Phenotype of White Sika Deer Due to SCF Gene Structural Variation
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Whole-Genome Resequencing and Sample Comparison
2.3. Variation Detection and Annotation
2.4. Establishment of Candidate Genome
2.5. Screening of Coat Color Gene Variation
2.6. Preparation and Staining of Paraffin Skin Tissue Sections
2.6.1. Skin Tissue Sectioning
2.6.2. HE Staining
2.6.3. L-DOPA Staining
2.7. Phenotypic Genotyping and Sanger Sequencing
3. Results
3.1. Resequencing Data and Genetic Variation
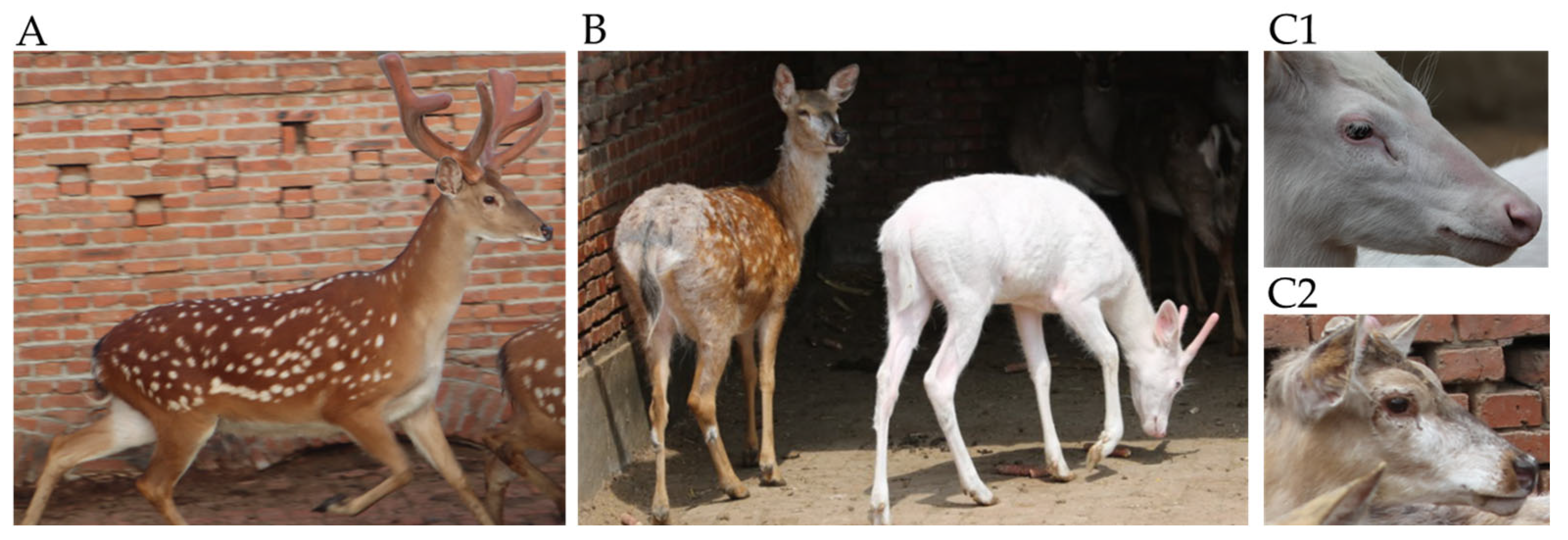
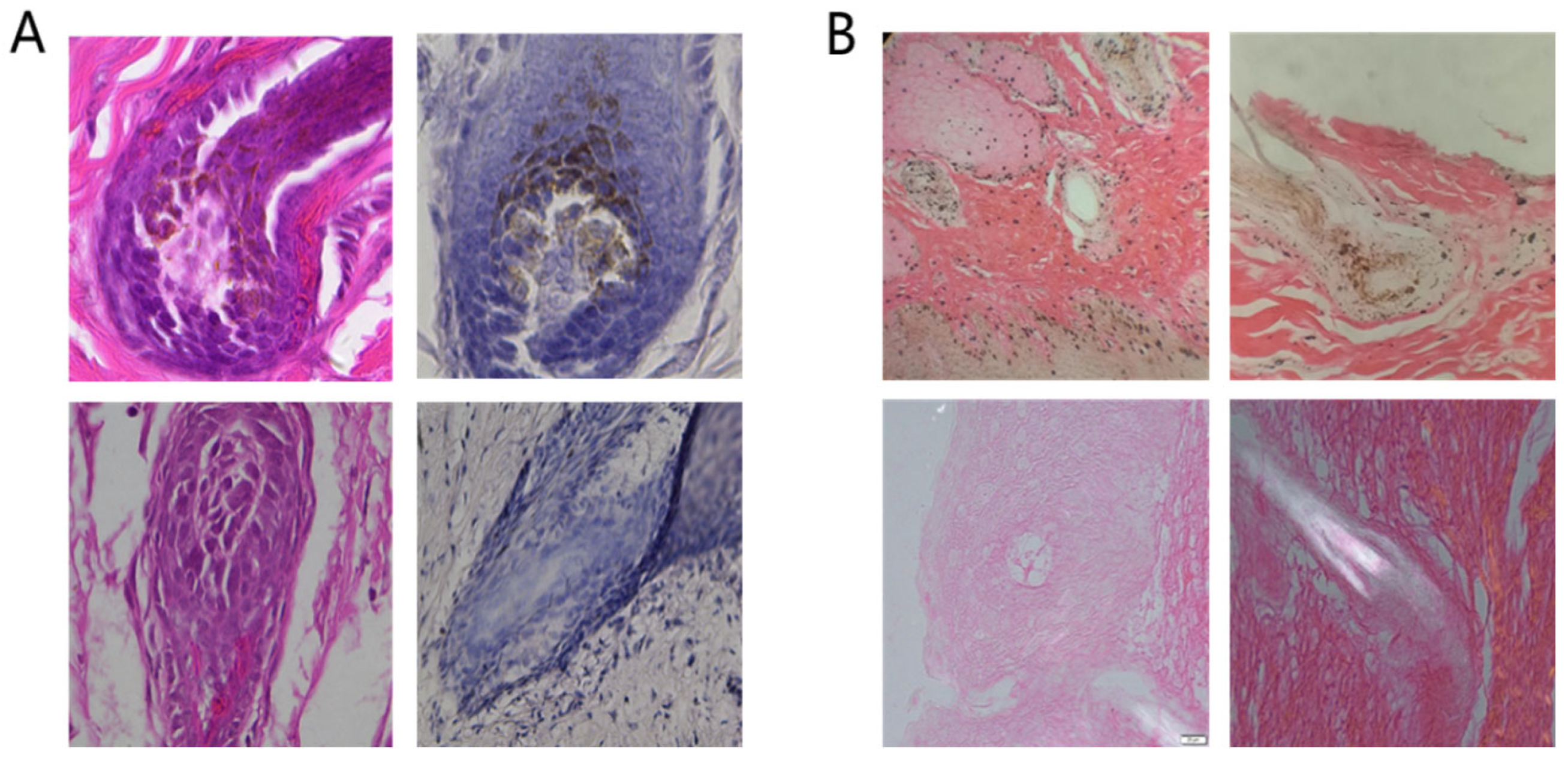
3.2. Staining Results for Tissue Sections
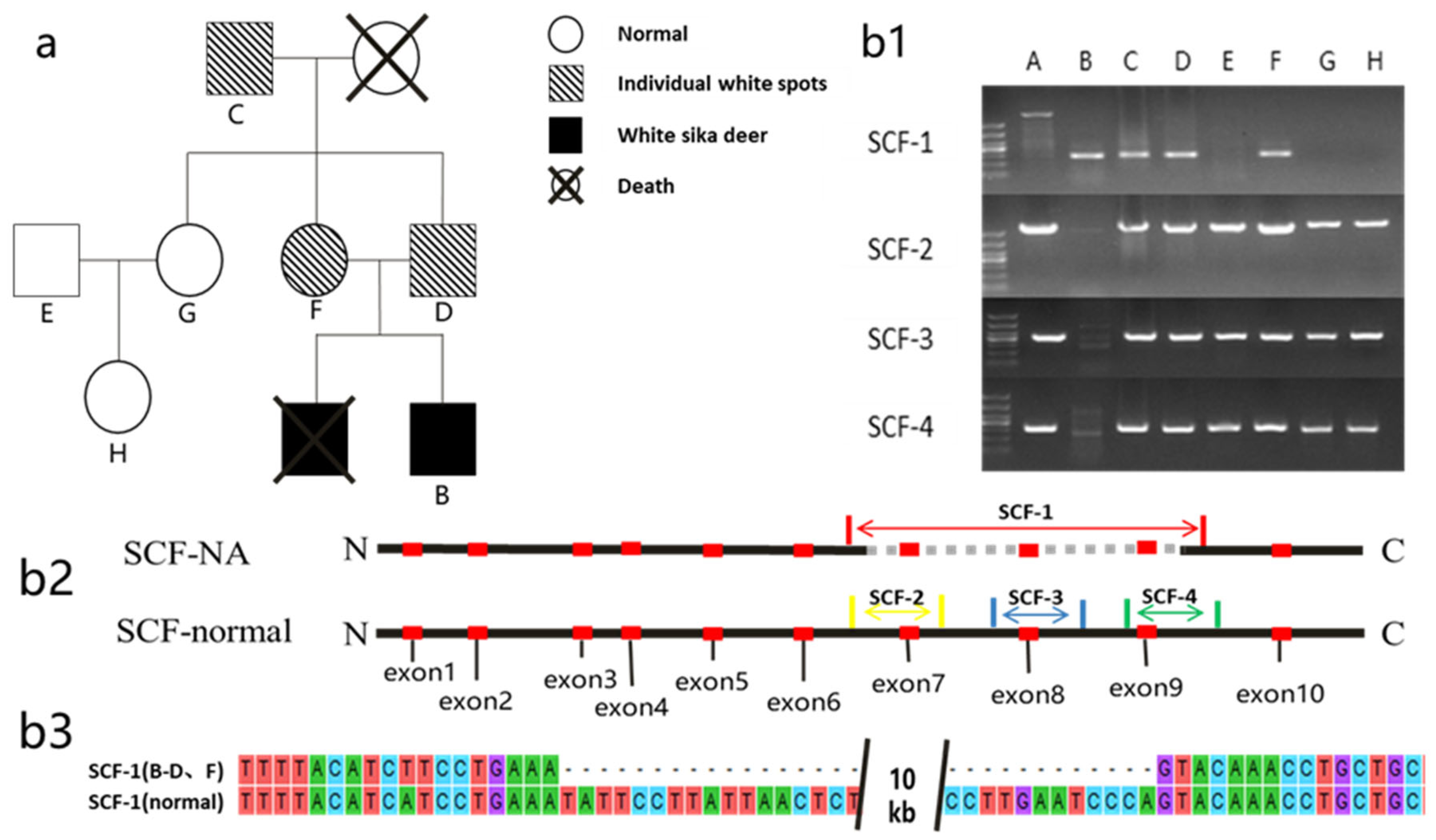
3.3. Analysis of SCF Genotype and Phenotype in White Sika Deer Family
3.4. Sanger Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Reiner, G.; Tramberend, K.; Nietfeld, F.; Volmer, K.; Wurmser, C.; Fries, R.; Willems, H. A genome-wide scan study identifies a single nucleotide substitution in the tyrosinase gene associated with white coat colour in a red deer (Cervus elaphus) population. BMC Genet. 2020, 21, 14. [Google Scholar] [CrossRef]
- Reiner, G.; Weber, T.; Nietfeld, F.; Fischer, D.; Wurmser, C.; Fries, R.; Willems, H. A genome-wide scan study identifies a single nucleotide substitution in MC1R gene associated with white coat colour in fallow deer (Dama dama). BMC Genet. 2020, 21, 126. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wu, X.; Li, Y.; Han, H.; Zhang, Y.; Yang, J.; Liu, Y. Effect of polymorphisms in the 5’-flanking sequence of MC1R on feather color in Taihang chickens. Poult. Sci. 2022, 101, 102192. [Google Scholar] [CrossRef]
- Charlier, C.; Denys, B.; Belanche, J.I.; Coppieters, W.; Grobet, L.; Mni, M.; Womack, J.; Hanset, R.; Georges, M. Microsatellite mapping of the bovine roan locus: A major determinant of White Heifer disease. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 1996, 7, 138–142. [Google Scholar] [CrossRef] [PubMed]
- Talenti, A.; Bertolini, F.; Williams, J.; Moaeen-Ud-Din, M.; Frattini, S.; Coizet, B.; Pagnacco, G.; Reecy, J.; Rothschild, M.F.; Crepaldi, P.; et al. Genomic Analysis Suggests KITLG is Responsible for a Roan Pattern in two Pakistani Goat Breeds. J. Hered. 2018, 109, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Besmer, P. The kit ligand encoded at the murine Steel locus: A pleiotropic growth and differentiation factor. Curr. Opin. Cell Biol. 1991, 3, 939–946. [Google Scholar] [CrossRef]
- Lundie, R.S. The genetics of colour in fat-tailed sheep: A review. Trop. Anim. Health Prod. 2011, 43, 1245–1265. [Google Scholar] [CrossRef]
- Aasland, M.; Klungland, H.; Lien, S. Two polymorphisms in the bovine mast cell growth factor gene (MGF). Anim. Genet. 2000, 31, 345. [Google Scholar] [CrossRef]
- Seitz, J.J.; Schmutz, S.M.; Thue, T.D.; Buchanan, F.C. A missense mutation in the bovine MGF gene is associated with the roan phenotype in Belgian Blue and Shorthorn cattle. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 1999, 10, 710–712. [Google Scholar] [CrossRef]
- Pillaiyar, T.; Manickam, M.; Jung, S.H. Recent development of signaling pathways inhibitors of melanogenesis. Cell Signal. 2017, 40, 99–115. [Google Scholar] [CrossRef]
- Annese, T.; Tamma, R.; Bozza, M.; Zito, A.; Ribatti, D. Autocrine/Paracrine Loop Between SCF+/c-Kit+ Mast Cells Promotes Cutaneous Melanoma Progression. Front. Immunol. 2022, 13, 794974. [Google Scholar] [CrossRef]
- Kawaguchi, M.; Hearing, V.J. The Roles of ADAMs Family Proteinases in Skin Diseases. Enzym. Res. 2011, 2011, 482498. [Google Scholar] [CrossRef]
- Imokawa, G.; Ishida, K. Inhibitors of intracellular signaling pathways that lead to stimulated epidermal pigmentation: Perspective of anti-pigmenting agents. Int. J. Mol. Sci. 2014, 15, 8293–8315. [Google Scholar] [CrossRef]
- Serre, C.; Busuttil, V.; Botto, J.M. Intrinsic and extrinsic regulation of human skin melanogenesis and pigmentation. Int. J. Cosmet. Sci. 2018, 40, 328–347. [Google Scholar] [CrossRef]
- Greenhill, E.R.; Kelsh, R.N. A pigment evolution Kitlg. Pigment Cell Melanoma Res. 2008, 21, 113–114. [Google Scholar] [CrossRef]
- Martin, F.H.; Suggs, S.V.; Langley, K.E.; Lu, H.S.; Ting, J.; Okino, K.H.; Morris, C.F.; McNiece, I.K.; Jacobsen, F.W.; Mendiaz, E.A. Primary structure and functional expression of rat and human stem cell factor DNAs. Cell 1990, 63, 203–211. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Amyere, M.; Vogt, T.; Hoo, J.; Brandrup, F.; Bygum, A.; Boon, L.; Vikkula, M. KITLG mutations cause familial progressive hyper- and hypopigmentation. J. Investig. Dermatol. 2011, 131, 1234–1239. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Karim, S.; Saharti, S.; Alganmi, N.; Mirza, Z.; Alfares, A.; Turkistany, S.; Al-Attas, M.; Noureldin, H.; Al Sakkaf, K.; Abusamra, H.; et al. Two Novel Homozygous HPS6 Mutations (Double Mutant) Identified by Whole-Exome Sequencing in a Saudi Consanguineous Family Suspected for Oculocutaneous Albinism. Life 2021, 12, 14. [Google Scholar] [CrossRef]
- Xu, X.; Dong, G.X.; Schmidt-Küntzel, A.; Zhang, X.L.; Zhuang, Y.; Fang, R.; Sun, X.; Hu, X.S.; Zhang, T.Y.; Yang, H.D.; et al. The genetics of tiger pelage color variations. Cell Res. 2017, 27, 954–957. [Google Scholar] [CrossRef] [PubMed]
- Garrido, G.; Fernández, A.; Montoliu, L. HPS11 and OCA8: Two new types of albinism associated with mutations in BLOC1S5 and DCT genes. Pigment Cell Melanoma Res. 2021, 34, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Fisher, D.E. Melanocyte biology and skin pigmentation. Nature 2007, 445, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Prota, G.; Hu, D.-N.; Vincensi, M.R.; McCormick, S.A.; Napolitano, A. Characterization of melanins in human irides and cultured uveal melanocytes from eyes of different colors. Exp. Eye Res. 1998, 67, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Nicholls, R.D.; Jong, M.T.; Fukai, K.; Spritz, R.A. Organization and sequence of the human P gene and identification of a new family of transport proteins. Genomics 1995, 26, 354–363. [Google Scholar] [CrossRef]
- Kunieda, T.; Nakagiri, M.; Takami, M.; Ide, H.; Ogawa, H. Cloning of bovine LYST gene and identification of a missense mutation associated with Chediak-Higashi syndrome of cattle. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 1999, 10, 1146–1149. [Google Scholar] [CrossRef]
- Hedberg-Oldfors, C.; Darin, N.; Oldfors, A. Muscle pathology in Vici syndrome-A case study with a novel mutation in EPG5 and a summary of the literature. Neuromuscul. Disord. NMD 2017, 27, 771–776. [Google Scholar] [CrossRef]
- Takeuchi, S.; Yamamoto, H.; Takeuchi, T. Expression of tyrosinase gene in amelanotic mutant mice. Biochem. Biophys. Res. Commun. 1988, 155, 470–475. [Google Scholar] [CrossRef]
- Brannan, C.I.; Lyman, S.D.; Williams, D.E.; Eisenman, J.; Anderson, D.M.; Cosman, D.; Bedell, M.A.; Jenkins, N.A.; Copeland, N.G. Steel-Dickie mutation encodes a c-kit ligand lacking transmembrane and cytoplasmic domains. Proc. Natl. Acad. Sci. USA 1991, 88, 4671–4674. [Google Scholar] [CrossRef]
- Mica, Y.; Lee, G.; Chambers, S.M.; Tomishima, M.J.; Studer, L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hESCs and patient-specific iPSCs. Cell Rep. 2013, 3, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
| Genes | AF | Mutation Position | Chromosome | Type |
|---|---|---|---|---|
| CIITA | 0.819079 | exon10:c.C1209T:p.S403S | MHL_chr10 | Synonymous SNV |
| CIITA | 0.8125 | exon10:c.G1644T:p.S548S | MHL_chr10 | Synonymous SNV |
| AP3B1 | 0.9471831 | exon23:c.A2754G:p.Q918Q | MHL_chr12 | Synonymous SNV |
| ICAT | 0.822368 | exon2:c.C268G:p.P90A | MHL_chr14 | Nonsynonymous SNV |
| LYST | 0.832237 | exon9:c.C1667T:p.S556L | MHL_chr15 | Nonsynonymous SNV |
| HPS3 | 0.834507 | exon9:c.G1590A:p.A530A | MHL_chr19 | Synonymous SNV |
| GSK3-β | 0.835526 | exon10:c.G1047A:p.L349L | MHL_chr19 | Synonymous SNV |
| DKK3 | 0.888158 | exon4:c.G510A:p.P170P | MHL_chr1 | Synonymous SNV |
| DKK3 | 0.9205298 | exon4:c.C513T:p.C171C | MHL_chr1 | Synonymous SNV |
| EPG5 | 0.9276316 | exon11:c.C2127T:p.P709P | MHL_chr27 | Synonymous SNV |
| EPG5 | 0.9337748 | exon27:c.A4762G:p.S1588G | MHL_chr27 | Nonsynonymous SNV |
| EPG5 | 0.807947 | exon29:c.C5040T:p.D1680D | MHL_chr27 | Synonymous SNV |
| EPG5 | 0.9342105 | exon32:c.T5643C:p.V1881V | MHL_chr27 | Synonymous SNV |
| TYRP1 | 0.838816 | exon1:c.C137T:p.S46F | MHL_chr29 | Nonsynonymous SNV |
| WRN | 0.809211 | exon12:c.T1529C:p.I510T | MHL_chr32 | Nonsynonymous SNV |
| SLC38A8 | 0.894737 | exon1:c.C24T:p.G8G | MHL_chr4 | Synonymous SNV |
| SLC38A8 | 0.888158 | exon6:c.C696T:p.H232H | MHL_chr4 | Synonymous SNV |
| HPS4 | 0.9212329 | exon9:c.G813A:p.A271A | MHL_chr5 | Synonymous SNV |
| VPS33A | 0.886986 | exon6:c.G735T:p.T245T | MHL_chr5 | Synonymous SNV |
| SCF | - | c.16444201-16454300 | MHL_chr3 | CNV |
| Phenotype | Genotype | Individuals | Amount |
|---|---|---|---|
| White | SCF789/SCF789 | B | 1 |
| White spots | SCF789/SCF1–9 | C, D, and F | 3 |
| Normal | SCF1–9/SCF1–9 | E, G, and H | 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Dong, S.; Liu, X.; Ding, N.; Xing, X. Phenotype of White Sika Deer Due to SCF Gene Structural Variation. Genes 2023, 14, 1035. https://doi.org/10.3390/genes14051035
Chen X, Dong S, Liu X, Ding N, Xing X. Phenotype of White Sika Deer Due to SCF Gene Structural Variation. Genes. 2023; 14(5):1035. https://doi.org/10.3390/genes14051035
Chicago/Turabian StyleChen, Xu, Shiwu Dong, Xin Liu, Ning Ding, and Xiumei Xing. 2023. "Phenotype of White Sika Deer Due to SCF Gene Structural Variation" Genes 14, no. 5: 1035. https://doi.org/10.3390/genes14051035
APA StyleChen, X., Dong, S., Liu, X., Ding, N., & Xing, X. (2023). Phenotype of White Sika Deer Due to SCF Gene Structural Variation. Genes, 14(5), 1035. https://doi.org/10.3390/genes14051035





