Japanese Flounder pol-miR-155 Is Involved in Edwardsiella tarda Infection via ATG3
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Lines
2.3. In Vivo Infection of E. tarda
2.4. Intracellular Replication Assay of E. tarda
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Luciferase Reporter Assay
2.7. Western Blot
2.8. Effect of pol-miR-155 and ATG3 on E. tarda Infection
2.9. MiRNA Mimic and siRNA
2.10. Statistical Analysis
3. Results
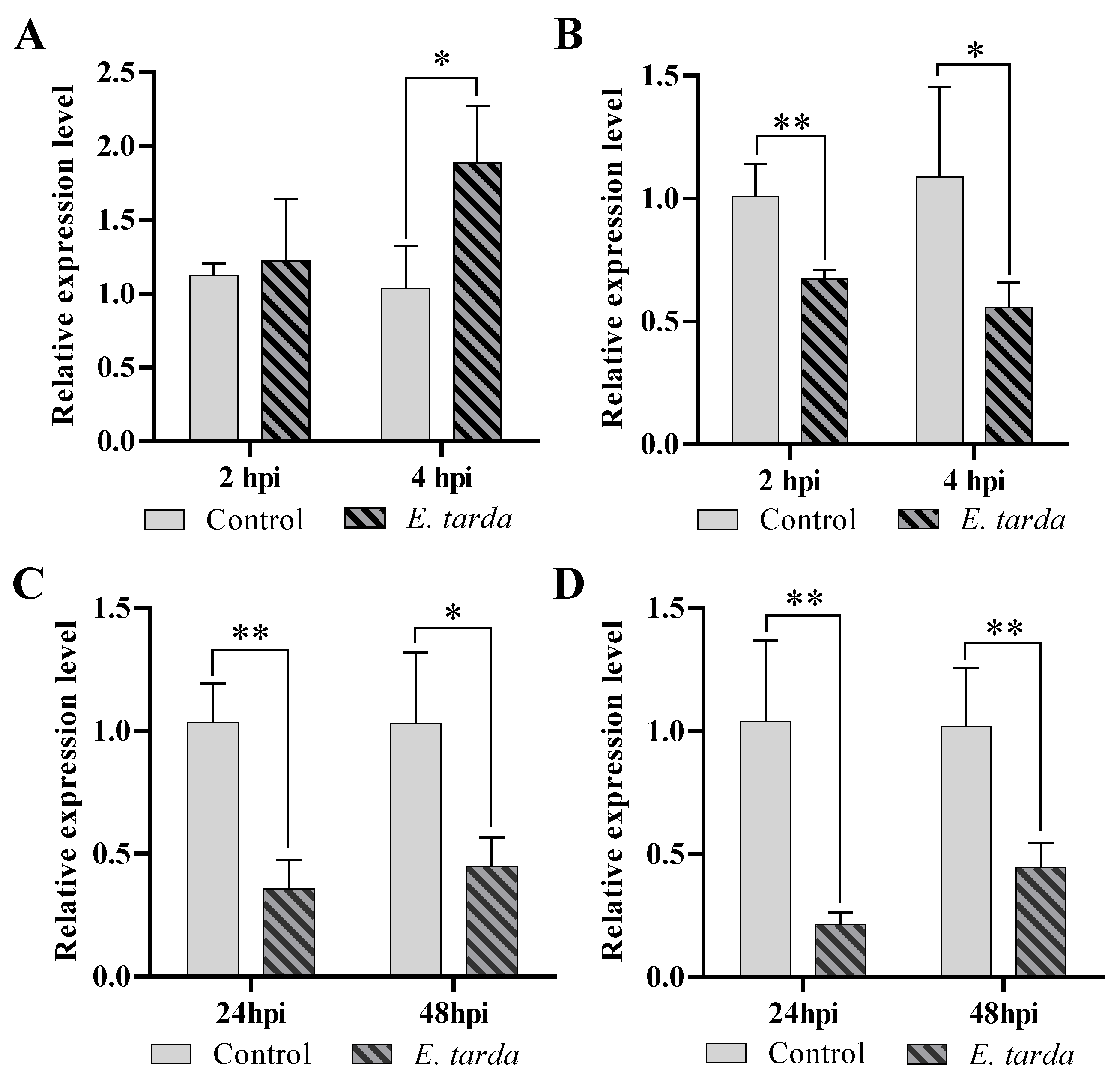
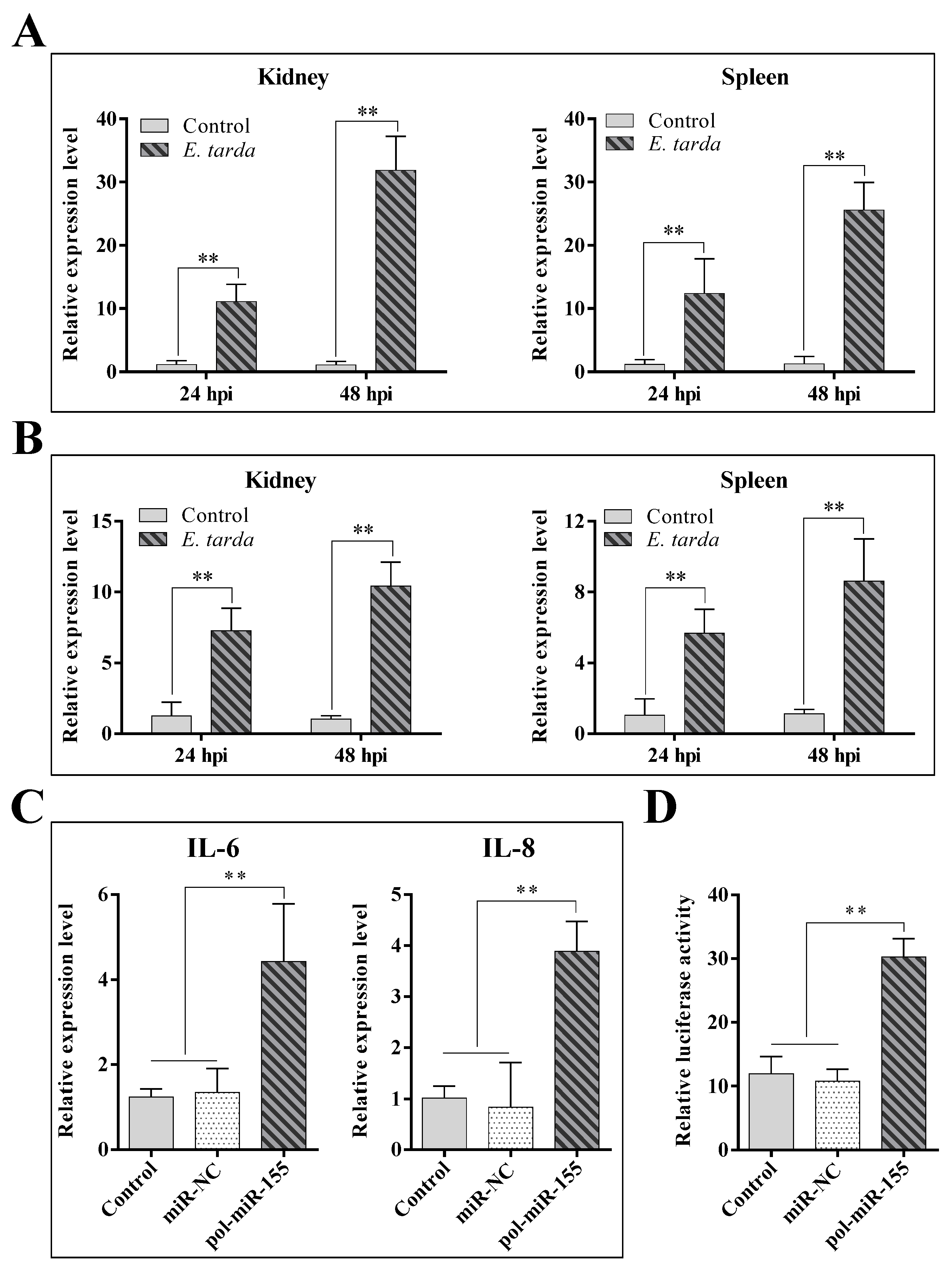
3.1. The Expression of pol-miR-155 and ATG3 during E. tarda Infection
3.2. Effects of pol-miR-155 and ATG3 on E. tarda Infection
3.3. Identification of ATG3 as a Target Gene of pol-miR-155
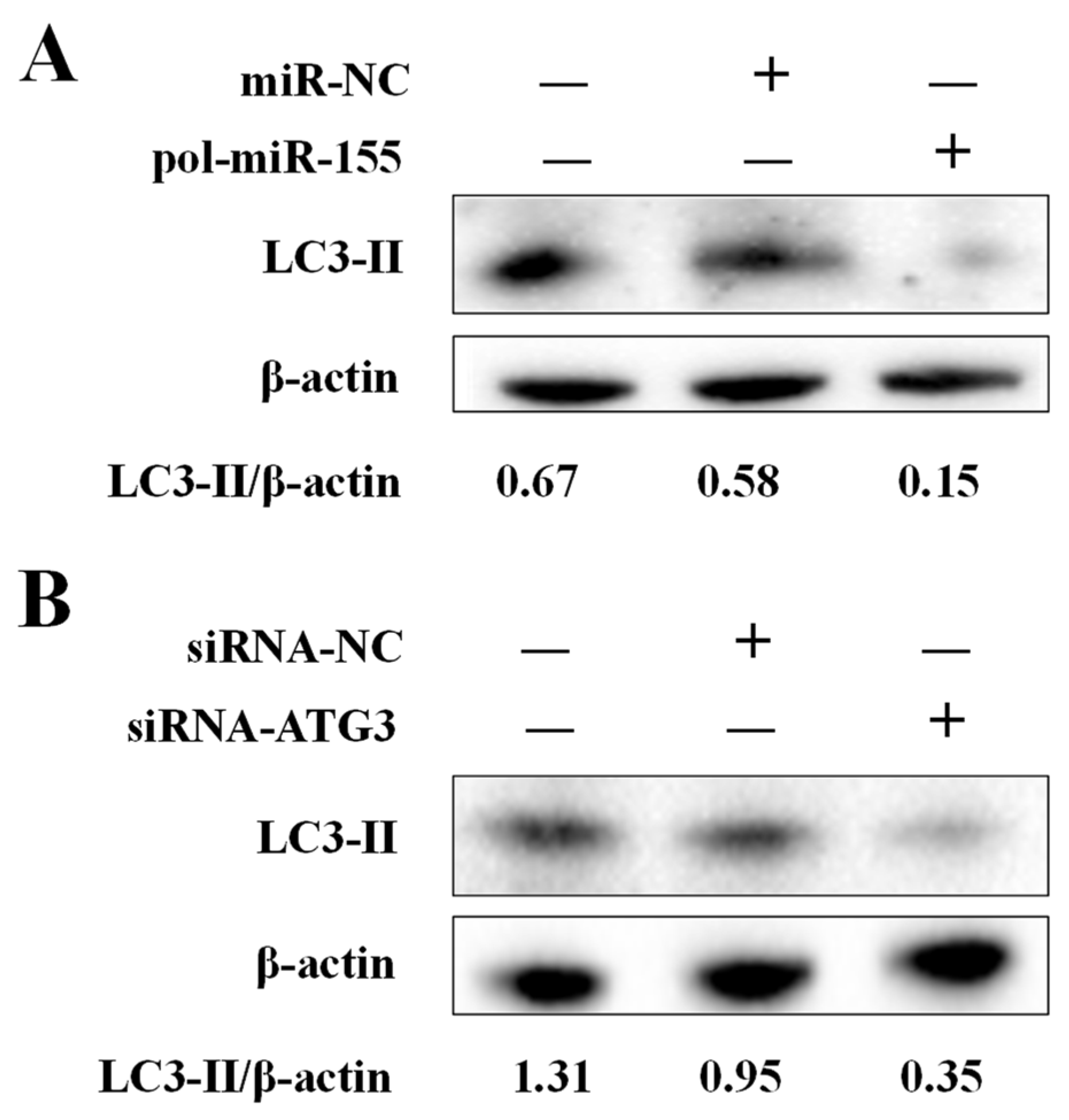
3.4. Effects of ATG3 and pol-miR-155 on Autophagy
3.5. The Influence of E. tarda and pol-miR-155 on the Activity of the NF-κB Pathway
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Correia de Sousa, M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef][Green Version]
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14, dmm047662. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu Gao, M.Y.; Zhang, L.; He, F.L.; Shi, Y.K.; Pan, X.H.; Wang, H. MicroRNA-155 affects oxidative damage through regulating autophagy in endothelial cells. Oncol. Lett. 2019, 17, 2237–2243. [Google Scholar] [CrossRef]
- Cui, Z.; Liu, L.; Kwame Amevor, F.; Zhu, Q.; Wang, Y.; Li, D.; Shu, G.; Tian, Y.; Zhao, X. High Expression of miR-204 in Chicken Atrophic Ovaries Promotes Granulosa Cell Apoptosis and Inhibits Autophagy. Front. Cell Dev. Biol. 2020, 8, 580072. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Pan, C.; Li, L.; Bian, Z.; Lv, Z.; Shi, L.; Zhang, J.; Li, D.; Gu, H.; Zhang, C.Y.; et al. MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein α. J. Allergy Clin. Immunol. 2013, 132, 426–436. [Google Scholar] [CrossRef][Green Version]
- Rao, L.; Meng, F.L.; Fang, R.; Cai, C.Y.; Zhao, X.L. Molecular mechanism of microRNA in regulating cochlear hair cell development. Yi Chuan Hered. 2019, 41, 994–1008. [Google Scholar]
- Cazzanelli, P.; Wuertz-Kozak, K. MicroRNAs in Intervertebral Disc Degeneration, Apoptosis, Inflammation, and Mechanobiology. Int. J. Mol. Sci. 2020, 21, 3601. [Google Scholar] [CrossRef]
- Zhao, Z.; Sun, W.; Guo, Z.; Zhang, J.; Yu, H.; Liu, B. Mechanisms of lncRNA/microRNA interactions in angiogenesis. Life Sci. 2020, 254, 116900. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA Functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef]
- Mishra, R.; Krishnamoorthy, P.; Kumar, H. MicroRNA-30e-5p Regulates SOCS1 and SOCS3 during Bacterial Infection. Front. Cell. Infect. Microbiol. 2021, 10, 604016. [Google Scholar] [CrossRef]
- Cui, J.; Li, Z.; Cui, K.; Gao, Y.; Zhang, B.; Niu, J.; Wang, Y. MicroRNA-20a-3p regulates the host immune response to facilitate the mycobacterium tuberculosis infection by targeting IKKβ/NF-κB pathway. Int. Immunopharmacol. 2021, 91, 107286. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Cui, H.; Ni, S.; Yan, Y.; Qin, Q. Comprehensive identification and profiling of host miRNAs in response to Singapore grouper iridovirus (SGIV) infection in grouper (Epinephelus coioides). Dev. Comp. Immunol. 2015, 52, 226–235. [Google Scholar] [CrossRef]
- Zhang, B.-C.; Zhou, Z.-J.; Sun, L. pol-miR-731, a teleost miRNA upregulated by megalocytivirus, negatively regulates virus-induced type I interferon response, apoptosis and cell cycle arrest. Sci. Rep. 2016, 6, 28354. [Google Scholar] [CrossRef] [PubMed]
- Nie, L.; Cai, S.-Y.; Sun, J.; Chen, J. MicroRNA-155 promotes pro-inflammatory functions and augments apoptosis of monocytes/macrophages during Vibrio anguillarum infection in ayu, Plecoglossus altivelis. Fish Shellfish Immunol. 2019, 86, 70–81. [Google Scholar] [CrossRef]
- Li, W.-R.; Guan, X.-L.; Jiang, S.; Sun, L. The novel fish miRNA pol-miR-novel_171 and its target gene FAM49B play a critical role in apoptosis and bacterial infection. Dev. Comp. Immunol. 2020, 106, 103616. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wang, L.; He, J.; Zhu, X.; Huang, W.; Wang, S.; Qin, Q.; Sun, H. MicroRNA-124 Promotes Singapore Grouper Iridovirus Replication and Negatively Regulates Innate Immune Response. Front. Immunol. 2021, 12, 767813. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, Y. AMPK and Autophagy. Adv. Exp. Med. Biol. 2019, 1206, 85–108. [Google Scholar]
- White, E.; Mehnert, J.M.; Chan, C.S. Autophagy, Metabolism, and Cancer. Clin. Cancer Res. 2015, 21, 5037–5046. [Google Scholar] [CrossRef][Green Version]
- Mizushima, N. Autophagy: Process and function. Genes Dev. 2007, 21, 2861–2873. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wong, S.Q.; Kumar, A.V.; Mills, J.; Lapierre, L.R. Autophagy in aging and longevity. Hum. Genet. 2020, 139, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Lin, E.; He, L.; Yu, J.; Tan, P.; Zhou, Y. Autophagy and Viral Infection. Adv. Exp. Med. Biol. 2019, 1209, 55–78. [Google Scholar]
- Keller, M.D.; Torres, V.J.; Cadwell, K. Autophagy and microbial pathogenesis. Cell Death Differ. 2020, 27, 872–886. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V. Autophagy in infection. Curr. Opin. Cell Biol. 2010, 22, 252–262. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Shen, Y.; Zhang, S.; Xiao, Y.; Shi, S. Salmonella Interacts with Autophagy to Offense or Defense. Front. Microbiol. 2020, 11, 721. [Google Scholar] [CrossRef][Green Version]
- Wang, M.; Fan, Z.; Han, H. Autophagy in Staphylococcus aureus Infection. Front. Cell. Infect. Microbiol. 2021, 11, 750222. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.; Tian, M.; Chang, J.; Li, F.; Zhang, G. MiRNA-192-5p attenuates airway remodeling and autophagy in asthma by targeting MMP-16 and ATG7. Biomed. Pharmacother. Biomed. Pharmacother. 2020, 122, 109692. [Google Scholar] [CrossRef]
- Lu, X.; Zhang, Y.; Zheng, Y.; Chen, B. The miRNA-15b/USP7/KDM6B axis engages in the initiation of osteoporosis by modulating osteoblast differentiation and autophagy. J. Cell. Mol. Med. 2021, 25, 2069–2081. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, J.; Chen, N. The Potential Role of miRNA-Regulated Autophagy in Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 7789. [Google Scholar] [CrossRef]
- Chen, D.; Chen, Y.; Lin, C.; Lo, C.; Liu, H.; Liao, T. MicroRNA-889 Inhibits Autophagy To Maintain Mycobacterial Survival in Patients with Latent Tuberculosis Infection by Targeting TWEAK. mBio 2020, 11, e03045-19. [Google Scholar] [CrossRef][Green Version]
- Jia, P.; Pan, H.; Cui, K.; Jia, K.; Yi, M. MicroRNA expression profiling of sea perch brain cells reveals the roles of microRNAs in autophagy induced by RGNNV infection. J. Fish Dis. 2021, 44, 1305–1314. [Google Scholar] [CrossRef]
- Li, W.; Guan, X. PUF60 of Japanese flounder is regulated by pol-miR-novel_395 and involved in pathogen infection, autophagy, and apoptosis. Dev. Comp. Immunol. 2021, 123, 104170. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Tooze, S.A. Emerging roles of ATG proteins and membrane lipids in autophagosome formation. Cell Discov. 2020, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Wesselborg, S.; Stork, B. Autophagy signal transduction by ATG proteins: From hierarchies to networks. Cell. Mol. Life Sci. 2015, 72, 4721–4757. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993, 333, 169–174. [Google Scholar] [CrossRef][Green Version]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Mizushima, N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020, 63, 1–10. [Google Scholar] [CrossRef]
- Vergne, I.; Lafont, F.; Espert, L.; Esclatine, A.; Biard-Piechaczyk, M. Autophagy, ATG proteins and infectious diseases. Med. Sci. 2017, 33, 312–318. [Google Scholar]
- Fang, D.; Xie, H.; Hu, T.; Shan, H.; Li, M. Binding Features and Functions of ATG3. Front. Cell Dev. Biol. 2021, 9, 685625. [Google Scholar] [CrossRef]
- Qiu, Y.; Zheng, Y.; Grace, C.R.R.; Liu, X.; Klionsky, D.J.; Schulman, B.A. Allosteric regulation through a switch element in the autophagy E2, Atg3. Autophagy 2020, 16, 183–184. [Google Scholar] [CrossRef]
- Sou, Y.S.; Waguri, S.; Iwata, J.; Ueno, T.; Fujimura, T.; Hara, T.; Sawada, N.; Yamada, A.; Mizushima, N.; Uchiyama, Y.; et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell 2008, 19, 4762–4775. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Radoshevich, L.; Murrow, L.; Chen, N.; Fernandez, E.; Roy, S.; Fung, C.; Debnath, J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 2010, 142, 590–600. [Google Scholar] [CrossRef][Green Version]
- Altman, B.J.; Jacobs, S.R.; Mason, E.F.; Michalek, R.D.; MacIntyre, A.N.; Coloff, J.L.; Ilkayeva, O.; Jia, W.; He, Y.W.; Rathmell, J.C. Autophagy is essential to suppress cell stress and to allow BCR-Abl-mediated leukemogenesis. Oncogene 2011, 30, 1855–1867. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Choi, J.; Park, S.; Biering, S.B.; Selleck, E.; Liu, C.Y.; Zhang, X.; Fujita, N.; Saitoh, T.; Akira, S.; Yoshimori, T.; et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 2014, 40, 924–935. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, T.; Yu, N.; Qian, M.; Feng, J.; Cao, S.; Yin, J.; Zhang, Q. ERK-mediated autophagy promotes inactivated Sendai virus (HVJ-E)-induced apoptosis in HeLa cells in an Atg3-dependent manner. Cancer Cell Int. 2018, 18, 200. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Fu, W.; Tang, M.; Zhang, C.; Hou, T.; Li, R.; Lu, X.; Wang, Y.; Zhou, J.; Li, X.; et al. PTK2-mediated degradation of ATG3 impedes cancer cells susceptible to DNA damage treatment. Autophagy 2017, 13, 579–591. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, S.B.; Aoki, T.; Jung, T.S. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet. Res. 2012, 43, 67. [Google Scholar] [CrossRef][Green Version]
- Guan, X.L.; Zhang, B.C.; Sun, L. Japanese flounder pol-miR-3p-2 suppresses Edwardsiella tarda infection by regulation of autophagy via p53. Dev. Comp. Immunol. 2020, 103, 103531. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Ning, X.; Sun, L. Megalocytivirus Induces Complicated Fish Immune Response at Multiple RNA Levels Involving mRNA, miRNA, and circRNA. Int. J. Mol. Sci. 2021, 22, 3156. [Google Scholar] [CrossRef]
- Wang, H.R.; Hu, Y.H.; Zhang, W.W.; Sun, L. Construction of an attenuated Pseudomonas fluorescens strain and evaluation of its potential as a cross-protective vaccine. Vaccine 2009, 27, 4047–4055. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.-L.; Li, H.; Miao, H.-Z. The establishment and partial characterization of a continuous fish cell line FG-9307 from the gill of flounder Paralichthys olivaceus. Aquaculture 1997, 156, 327–333. [Google Scholar] [CrossRef]
- Sun, K.; Wang, H.-L.; Zhang, M.; Xiao, Z.-Z.; Sun, L. Genetic mechanisms of multi-antimicrobial resistance in a pathogenic Edwardsiella tarda strain. Aquaculture 2009, 289, 134–139. [Google Scholar] [CrossRef]
- Staedel, C.; Darfeuille, F. MicroRNAs and bacterial infection. Cell Microbiol. 2013, 15, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Cullen, B.R. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe 2008, 3, 375–387. [Google Scholar] [CrossRef][Green Version]
- Huang, C.W.; Tsai, K.N.; Chen, Y.S.; Chang, R.Y. Differential miRNA Expression Profiling Reveals Correlation of miR125b-5p with Persistent Infection of Japanese Encephalitis Virus. Int. J. Mol. Sci. 2021, 22, 4218. [Google Scholar] [CrossRef]
- Andreassen, R.; Høyheim, B. miRNAs associated with immune response in teleost fish. Dev. Comp. Immunol. 2017, 75, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, C.; Mano, M.; Eulalio, A. MicroRNAs at the Host-Bacteria Interface: Host Defense or Bacterial Offense. Trends Microbiol. 2019, 27, 206–218. [Google Scholar] [CrossRef]
- Johnston, D.G.W.; Kearney, J.; Zaslona, Z.; Williams, M.A.; O’Neill, L.A.J.; Corr, S.C. MicroRNA-21 Limits Uptake of Listeria monocytogenes by Macrophages to Reduce the Intracellular Niche and Control Infection. Front. Cell. Infect. Microbiol. 2017, 7, 201. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, T.; Yu, J.; Zhang, Y.; Li, L.; Chen, Y.; Li, D.; Liu, F.; Zhang, C.Y.; Gu, H.; Zen, K. Salmonella enterica serovar enteritidis modulates intestinal epithelial miR-128 levels to decrease macrophage recruitment via macrophage colony-stimulating factor. J. Infect. Dis. 2014, 209, 2000–2011. [Google Scholar] [CrossRef]
- Khan, M.; Harms, J.S.; Liu, Y.; Eickhoff, J.; Tan, J.W.; Hu, T.; Cai, F.; Guimaraes, E.; Oliveira, S.C.; Dahl, R.; et al. Brucella suppress STING expression via miR-24 to enhance infection. PLoS Pathog. 2020, 16, e1009020. [Google Scholar] [CrossRef]
- Li, M.; Wu, M.; Sun, Y.; Sun, L. Edwardsiella tarda TraT is an anti-complement factor and a cellular infection promoter. Commun. Biol. 2022, 5, 637. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.Y.; Siame, B.A.; Tenkink, B.J.; Noort, R.J.; Mok, Y.K. Edwardsiella tarda—Virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012, 14, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Li, H.; Lu, K. Autophagy Regulation of Bacterial Pathogen Invasion. Adv. Exp. Med. Biol. 2019, 1209, 43–54. [Google Scholar]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004, 36, 2503–2518. [Google Scholar] [CrossRef]
- Hu, J.; Huang, S.; Liu, X.; Zhang, Y.; Wei, S.; Hu, X. miR-155: An Important Role in Inflammation Response. J. Immunol. Res. 2022, 2022, 7437281. [Google Scholar] [CrossRef]
- Maciak, K.; Dziedzic, A.; Miller, E.; Saluk-Bijak, J. miR-155 as an Important Regulator of Multiple Sclerosis Pathogenesis. A Review. Int. J. Mol. Sci. 2021, 22, 4332. [Google Scholar] [CrossRef]
- Park, M.; Choi, S.; Kim, S.; Kim, J.; Lee, D.K.; Park, W.; Kim, T.; Jung, J.; Hwang, J.Y.; Won, M.H.; et al. NF-kappaB-responsive miR-155 induces functional impairment of vascular smooth muscle cells by downregulating soluble guanylyl cyclase. Exp. Mol. Med. 2019, 51, 1–12. [Google Scholar] [PubMed][Green Version]
- Jiang, K.; Yang, J.; Guo, S.; Zhao, G.; Wu, H.; Deng, G. Peripheral Circulating Exosome-Mediated Delivery of miR-155 as a Novel Mechanism for Acute Lung Inflammation. Mol. Ther. J. Am. Soc. Gene Ther. 2019, 27, 1758–1771. [Google Scholar] [CrossRef]
- Singh, A.; Srivastava, N.; Yadav, A.; Ateeq, B. Targeting AGTR1/NF-kappaB/CXCR4 axis by miR-155 attenuates oncogenesis in glioblastoma. Neoplasia 2020, 22, 497–510. [Google Scholar] [CrossRef]


| Primers | Sequence (5′-3′) a |
|---|---|
| 3UTR-ATG3-F | TCTAGTTGTTTAAACGAGCTCACACATAGAGATGAAACT |
| 3UTR-ATG3-R | CCTGCAGGTCGACTCTAGAGTCACAGTCTGTACAGAC |
| pol-miR-155-RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCCT |
| pol-miR-155-F | CGCGTTAATGCTAATCGTGAT |
| pol-miR-155-R | AGTGCAGGGTCCGAGGTATT |
| ATG3-qRT-F | AAACAGATGAGGCGACCCTG |
| ATG3-qRT-R | GAGTCGAGGGGTCTGGTAGT |
| IL-6-qRT-F | CTCCAGTCGAATACGAGCCC |
| IL-6-qRT-R | ACTCTTTCTGGTGGTGAGCG |
| IL-8-qRT-F | GCCTGAGAAGCCTAGGAGTG |
| IL-8-qRT-R | TGACTCTCTTCACCCACGGA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Guan, X. Japanese Flounder pol-miR-155 Is Involved in Edwardsiella tarda Infection via ATG3. Genes 2023, 14, 958. https://doi.org/10.3390/genes14050958
Zhang Z, Guan X. Japanese Flounder pol-miR-155 Is Involved in Edwardsiella tarda Infection via ATG3. Genes. 2023; 14(5):958. https://doi.org/10.3390/genes14050958
Chicago/Turabian StyleZhang, Zhanwei, and Xiaolu Guan. 2023. "Japanese Flounder pol-miR-155 Is Involved in Edwardsiella tarda Infection via ATG3" Genes 14, no. 5: 958. https://doi.org/10.3390/genes14050958
APA StyleZhang, Z., & Guan, X. (2023). Japanese Flounder pol-miR-155 Is Involved in Edwardsiella tarda Infection via ATG3. Genes, 14(5), 958. https://doi.org/10.3390/genes14050958






