Transcriptome Analysis Reveals the Age-Related Developmental Dynamics Pattern of the Longissimus Dorsi Muscle in Ningxiang Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Hematoxylin and Eosin (HE) Staining
2.3. RNA Extraction and Sequencing Analysis
2.4. Quantitative Real-Time PCR (qRT-PCR) Validation
2.5. Statistical Analysis
3. Results
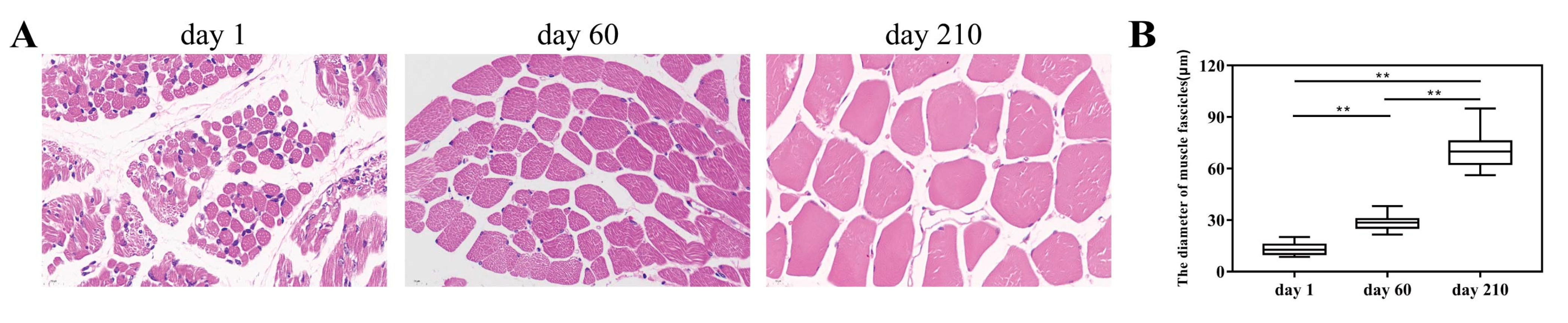
3.1. Morphological Changes of Longissimus Dorsi Muscle Tissues and Overview of Transcriptomics Data
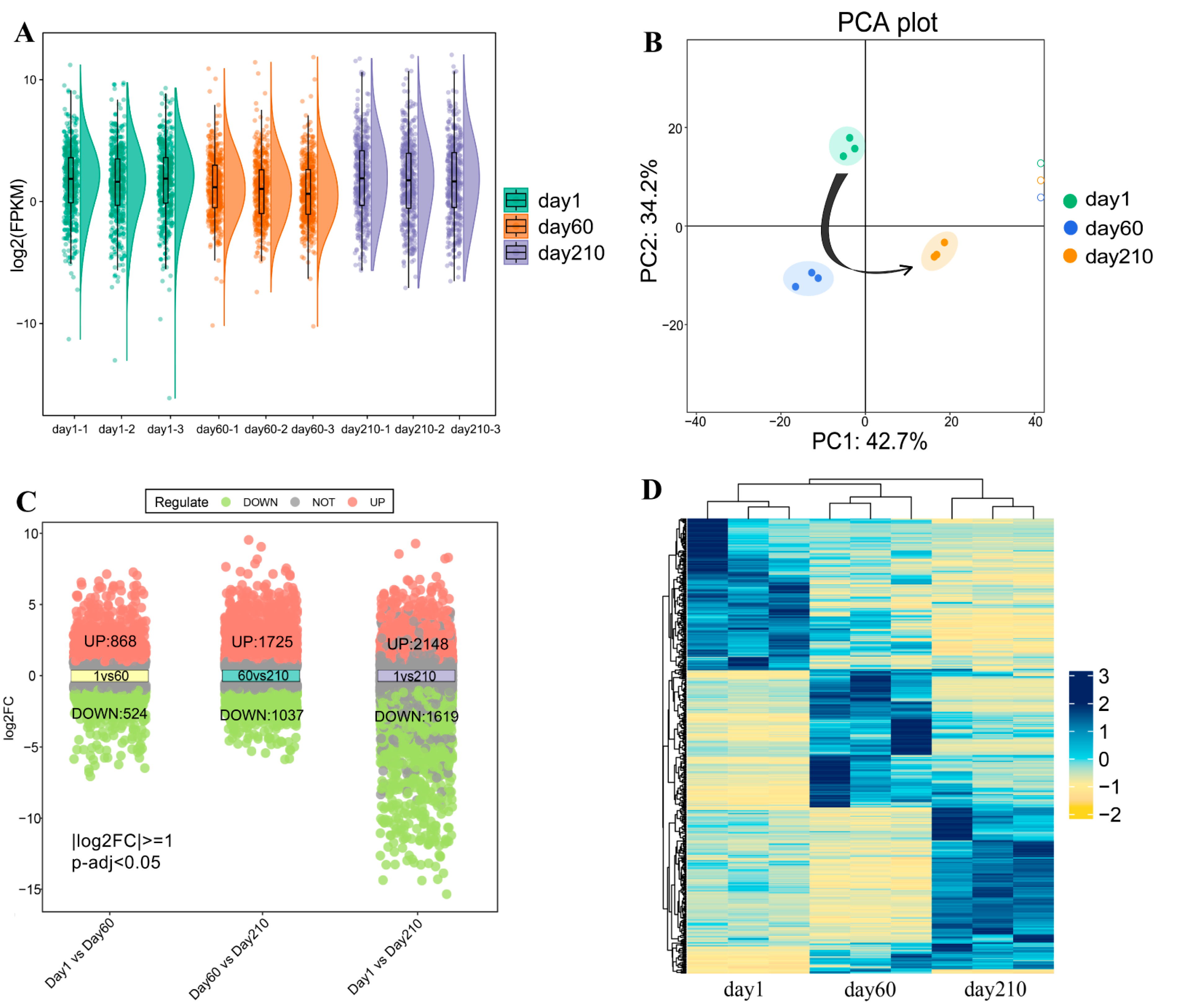
3.2. Identification of DEGs in the Transcriptome of Longissimus Dorsi Muscle at Three Different Developmental Stages
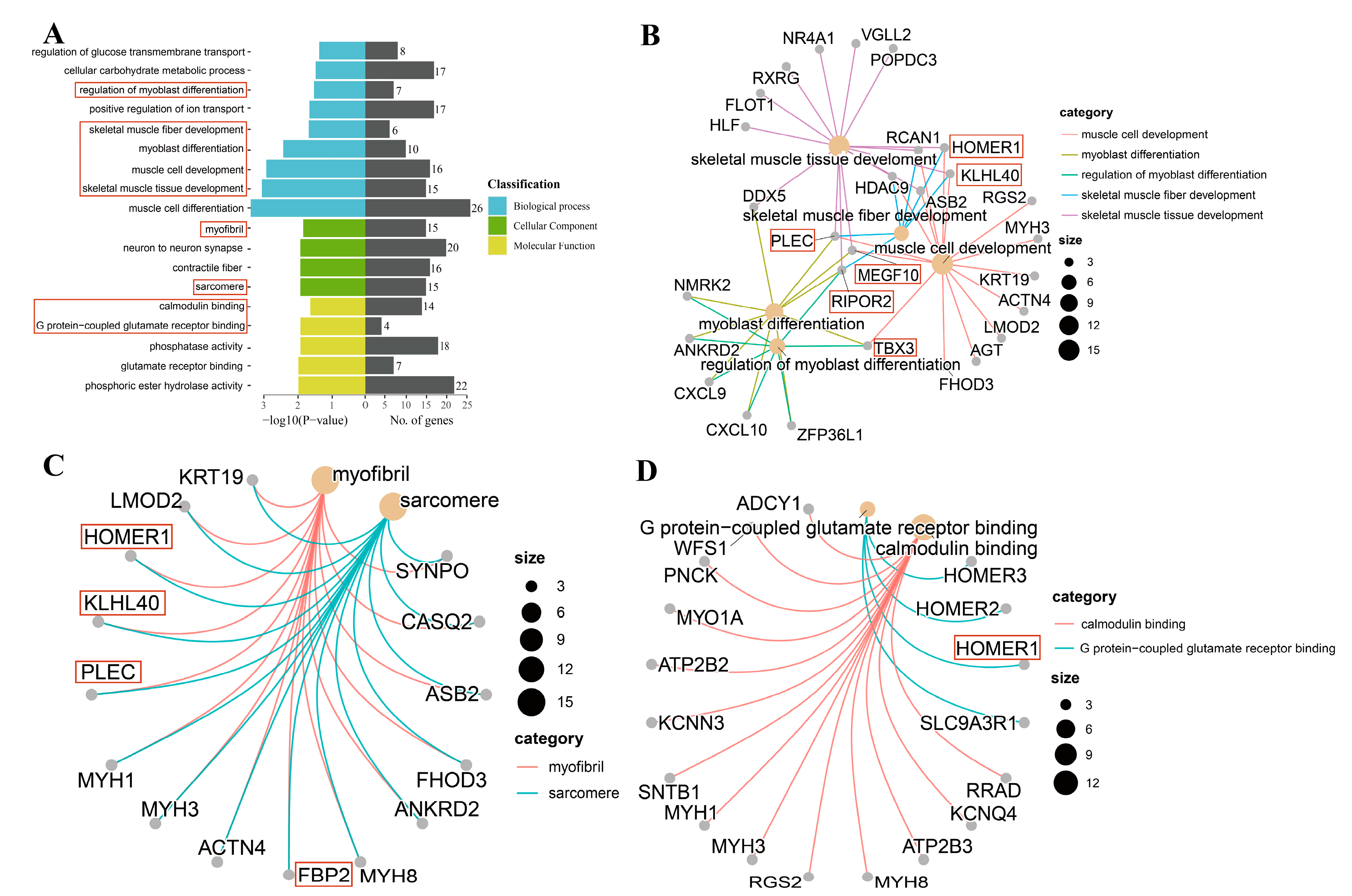
3.3. GO Enrichment Analysis
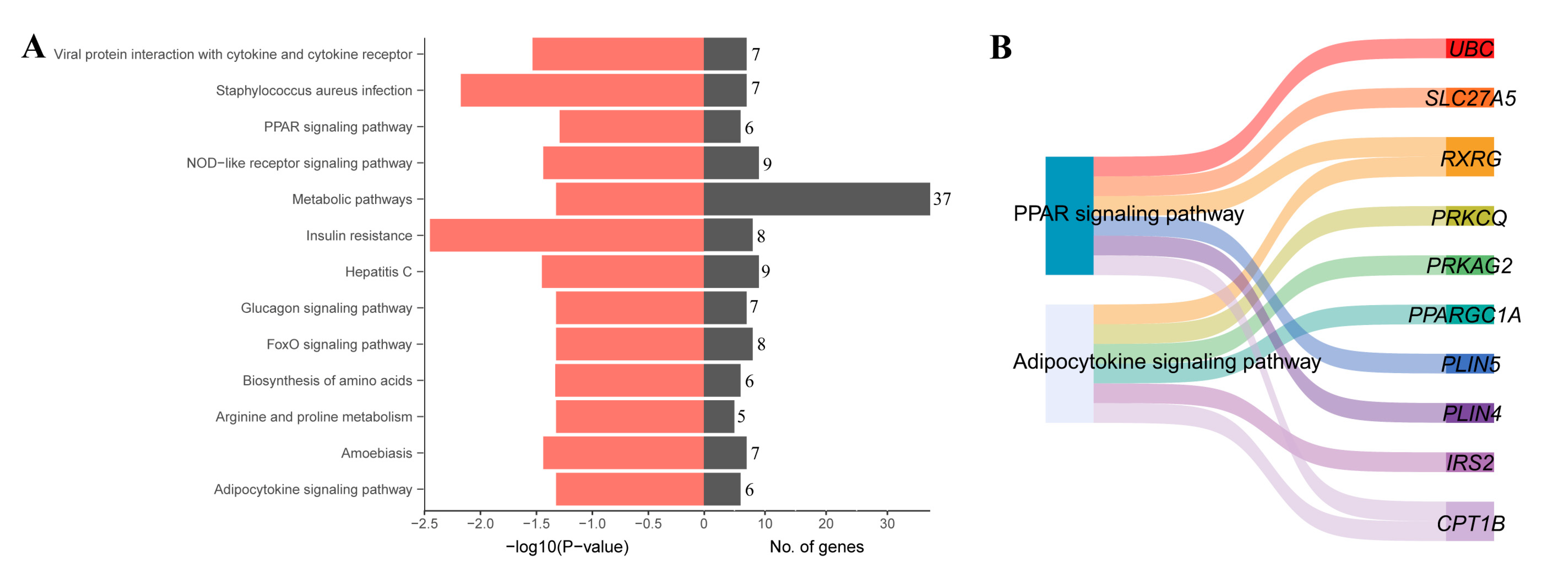
3.4. KEGG Enrichment Analysis
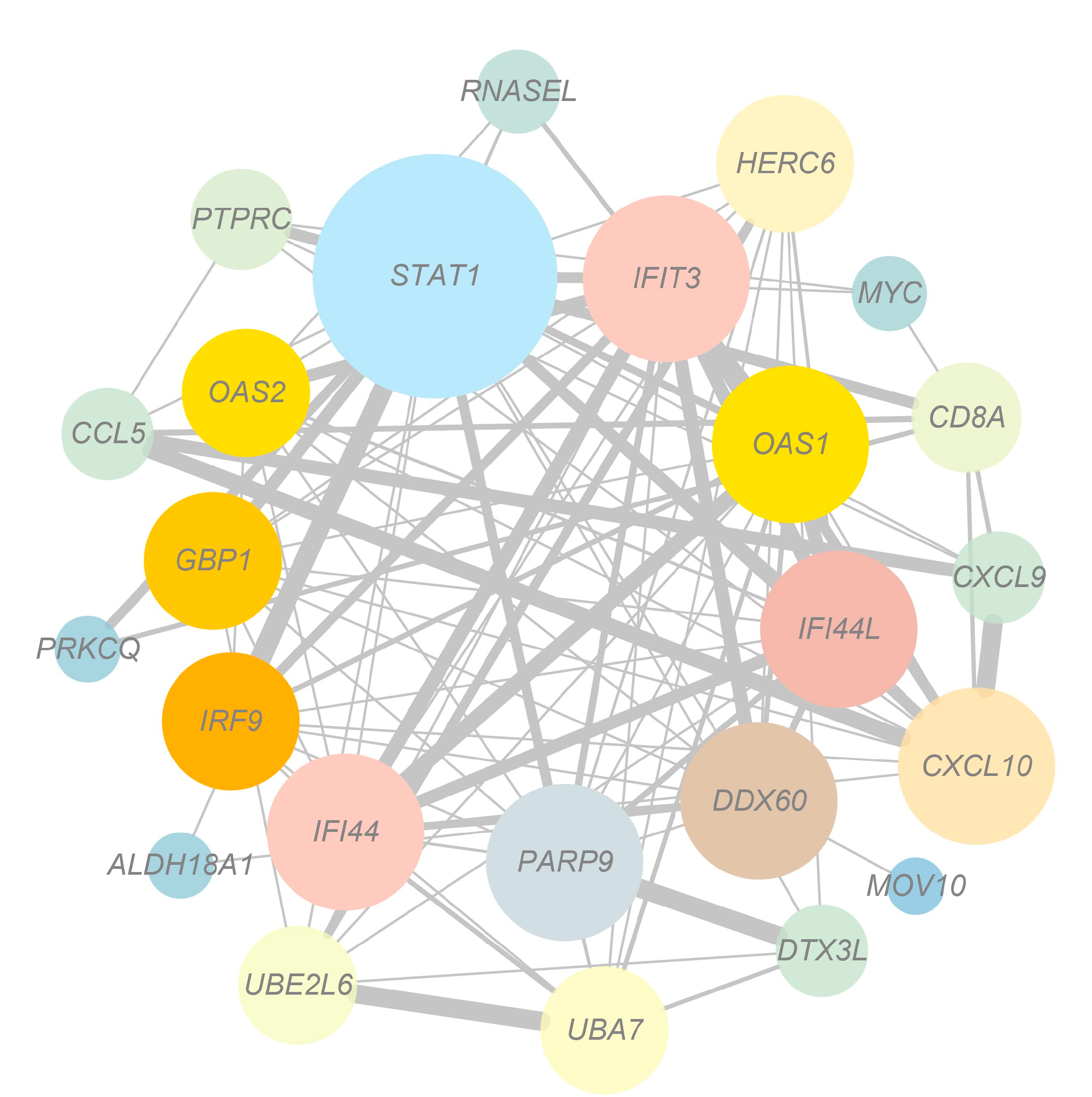
3.5. Network Analysis of Target Genes
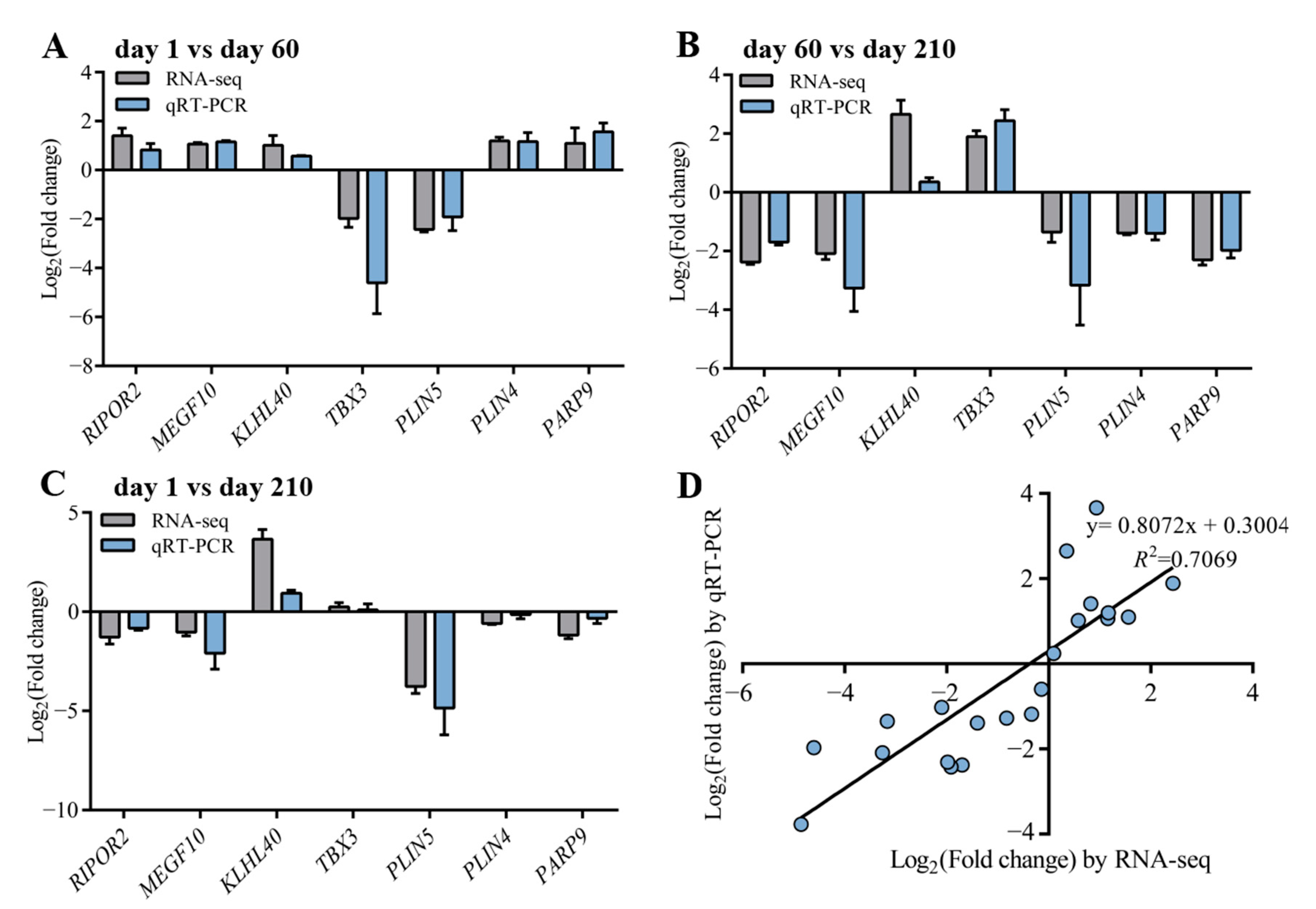
3.6. Validation of the Results by qRT-pCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nonneman, D.J.; Shackelford, S.D.; King, D.A.; Wheeler, T.L.; Wiedmann, R.T.; Snelling, W.M.; Rohrer, G.A. Genome-wide association of meat quality traits and tenderness in swine. J. Anim. Sci. 2013, 91, 4043–4050. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Z.; Oyelami, F.O.; Sun, H.; Xu, Z.; Ma, P.; Wang, Q.; Pan, Y. Identification of genes related to intramuscular fat independent of backfat thickness in Duroc pigs using single-step genome-wide association. Anim. Genet. 2021, 52, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, J.; Gong, H.; Cui, L.; Zhang, W.; Ma, J.; Chen, C.; Ai, H.; Xiao, S.; Huang, L.; et al. Genetic correlation of fatty acid composition with growth, carcass, fat deposition and meat quality traits based on GWAS data in six pig populations. Meat. Sci. 2019, 150, 47–55. [Google Scholar] [CrossRef]
- Rosenvold, K.; Andersen, H.J. Factors of significance for pork quality-a review. Meat. Sci. 2003, 64, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xiong, X.; Yang, J.; Zhou, L.; Yang, B.; Ai, H.; Ma, H.; Xie, X.; Huang, Y.; Fang, S.; et al. Genome-wide association analyses for meat quality traits in Chinese Erhualian pigs and a Western Duroc × (Landrace × Yorkshire) commercial population. Genet. Sel. Evol. 2015, 47, 44. [Google Scholar] [CrossRef]
- Ma, H.; Jiang, J.; He, J.; Liu, H.; Han, L.; Gong, Y.; Li, B.; Yu, Z.; Tang, S.; Zhang, Y.; et al. Long-read assembly of the Chinese indigenous Ningxiang pig genome and identification of genetic variations in fat metabolism among different breeds. Mol. Ecol. Resour. 2022, 22, 1508–1520. [Google Scholar] [CrossRef]
- Miao, W.; Ma, Z.; Tang, Z.; Yu, L.; Liu, S.; Huang, T.; Wang, P.; Wu, T.; Song, Z.; Zhang, H.; et al. Integrative ATAC-seq and RNA-seq Analysis of the Longissimus Muscle of Luchuan and Duroc Pigs. Front. Nutr. 2021, 8, 42672. [Google Scholar] [CrossRef]
- Song, S.Q.; Ma, W.W.; Zeng, S.X.; Zhang, C.L.; Yan, J.; Sun, C.C.; Li, X.; Wang, R.M.; Li, Z.Q. Transcriptome analysis of differential gene expression in the longissimus dorsi muscle from Debao and landrace pigs based on RNA-sequencing. Biosci. Rep. 2019, 39, BSR20192144. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, Y.; Zhang, B.; Zhong, H.; Lu, Y.; Zhang, H. Candidate gene screening for lipid deposition using combined transcriptomic and proteomic data from Nanyang black pigs. BMC Genomics 2021, 22, 441. [Google Scholar] [CrossRef]
- Cheng, F.; Liang, J.; Yang, L.; Lan, G.; Wang, L.; Wang, L. Systematic identification and comparison of the expressed profiles of lncRNAs, miRNAs, circRNAs, and mRNAs with associated co-expression networks in pigs with low and high intramuscular fat. Animals 2021, 11, 3212. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K.; Van Goor, A.; Walker, K.E.; Hailstock, T.; Franklin, J.; Dai, C. Importance of the pig as a human biomedical model. Sci. Transl. Med. 2021, 13, eabd5758. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Lei, L.; Wang, Z.; Yin, Y.; Yang, H.; Yang, Z.; Chen, J. Differentially expressed genes in the longissimus dorsi muscle between the Chinese indigenous Ningxiang pig and Large White breed using RNA sequencing. Czech. J. Anim. Sci. 2022, 67, 442–453. [Google Scholar] [CrossRef]
- Hirayama, E.; Kim, J. Identification and characterization of a novel neural cell adhesion molecule (NCAM)-associated protein from quail myoblasts: Relationship to myotube formation and induction of neurite-like protrusions. Differentiation 2008, 76, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Vargas-Franco, D.; Saha, M.; Davis, R.M.; Manko, K.A.; Draper, I.; Pacak, C.A.; Kang, P.B. Megf10 deficiency impairs skeletal muscle stem cell migration and muscle regeneration. FEBS Open Bio. 2021, 11, 114–123. [Google Scholar] [CrossRef]
- Ravenscroft, G.; Miyatake, S.; Lehtokari, V.L.; Todd, E.J.; Vornanen, P.; Yau, K.S.; Hayashi, Y.K.; Miyake, N.; Tsurusaki, Y.; Doi, H.; et al. Mutations in KLHL40 are a frequent cause of severe autosomal-recessive nemaline myopathy. Am. J. Hum. Genet. 2013, 93, 6–18. [Google Scholar] [CrossRef]
- Zrelski, M.M.; Kustermann, M.; Winter, L. Muscle-Related Plectinopathies. Cells 2021, 10, 2480. [Google Scholar] [CrossRef]
- Lee, H.S.; Cho, H.H.; Kim, H.K.; Bae, Y.C.; Baik, H.S.; Jung, J.S. Tbx3, a transcriptional factor, involves in proliferation and osteogenic differentiation of human adipose stromal cells. Mol. Cell. Biochem. 2007, 296, 129–136. [Google Scholar] [CrossRef]
- Oh, T.J.; Adhikari, A.; Mohamad, T.; Althobaiti, A.; Davie, J. TBX3 represses TBX2 under the control of the PRC2 complex in skeletal muscle and rhabdomyosarcoma. Oncogenesis 2019, 8, 27. [Google Scholar] [CrossRef]
- Wiśniewski, J.; Piróg, M.; Hołubowicz, R.; Dobryszycki, P.; McCubrey, J.A.; Rakus, D.; Gizak, A. Dimeric and tetrameric forms of muscle fructose-1,6-bisphosphatase play different roles in the cell. Oncotarget 2017, 8, 115420–115433. [Google Scholar] [CrossRef]
- Michel, R.N.; Dunn, S.E.; Chin, E.R. Calcineurin and skeletal muscle growth. Proc. Nutr. Soc. 2002, 4, 46–47. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hao, X.; Zhang, M.; Wang, K.; Li, S.; Chen, X.; Yang, L.; Hu, L.; Zhang, S. Polymorphisms of HOMER1 gene are associated with piglet splay leg syndrome and one significant SNP can affect its intronic promoter activity in vitro. BMC Genet. 2018, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, K.H.; Ødegård, J.; Olsen, D.; Meuwissen, T.H. Genetic variation in efficiency to deposit fat and lean meat in Norwegian Landrace and Duroc pigs. J. Anim. Sci. 2015, 93, 3794–3800. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.M.; Qin, J.; Liu, S.G.; Cai, R.; Chen, X.C.; Wang, X.M.; Pang, W.J. PDGFRα Regulated by miR-34a and FoxO1 Promotes Adipogenesis in Porcine Intramuscular Preadipocytes through Erk Signaling Pathway. Int. J. Mol. Sci. 2017, 18, 2424. [Google Scholar] [CrossRef]
- Wang, J.; Chen, M.Y.; Chen, J.F.; Ren, Q.L.; Zhang, J.Q.; Cao, H.; Xing, B.S.; Pan, C.Y. LncRNA IMFlnc1 promotes porcine intramuscular adipocyte adipogenesis by sponging miR-199a-5p to up-regulate CAV-1. BMC Mol. Cell. Biol. 2020, 21, 77. [Google Scholar] [CrossRef]
- Li, B.; Yang, J.; Gong, Y.; Xiao, Y.; Chen, W.; Zeng, Q.; Xu, K.; Duan, Y.; Ma, H. Effects of age on subcutaneous adipose tissue proteins in Chinese indigenous Ningxiang pig by TMT-labeled quantitative proteomics. J. Proteom. 2022, 265, 104650. [Google Scholar] [CrossRef]
- Sjöström, M.; Lexell, J.; Downham, D.Y. Differences in fiber number and fiber type proportion within fascicles. A quantitative morphological study of whole vastus lateralis muscle from childhood to old age. Anat. Rec. 1992, 234, 183–189. [Google Scholar] [CrossRef]
- Lv, Z.; Ding, Y.; Cao, W.; Wang, S.; Gao, K. Role of RHO family interacting cell polarization regulators (RIPORs) in health and disease: Recent advances and prospects. Int. J. Biol. Sci. 2022, 18, 800–808. [Google Scholar] [CrossRef]
- Xu, F.; Zou, C.; Gao, Y.; Shen, J.; Liu, T.; He, Q.; Li, S.; Xu, S. Comprehensive analyses identify RIPOR2 as a genomic instability-associated immune prognostic biomarker in cervical cancer. Front. Immunol. 2022, 13, 930488. [Google Scholar] [CrossRef]
- Park, S.Y.; Yun, Y.; Kim, M.J.; Kim, I.S. Myogenin is a positive regulator of MEGF10 expression in skeletal muscle. Biochem. Biophys. Res. Commun. 2014, 450, 1631–1637. [Google Scholar] [CrossRef]
- Saha, M.; Rizzo, S.A.; Ramanathan, M.; Hightower, R.M.; Santostefano, K.E.; Terada, N.; Finkel, R.S.; Berg, J.S.; Chahin, N.; Pacak, C.A.; et al. Selective serotonin reuptake inhibitors ameliorate MEGF10 myopathy. Hum. Mol. Genet. 2019, 28, 2365–2377. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.; O’Rourke, J.; Long, C.; Doering, J.; Ravenscroft, G.; Bezprozvannaya, S.; Nelson, B.R.; Beetz, N.; Li, L.; Chen, S.; et al. KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy. J. Clin. Investig. 2014, 124, 3529–3539. [Google Scholar] [CrossRef] [PubMed]
- Benito, D.N.-d.; Nascimento, A.; Abicht, A.; Ortez, C.; Jou, C.; Müller, J.S.; Evangelista, T.; Töpf, A.; Thompson, R.; Jimenez-Mallebrera, C.; et al. KLHL40-related nemaline myopathy with a sustained, positive response to treatment with acetylcholinesterase inhibitors. J. Neurol. 2016, 263, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Han, S.; Cui, C.; Wang, Y.; Li, D.; Zhu, Q. Plectin regulates Wnt signaling mediated-skeletal muscle development by interacting with Dishevelled-2 and antagonizing autophagy. Gene 2021, 783, 145562. [Google Scholar] [CrossRef]
- Frank, D.U.; Emechebe, U.; Thomas, K.R.; Moon, A.M. Mouse TBX3 mutants suggest novel molecular mechanisms for Ulnar-mammary syndrome. PLoS ONE 2013, 8, e67841. [Google Scholar] [CrossRef]
- Pietras, Ł.; Stefanik, E.; Rakus, D.; Gizak, A. FBP2-A New Player in Regulation of Motility of Mitochondria and Stability of Microtubules in Cardiomyocytes. Cells 2022, 11, 1710. [Google Scholar] [CrossRef]
- Osipova, E.; Barsacchi, R.; Brown, T.; Sadanandan, K.; Gaede, A.H.; Monte, A.; Jarrells, J.; Moebius, C.; Pippel, M.; Altshuler, D.L.; et al. Loss of a gluconeogenic muscle enzyme contributed to adaptive metabolic traits in hummingbirds. Science 2023, 379, 185–190. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, L.; Shi, C.; Wei, R.; Yin, C. Interaction of the Homer1 EVH1 domain and skeletal muscle ryanodine receptor. Biochem. Biophys. Res. Commun. 2019, 514, 720–725. [Google Scholar] [CrossRef]
- Cui, X.; Cui, H.; Liu, L.; Zhao, G.; Liu, R.; Li, Q.; Zheng, M.; Wen, J. Decreased testosterone levels after caponization leads to abdominal fat deposition in chickens. BMC Genomics. 2018, 19, 344. [Google Scholar] [CrossRef]
- Wei, D.; Sun, Q.; Li, Y.; Li, C.; Li, X.; Sun, C. Leptin Reduces Plin5 m6A Methylation through FTO to Regulate Lipolysis in Piglets. Int. J. Mol. Sci. 2021, 22, 10610. [Google Scholar] [CrossRef]
- Han, X.; Zhu, J.; Zhang, X.; Song, Q.; Ding, J.; Lu, M.; Sun, S.; Hu, G. Plin4-Dependent Lipid Droplets Hamper Neuronal Mitophagy in the MPTP/p-Induced Mouse Model of Parkinson’s Disease. Front. Neurosci. 2018, 12, 397. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.M.; Ren, L.J.; Chen, L.; Zhang, X.; Cheng, M.L.; Li, W.Z.; Zhang, Y.Y.; Gao, S.Z. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat deposition. Lipids. 2009, 44, 1029–1037. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Zhang, Y.; Liu, Y.; Chen, J.; Zong, C.; Yu, C.; Cui, S.; Gao, W.; Qin, D.; Sun, W.; et al. Signal transducer and activator of transcription 5B (STAT5B) modulates adipocyte differentiation via MOF. Cell Signal. 2015, 27, 2434–2443. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xi, Y.; Chen, J.; Zhu, P.; Kang, J.; Zou, Z.; Wang, F.; Bu, S. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 1360–1366. [Google Scholar] [CrossRef]
- Stephens, J.M.; Morrison, R.F.; Pilch, P.F. The expression and regulation of STATs during 3T3-L1 adipocyte differentiation. J. Biol. Chem. 1996, 271, 10441–10444. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Zhang, Z.M.; Zhou, L.T.; Zhu, J.; Zhang, X.H.; Qi, W.; Ding, S.; Xu, Q.; Han, X.; Zhao, Y.M.; et al. Di (2-ethylhexyl) phthalate Disorders Lipid Metabolism via TYK2/STAT1 and Autophagy in Rats. Biomed. Environ. Sci. 2019, 32, 406–418. [Google Scholar]
- Gao, Y.; Li, Y.; Guo, X.; Wu, Z.; Zhang, W. Loss of STAT1 in bone marrow-derived cells accelerates skeletal muscle regeneration. PLoS ONE 2012, 7, e37656. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Dong, M.; Wang, Q.; Kumar, S.; Zhang, R.; Cheng, W.; Xiang, J.; Wang, G.; Ouyang, K.; Zhou, R.; et al. HIMF deletion ameliorates acute myocardial ischemic injury by promoting macrophage transformation to reparative subtype. Basic. Res. Cardiol. 2021, 116, 30. [Google Scholar] [CrossRef]
| Gene | Primer Sequence (5′-3′) | Annealing Temperature (°C) | Amplicon Length (bp) | GenBank No |
|---|---|---|---|---|
| RIPOR2 | F: CACGCCTTCTTTTCTAATCTACCG | 60 | 86 | XM_047419593.1 |
| R: CTGAAGCTGAGGGACAGTGG | ||||
| MEGF10 | F: AATCTCAGGCTCTCGGGTTG | 60 | 139 | XM_021084704.1 |
| R: GGCTGCACACATTAGGGTCT | ||||
| KLHL40 | F: CCACCAATGTGGTCATACACG | 60 | 107 | NM_001165887.2 |
| R: TGCCCCTGTAGTGTATCCTT | ||||
| TBX3 | F: CGGACAAACACGGATTTACTATCT | 60 | 151 | XM_001927997.5 |
| R: TATGCCGTCACAGCGATGAA | ||||
| PLIN5 | F: CACTCTGTCCTACCCACAGG | 60 | 113 | XM_021076783.1 |
| R: CTGCACTGCGTTCTGCTGG | ||||
| PLIN4 | F: ATAAGGCCAGGGAGCTGAGA | 60 | 162 | XM_021084054.1 |
| R: AGAAGCTGCTCAGGGTCTTG | ||||
| PARP9 | F: GCTTCCTTTGTAGGCGGGTG | 60 | 254 | XM_021070264.1 |
| R: ACAGCAGCCCCTTTCTGG | ||||
| GAPDH | F: GGCAAATTCAACGGCACAGTCAAG | 60 | 81 | NM_001256799.3 |
| R: TCGCTCCTGGAAGATGGTGATGG |
| Terms | Raw Reads Number | Clean Reads Number | Clean Reads Rate | Clean Q30 Bases Rate | Mapped Reads | Mapping Rate |
|---|---|---|---|---|---|---|
| Day 1-1 | 47582910 | 46885968 | 98.53% | 90.99% | 48173005 | 95.58% |
| Day 1-2 | 45499772 | 44813742 | 98.49% | 91.02% | 45051239 | 95.35% |
| Day 1-3 | 46522890 | 45826386 | 98.50% | 90.99% | 46504851 | 95.51% |
| Day 60-1 | 47532094 | 46830986 | 98.53% | 91.41% | 50456613 | 95.98% |
| Day 60-2 | 44687256 | 44022350 | 98.51% | 90.69% | 47205558 | 95.68% |
| Day 60-3 | 40913650 | 40292170 | 98.48% | 90.52% | 44041267 | 95.72% |
| Day 210-1 | 44536574 | 44217290 | 99.28% | 92.30% | 46642037 | 95.85% |
| Day 210-2 | 45793204 | 45430838 | 99.21% | 92.14% | 47663400 | 95.53% |
| Day 210-3 | 45371844 | 45096524 | 99.39% | 93.08% | 47805991 | 96.03% |
| Gene | Enriched Pathway | Function | Reference |
|---|---|---|---|
| RIPOR2 | Regulation of myoblast differentiation; muscle cell development; and skeletal muscle tissue development. | An important factor for myogenic cell fusion and differentiation. | [14] |
| MEGF10 | Muscle cell development; myoblast differentiation; and skeletal muscle tissue development. | MEGF10 deficiency impairs skeletal muscle stem cell migration and muscle regeneration. | [15] |
| KLHL40 | Skeletal muscle tissue development and muscle cell development. | As a striated-muscle-specific protein, it plays a key role in muscle development and function. | [16] |
| PLEC | Myoblast differentiation; muscle cell development; and skeletal muscle tissue development. | PLEC deficiency leads to a disorganization of myofibrils and sarcomeres. | [17] |
| TBX3 | Regulation of myoblast differentiation; myoblast differentiation; and muscle cell development. | TBX3, an essential regulatory factor for muscle and its accessory tissues, plays an important role in muscle and skeletal development. | [18,19] |
| FBP2 | Metabolic pathways and glycolysis/gluconeogenesis. | FBP2, besides being a regulatory enzyme of glyconeogenesis, also protects mitochondria against calcium stress and plays a key role in the regulation of the cell cycle. | [20] |
| HOMER1 | Skeletal muscle tissue development and muscle cell development. | HOMER1 may influence skeletal muscle function via Ca2+. | [21,22] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liufu, S.; Lan, Q.; Liu, X.; Chen, B.; Xu, X.; Ai, N.; Li, X.; Yu, Z.; Ma, H. Transcriptome Analysis Reveals the Age-Related Developmental Dynamics Pattern of the Longissimus Dorsi Muscle in Ningxiang Pigs. Genes 2023, 14, 1050. https://doi.org/10.3390/genes14051050
Liufu S, Lan Q, Liu X, Chen B, Xu X, Ai N, Li X, Yu Z, Ma H. Transcriptome Analysis Reveals the Age-Related Developmental Dynamics Pattern of the Longissimus Dorsi Muscle in Ningxiang Pigs. Genes. 2023; 14(5):1050. https://doi.org/10.3390/genes14051050
Chicago/Turabian StyleLiufu, Sui, Qun Lan, Xiaolin Liu, Bohe Chen, Xueli Xu, Nini Ai, Xintong Li, Zonggang Yu, and Haiming Ma. 2023. "Transcriptome Analysis Reveals the Age-Related Developmental Dynamics Pattern of the Longissimus Dorsi Muscle in Ningxiang Pigs" Genes 14, no. 5: 1050. https://doi.org/10.3390/genes14051050
APA StyleLiufu, S., Lan, Q., Liu, X., Chen, B., Xu, X., Ai, N., Li, X., Yu, Z., & Ma, H. (2023). Transcriptome Analysis Reveals the Age-Related Developmental Dynamics Pattern of the Longissimus Dorsi Muscle in Ningxiang Pigs. Genes, 14(5), 1050. https://doi.org/10.3390/genes14051050





