis between PCSK3 SNPs and Sjögren Syndrome.
As shown, the variant homozygous genotype (TT) of rs4932178 SNP was associated with SS susceptibility (p = 0.016 and OR = 1.71). Conversely, no statistically significant associations were observed for rs4702. To complete the association analysis, we also evaluated the recessive genetic model, but in this case, we did not observe any significant differences.
4. Discussion
Increasing evidence supports the role of the Furin enzyme in the regulation of peripheral immune tolerance by processing many substrates with immunoregulatory functions, including cytokines and integrins [7]. PCSK3 expression is indeed upregulated in activated T cells and previous studies have shown that it is highly expressed in patients with autoimmune diseases, such as SLE [5] or RA [24].
In this study, in agreement with data previously reported by Ranta et al. [13], we observed an increase of PCSK3 levels in PBMC of SS patients. Functional studies have demonstrated that PCSK3 is indispensable in maintaining peripheral tolerance, and alteration of its expression levels could affect Treg cell function [10]. Indeed, inhibition of PCSK3 was found to reduce the expression and activation of cytokines and growth factors, such as TGFβ [10]. We also reported a positive correlation between PCSK3 and IFN-γ expression levels. The main pathophysiological mechanisms involved in SS are mediated through the interferon IFN I and IFN II pathways. In particular, in vivo studies have shown that reduction of IFN-γ seems to inhibit the development of SS [25]. Another study showed that PCSK3 is one of the genes most consistently activated by IL-12, the main mediator of T-Helpers [26]. The authors also demonstrated that the secretion of IFN-γ was enhanced by Furin expression, while the inhibition of this enzyme interfered with IFN-γ production. However, it is not yet clear how this regulation takes place. Indeed, the authors had initially identified a potential Furin-target amino acid sequence on IFN-γ and hypothesized that the enzyme could directly cleave IFN-γ to promote its maturation. The introduction of amino acid mutations in this IFN-γ sequence did not alter the Furin enhance activity, suggesting a different form of regulation [26].
In the present work, we also described an association between rs4932178 SNP located in the PCSK3 promoter region and the expression level of this gene. Specifically, patients with the homozygous variant genotype showed an increase of PCSK3 expression and this genotype turned out to be also associated with a higher risk of developing SS. The PCSK3 gene is located at position 15q26.1, and its transcription is mediated by three distinct promoters. Rs4932178 SNP is located in the most active promoter region, at −229 nucleotides from the start codon AUG. Lei et al. demonstrated using a luciferase reporter gene assay that transcription activity in variant allele carriers is about three times higher than in wild-type allele carriers of this SNP [19]. Indeed, they showed that the promoter with variant allele binds more efficiently the transcription factor NF-E2 [nuclear factor (erythroid-derived 2)] and induces a higher transcription activity of PCSK3 gene.
Moreover, we observed a different genotypic distribution of rs4932178 polymorphism in SS patients than reported in the 1000 Genomes Project database for the European non-Finnish population. In particular, the variant homozygous genotype (TT) seems to be more frequent in the SS cohort compared to the general population. Since this genotype appears to be associated with increased expression levels of PCSK3, it is not surprising that it is more frequent in SS patients, who, in fact, express higher levels of this enzyme.
During the last two years, Furin activity has also been extensively evaluated with regard to SARS-CoV-2 infection. Indeed, several studies demonstrated that the spike (S) protein of this virus contains a functional Furin-cleavage sequence (RRAR), essential for the membrane-fusion process and for the viral entry into the cell [27]. Moreover, genetic variants in the PCSK3 gene have been described as possibly associated with SARS-CoV-2 infection susceptibility [28]. COVID-19 shares clinical manifestations and pathogenic mechanisms with autoimmune diseases and several immune reactions participate in the pathogenesis of both conditions [29,30]. Indeed, numerous immunity genes were found associated with COVID-19 susceptibility and severity [31,32]. Our study now suggests that PCSK3, already described as associated to COVID19, could also be included in the IFN-γ network and considered a novel susceptibility gene for autoimmune disorders.
5. Conclusions
In conclusion, we observed an increase of PCSK3 mRNA levels in SS patients compared to control subjects and an association of variant homozygous genotype (TT) of rs4932178 SNP, both with a higher expression of this gene and with the SS susceptibility. Moreover, we observed a positive correlation between PCSK3 and IFN-γ expression levels. This study has some limitations: the absence of data regarding the measurement of Furin protein levels by ELISA and the small size of the analysed cohort. Although replication studies on a larger cohort will be necessary to confirm our data, these results suggest for the first time that Furin could play a role in SS development. Functional studies are necessary to clarify the contribution of Furin to the SS pathogenesis.
Author Contributions
Conceptualization, P.B. and C.C.; methodology, G.D.B. and A.L.; formal analysis, G.D.B. and A.L.; investigation, S.C., R.P. and C.P.; resources, S.C., R.P. and C.P.; data curation, G.D.B.; writing—original draft preparation, G.D.B., A.L. and C.C.; writing—review and editing, L.N. and P.B.; supervision, P.B. and C.C.; funding acquisition, G.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded primarily by the MUR-PNRR M4-C2-I1.3 PE6 project PE00000019 Heal Italia, and partially by EU-Horizon-HLTH-2021-ID: 101057100 (UNDINE) to G.N.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of Sapienza University of ROME (protocol code 4688/2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All available data may be found in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chivasso, C.; Sarrand, J.; Perret, J.; Delporte, C.; Soyfoo, M.S. The Involvement of Innate and Adaptive Immunity in the Initiation and Perpetuation of Sjögren’s Syndrome. Int. J. Mol. Sci. 2021, 22, 658. [Google Scholar] [CrossRef]
- Imgenberg-Kreuz, J.; Rasmussen, A.; Sivils, K.; Nordmark, G. Genetics and epigenetics in primary Sjögren’s syndrome. Rheumatology 2021, 60, 2085–2098. [Google Scholar] [CrossRef]
- Turpeinen, H.; Raitoharju, E.; Oksanen, A.; Oksala, N.; Levula, M.; Lyytikäinen, L.P.; Järvinen, O.; Creemers, J.W.; Kähönen, M.; Laaksonen, R.; et al. Proprotein convertases in human atherosclerotic plaques: The overexpression of FURIN and its substrate cytokines BAFF and APRIL. Atherosclerosis 2011, 219, 799–806. [Google Scholar] [CrossRef]
- Lin, H.; Ah Kioon, M.D.; Lalou, C.; Larghero, J.; Launay, J.M.; Khatib, A.M.; Cohen-Solal, M. Protective role of systemic furin in immune response-induced arthritis. Arthritis Rheum. 2012, 64, 2878–2886. [Google Scholar] [CrossRef]
- Wu, T.; Ding, H.; Han, J.; Arriens, C.; Wei, C.; Han, W.; Pedroza, C.; Jiang, S.; Anolik, J.; Petri, M.; et al. Antibody-Array-Based Proteomic Screening of Serum Markers in Systemic Lupus Erythematosus: A Discovery Study. J. Proteome Res. 2016, 15, 2102–2114. [Google Scholar] [CrossRef]
- Garten, W. Characterization of Proprotein Convertases and Their Involvement in Virus Propagation. In Activation of Viruses by Host Proteases; Springer: Berlin/Heidelberg, Germany, 2018; pp. 205–248. [Google Scholar]
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073. [Google Scholar] [CrossRef]
- Cordova, Z.M.; Grönholm, A.; Kytölä, V.; Taverniti, V.; Hämäläinen, S.; Aittomäki, S.; Niininen, W.; Junttila, I.; Ylipää, A.; Nykter, M.; et al. Myeloid cell expressed proprotein convertase FURIN attenuates inflammation. Oncotarget 2016, 7, 54392–54404. [Google Scholar] [CrossRef]
- Ortutay, Z.; Oksanen, A.; Aittomäki, S.; Ortutay, C.; Pesu, M. Proprotein convertase FURIN regulates T cell receptor-induced transactivation. J. Leukoc. Biol. 2015, 98, 73–83. [Google Scholar] [CrossRef]
- Pesu, M.; Watford, W.T.; Wei, L.; Xu, L.; Fuss, I.; Strober, W.; Andersson, J.; Shevach, E.M.; Quezado, M.; Bouladoux, N.; et al. T-cell-expressed proprotein convertase furin is essential for maintenance of peripheral immune tolerance. Nature 2008, 455, 246–250. [Google Scholar] [CrossRef]
- Psianou, K.; Panagoulias, I.; Papanastasiou, A.D.; de Lastic, A.L.; Rodi, M.; Spantidea, P.I.; Degn, S.E.; Georgiou, P.; Mouzaki, A. Clinical and immunological parameters of Sjögren’s syndrome. Autoimmun. Rev. 2018, 17, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Sisto, M.; Lisi, S.; Lofrumento, D.D.; Ingravallo, G.; Mitolo, V.; D’Amore, M. Expression of pro-inflammatory TACE-TNF-α-amphiregulin axis in Sjögren’s syndrome salivary glands. Histochem. Cell. Biol. 2010, 134, 345–353. [Google Scholar] [CrossRef]
- Ranta, N.; Valli, A.; Grönholm, A.; Silvennoinen, O.; Isomäki, P.; Pesu, M.; Pertovaara, M. Proprotein convertase enzyme FURIN is upregulated in primary Sjögren’s syndrome. Clin. Exp. Rheumatol. 2018, 36 (Suppl. S112), 47–50. [Google Scholar] [PubMed]
- Ortutay, Z.; Grönholm, A.; Laitinen, M.; Keresztes-Andrei, M.; Hermelo, I.; Pesu, M. Identification of Novel Genetic Regulatory Region for Proprotein Convertase FURIN and Interferon Gamma in T Cells. Front. Immunol. 2021, 12, 630389. [Google Scholar] [CrossRef]
- Ivashkiv, L.B. IFNγ: Signalling, epigenetics and roles in immunity, metabolism, disease and cancer immunotherapy. Nat. Rev. Immunol. 2018, 18, 545–558. [Google Scholar] [CrossRef]
- Sebastian, A.; Madej, M.; Sebastian, M.; Łuczak, A.; Gajdanowicz, P.; Zemelka-Wiącek, M.; Wiland, P. The Clinical and Immunological Activity Depending on the Presence of Interferon γ in Primary Sjögren’s Syndrome-A Pilot Study. J. Clin. Med. 2021, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Arakaki, R.; Tawara, H.; Nagao, R.; Tanaka, H.; Tamura, K.; Kawahito, Y.; Otsuka, K.; Ushio, A.; Tsunematsu, T.; et al. Disturbed natural killer cell homeostasis in the salivary gland enhances autoimmune pathology via IFN-γ in a mouse model of primary Sjögren’s syndrome. Front. Med. 2022, 9, 1036787. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, X.; Shi, X.; Liu, Y.; Cheng, D.; Tian, Q.; Lin, N.; Wei, W.; Wu, H. CXCL9, 10, 11/CXCR3 Axis Contributes to the Progress of Primary Sjogren’s Syndrome by Activating GRK2 to Promote T Lymphocyte Migration. Inflammation 2023. Epub ahead of print. [Google Scholar] [CrossRef]
- Lei, R.X.; Shi, H.; Peng, X.M.; Zhu, Y.H.; Cheng, J.; Chen, G.H. Influence of a single nucleotide polymorphism in the P1 promoter of the furin gene on transcription activity and hepatitis B virus infection. Hepatology 2009, 50, 763–771. [Google Scholar] [CrossRef]
- Yang, S.; Fu, Z.Z.; Zhang, Y.Q.; Fu, B.H.; Dong, L. The G to A transformation of rs4702 polymorphism in 3’UTR of FURIN reduced the risk of radiotherapy-induced cognitive impairment in glioma patients. J. Cell. Mol. Med. 2022, 26, 684–692. [Google Scholar] [CrossRef]
- Hou, Y.; Liang, W.; Zhang, J.; Li, Q.; Ou, H.; Wang, Z.; Li, S.; Huang, X.; Zhao, C. Schizophrenia-associated rs4702 G allele-specific downregulation of FURIN expression by miR-338-3p reduces BDNF production. Schizophr. Res. 2018, 199, 176–180. [Google Scholar] [CrossRef]
- Shiboski, C.H.; Shiboski, S.C.; Seror, R.; Criswell, L.A.; Labetoulle, M.; Lietman, T.M.; Rasmussen, A.; Scofield, H.; Vitali, C.; Bowman, S.J.; et al. 2016 American College of Rheumatology/European League Against Rheumatism Classification Criteria for Primary Sjögren’s Syndrome: A Consensus and Data-Driven Methodology Involving Three International Patient Cohorts. Arthritis Rheumatol. 2017, 69, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Colafrancesco, S.; Ciccacci, C.; Priori, R.; Latini, A.; Picarelli, G.; Arienzo, F.; Novelli, G.; Valesini, G.; Perricone, C.; Borgiani, P. STAT4, TRAF3IP2, IL10, and HCP5 Polymorphisms in Sjögren’s Syndrome: Association with Disease Susceptibility and Clinical Aspects. J. Immunol. Res. 2019, 2019, 7682827. [Google Scholar] [CrossRef] [PubMed]
- Valli, A.; Ranta, N.; Grönholm, A.; Silvennoinen, O.; Pesu, M.; Isomäki, P. Increased expression of the proprotein convertase enzyme FURIN in rheumatoid arthritis. Scand. J. Rheumatol. 2019, 48, 173–177. [Google Scholar] [CrossRef]
- Nocturne, G.; Mariette, X. Advances in understanding the pathogenesis of primary Sjögren’s syndrome. Nat. Rev. Rheumatol. 2013, 9, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Pesu, M.; Muul, L.; Kanno, Y.; O’Shea, J.J. Proprotein convertase furin is preferentially expressed in T helper 1 cells and regulates interferon gamma. Blood 2006, 108, 983–985. [Google Scholar] [CrossRef] [PubMed]
- Coutard, B.; Valle, C.; de Lamballerie, X.; Canard, B.; Seidah, N.G.; Decroly, E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020, 176, 104742. [Google Scholar] [CrossRef] [PubMed]
- Latini, A.; Agolini, E.; Novelli, A.; Borgiani, P.; Giannini, R.; Gravina, P.; Smarrazzo, A.; Dauri, M.; Andreoni, M.; Rogliani, P.; et al. COVID-19 and Genetic Variants of Protein Involved in the SARS-CoV-2 Entry into the Host Cells. Genes 2020, 11, 1010. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Sawalha, A.H.; Lu, Q. COVID-19 and autoimmune diseases. Curr. Opin. Rheumatol. 2021, 33, 155–162. [Google Scholar] [CrossRef]
- Novelli, L.; Motta, F.; De Santis, M.; Ansari, A.A.; Gershwin, M.E.; Selmi, C. The JANUS of chronic inflammatory and autoimmune diseases onset during COVID-19-A systematic review of the literature. J. Autoimmun. 2021, 117, 102592. [Google Scholar] [CrossRef]
- Li, P.; Ke, Y.; Shen, W.; Shi, S.; Wang, Y.; Lin, K.; Guo, X.; Wang, C.; Zhang, Y.; Zhao, Z. Targeted screening of genetic associations with COVID-19 susceptibility and severity. Front. Genet. 2022, 13, 1073880. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; COVID Human Genetic Effort; Cobat, A.; Casanova, J.L. Human genetic and immunological determinants of critical COVID-19 pneumonia. Nature 2022, 603, 587–598. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).