Abstract
TNF α-induced protein 1 (TNFAIP1) was first identified in human umbilical vein endothelial cells and can be induced by tumor necrosis factor α (TNFα). Early studies have found that TNFAIP1 is involved in the development of many tumors and is closely associated with the neurological disorder Alzheimer’s disease. However, little is known about the expression pattern of TNFAIP1 under physiological conditions and its function during embryonic development. In this study, we used zebrafish as a model to illustrate the early developmental expression pattern of tnfaip1 and its role in early development. First, we examined the expression pattern of tnfaip1 during early zebrafish development using quantitative real-time PCR and whole mount in situ hybridization and found that tnfaip1 was highly expressed in early embryonic development and, subsequently, expression became localized to anterior embryonic structures. To investigate the function of tnfaip1 during early development, we constructed a model of a stably inherited tnfaip1 mutant using the CRISPR/Cas9 system. Tnfaip1 mutant embryos showed significant developmental delays as well as microcephaly and microphthalmia. At the same time, we found decreased expression of the neuronal marker genes tuba1b, neurod1, and ccnd1 in tnfaip1 mutants. Analysis of transcriptome sequencing data revealed altered expression of the embryonic development related genes dhx40, hspa13, tnfrsf19, nppa, lrp2b, hspb9, clul1, zbtb47a, cryba1a, and adgrg4a in the tnfaip1 mutants. These findings suggest an important role for tnfaip1 in the early development of zebrafish.
1. Introduction
TNF α-induced protein 1 (TNFAIP1) was first identified in human umbilical vein endothelial cells and can be induced by tumor necrosis factor α (TNFα) [1]. Human TNFAIP1 is located on chromosome 17 and encodes a protein consisting of 316 amino acids. TNFAIP1 is a member of the PDIP1 subfamily of the KCTD protein family, which contains a conserved BTB/POZ domain at the N-terminal and functions through protein–protein interactions to form homologous or heterodimers [2]. The KCTD protein family can participate in the generation and transmission of cAMP signals, regulate neuronal function [3,4], and participate in the regulation of neuron development [5] and synaptic transmission [6]. At the same time, members of this protein family play an important regulatory role in the cardiovascular development of early embryos [7,8,9]. TNFAIP1 mediates DNA replication, repair, and cell cycle regulation by binding PCNA and a small subunit of polymerase δ (p50) [10]. Numerous studies have shown that TNFAIP1 is associated with proliferation [11], apoptosis [12,13,14], metastasis [15,16,17,18], and inflammatory responses [19] in a variety of tumors. In addition, TNFAIP1 is involved in chronic hepatitis B virus (HBV) infection, mediating immune tolerance to HBV [20]. Early studies found that TNFAIP1 is highly expressed at low pathogenic sites in Caenorhabditis elegans transgenic models of Alzheimer’s disease, and aberrant expression of the TNFAIP1 was detected in the brains of Alzheimer’s disease patients after death [21]. Amyloid-β (Aβ) induces the upregulation of TNFAIP1 expression, promoting neuronal cell damage [22,23,24]. The inhibition of TNFAIP1 expression inhibits nerve injury induced by oxygen–glucose deprivation/reperfusion [25]. TNFAIP1 and p-TNFAIP1 (phosphorylation of TNFAIP1-Ser280 site) are upregulated in the cerebral cortex and hippocampal neurons of APP/PS1 double transgenic mice, suggesting a close relationship between TNFAIP1 and Alzheimer’s disease pathogenesis [26]. TNFAIP1 expression is increased in the rat chronic constriction injury (CCI) model and influences the development of neuropathic pain in CCI rats [27]. TNFAIP1 has been found to interact with Rnd2 and Rnd3 to influence the dynamics of neuronal cell migration, neuronal branching, and dendritic spines in the embryonic cerebral cortex [28,29]. These studies reveal important functions of TNFAIP1 in nervous system development and Alzheimer’s disease. Thus far, studies on the function of TNFAIP1 have mainly focused on pathological conditions such as tumors and Alzheimer’s disease, while there are few studies on the function of TNFAIP1 under physiological conditions, in particular, the role of TNFAIP1 in regulating embryonic development is still unknown.
Species homology analysis revealed that the TNFAIP1 gene is highly conserved across species, with 73% sequence homology between zebrafish and humans. Zebrafish are popular model organisms for development and disease research, and CRISPR/Cas9 gene editing of zebrafish is an important tool for functional genomics research [30]. Therefore, using the zebrafish model to study the role of tnfaip1 in embryonic development may better contribute to understanding the function of TNFAIP1 under physiological conditions.
In this study, we first detected the expression pattern of tnfaip1 at different stages of zebrafish embryonic development and found that tnfaip1 was highly expressed in early embryos, and, between 24 hpf and 72 hpf, the expression of tnfaip1 was mainly localized in the anterior organs of the embryo. The tnfaip1 mutant model was constructed using the CRISPR/Cas9 system. Phenotypic analysis revealed that tnfaip1 mutant embryos showed significant developmental delay as well as microcephaly and microphthalmia. In addition, expression of neuronal marker genes was reduced in the mutant embryos. Analysis of transcriptome sequencing data revealed that the abnormal embryonic development was due to altered expression of genes associated with development in the mutant. Taken together, our results demonstrate the importance of tnfaip1 in embryonic development, which provides a new theoretical basis for exploring the function of tnfaip1 under physiological conditions.
2. Materials and Methods
2.1. Zebrafish Husbandry
Zebrafish (Danio rerio) (TU strain) were obtained from the China Zebrafish Resource Center (Wuhan, China). All wild-type and mutant zebrafish were bred under 14 h/10 h day/night cycle at 28.5 °C. Adult male and female zebrafish were mated in a 1:1 ratio to obtain embryos and staged in hours post-fertilization (hpf) or days post-fertilization (dpf). Fertilized eggs were cultured at 28.5 °C in E3 water (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2·H2O, 0.33 mM MgSO4·7H2O). Zebrafish experiments were performed according to the zebrafish book [31].
2.2. Whole Mount In Situ Hybridization
Zebrafish in situ hybridization experiments were performed according to the standard procedure for in situ hybridization reported previously [32]. The T7 promoter sequence was added to the 5’ end of the reverse primer used to synthesize the antisense RNA probe template for PCR amplification (Q5® Hot Start High-Fidelity 2× Master Mix, M0494, NEB, Ipswich, MA, USA) with a cDNA template from 72 hpf wild-type zebrafish embryos. Digoxigenin-labelled antisense RNA probes were synthesized using T7 RNA polymerase (EP0111, Thermo, Waltham, MA, USA) and DIG RNA Labeling Mix (11277073910, Roche, Basel, Switzerland). After 24 hpf, the embryos were permeabilized with proteinase K: 24 hpf (2 min), 48 hpf (30 min), 72 hpf (45 min). DIG-labelled probes were detected using an alkaline-phosphatase-conjugated anti-DIG Fab fragments (11093274910, Roche, Switzerland), followed by staining with nitro blue tetrazolium (NBT; N6876, Sigma, Burlington, MA, USA) and 5-bromo-4-chloro-3-indolyl phosphate (BICP; B8503, Sigma, USA). All primer sequences used for the synthesis of antisense RNA probes are listed in Supplementary Table S1.
2.3. Generation of tnfaip1 Mutants
Generation of a tnfaip1 zebrafish mutant model was performed using CRISPR/Cas9 gene editing technology [30,33]. The tnfaip1 knockout target sites were designed using the CHOPCHOP online website (https://chopchop.rc.fas.harvard.edu/) (accessed on 20 July 2020), and the target sequences are listed in Supplementary Table S2. The sgRNA for the target sites was synthesized using an in vitro transcription kit (HiScribe™T7 Quick High Yield RNA Synthesis Kit, E2050S, NEB, USA) and, subsequently, purified using an RNA purification kit (RC201, Vazyme, Nanjing, China). TrueCut™ Cas9 protein v2 (A36498, Waltham, MA, USA) was purchased from Invitrogen. Subsequently, the ~1 nL of 150 ng/μL sgRNA and 0.5 μg/μL Cas9 protein complexes were microinjected into one-cell stage embryos. The homozygous mutant of tnfaip1 was obtained by crossing heterozygotes. Primer sequences to identify the genotype of the mutant are listed in Supplementary Table S2.
2.4. Transcriptome Sequencing and Data Analysis
Sibling and tnfaip1−/− embryos were harvested at 72 hpf, with 4 replicates in each group, frozen in liquid nitrogen, and sent to Majorbio (Shanghai, China) for RNA extraction and transcriptome sequencing. Clean data of each sample obtained by RNA-seq reached more than 7.3 Gb. The clean reads of each sample were mapped to the zebrafish reference genome (GRCz11, http://asia.ensembl.org/Danio_rerio/Info/Index;) (accessed on 25 December 2022). STAR-2.7.1a was used to map reads to reference genome [34]. Quantification of gene expression was performed using RSEM [35]. Differential gene expression analysis was then performed using the package “DESeq2” [36]. DEGs between groups (tnfaip1−/− vs. sibling) were identified using the criteria p-adjust < 0.05 and|log2 (fold change)| ≥ 1. Volcano plots were drawn using the R package “ggplot2”. Gene Ontology (GO) and KEGG Pathway enrichment analyzes were performed with the “clusterProfiler” R package, and corrected p-values < 0.05 were considered significantly enriched.
2.5. Quantitative Real-Time PCR
Embryonic samples were collected from zebrafish at various developmental stages, and total RNA was extracted using RNAiso Plus (9108, Takara, Okinawa, Japan). cDNA was, subsequently, synthesized using GoScript™ reverse transcriptase (A5003, Promega, Madison, WI, USA). In this study, 18SrRNA was used as an internal reference gene used for detecting the relative expression of genes, and all primer sequences are listed in Supplementary Table S3. Real-time PCR was performed on a QuantStudio™ 5 System Real-Time PCR Detection System (ABI, Foster City, CA, USA) using TB Green® Premix ExTaq™ II (RR820A, Takara, Japan), and all samples were performed in triplicate. Relative expression levels of mRNA were calculated using 2−ΔΔCT and expressed as mean ± SEM.
2.6. Protein Extraction and Western Blotting
Zebrafish embryos at 72 hpf wild-type and tnfaip1−/− were collected and lysed in RIPA lysis buffer (BL504A, Biosharp, Hefei, China) with a protease inhibitor cocktail (M5293, Abmole, Houston, TX, USA). After protein separation with 12% polyacrylamide gels, they were transferred to PVDF membranes, incubated with antibodies, and detected. The zebrafish Tnfaip1 antibody (1:200) was customized at BOSTER (Wuhan, China). The TUBULIN antibody was purchased from Affinity Biosciences (1:5000, AF7010). The secondary antibodies goat anti-Rabbit IgG was purchased from Affinity Biosciences (1:10,000, S0001).
2.7. Embryo Imaging
Embryos before 72 hpf were manually stripped of their chorion, and all photographed embryos were anesthetized with 0.003% tricaine (E10521, Sigma, USA). Embryos were fixed in biconvex slides with 1% low melting agarose. Image acquisition was performed with a fluorescent stereomicroscope (M205 FA, Leica, Wetzlar, Germany). All photographed embryos were subjected to body length and eye size measurements using Image J.
2.8. Statistical Analysis
All experiments were performed with at least three independent biological replicates. Data were analyzed with unpaired two-tailed t-tests using GraphPad Prism 6. All data are presented as mean ± SEM. p < 0.05 was considered statistically significant, p < 0.01 was considered highly statistically significant, and p > 0.05 was considered not statistically significant.
3. Results
3.1. Spatio-Temporal Expression Pattern of tnfaip1 in Zebrafish during Early Development
The expression pattern of a gene is the basis for its function. To investigate the role of tnfaip1 during early zebrafish development, we first examined its expression during different developmental stages. Embryos were collected at different developmental stages, total RNA was extracted and reverse transcribed into cDNA, the qPCR primers specific for the tnfaip1 gene were designed using the 18SrRNA as an internal control, and quantitative real-time PCR was performed to detect the expression levels of tnfaip1 mRNA at each stage of zebrafish embryo development. The results showed that tnfaip1 was highly expressed during early zebrafish development, from 1 cell to 72 hpf (Figure 1A). In situ hybridization showed that tnfaip1 was ubiquitously expressed during the cell period, and expression became localized to anterior embryonic structures between 11 hpf and 24 hpf (Figure 1B). Tnfaip1 expression remained localized to the anterior embryo between 24 hpf and 72 hpf (Figure 1B,C). The in situ hybridization and qPCR results indicated that tnfaip1 was a maternally expressed gene that functioned mainly through maternal expression in the early stage. Tnfaip1 expression became localized to anterior embryo sometime between 11 hpf and 24 hpf and continued to be expressed in the anterior embryo after 24 hpf, revealing its possible involvement in the early development of multiple tissues and organs in the anterior structures of the zebrafish embryo.
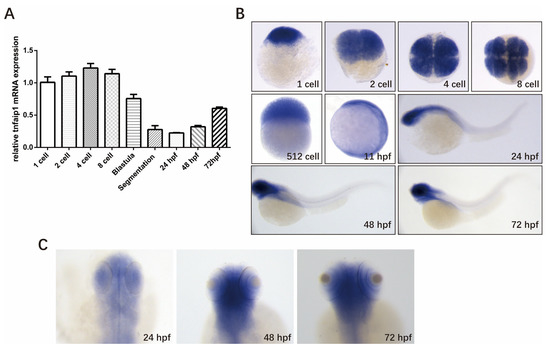
Figure 1.
Spatio-temporal expression pattern of tnfaip1 in zebrafish during early development. (A) qPCR detection of the relative expression level of tnfaip1 mRNA during zebrafish embryo development, with 18SrRNA as an internal control. (B) Whole mount in situ hybridization to detect tnfaip1 expression during zebrafish embryo development. n = 15 per group; scale bar, 500 μm. (C) High magnification of tnfaip1 expression in zebrafish 24, 48, and 72 hpf anterior organs. n = 15 per group; scale bar, 250 μm.
3.2. Generation of the tnfaip1 Mutant in Zebrafish Using the CRISPR/Cas9 System
To investigate the function of tnfaip1 during early development, we generated a zebrafish tnfaip1 mutant model using the CRISPR/Cas9 system. We designed two target sites on exon 2 of the tnfaip1 in zebrafish and considered targeting efficiency, target location, and off-target effects (Figure 2A). The target site sgRNA and Cas9 protein were co-microinjected into 1-cell stage wild-type zebrafish embryos. Targeting efficiency was characterized after microinjection, and, after crossing the F0 and F1 generations, a mutant type with a 160 bp deletion in exon 2 was finally screened (Figure 2B and Figure S1). Sequence alignment analysis revealed that this mutation resulted in a shift in the open reading frame of tnfaip1, prematurely terminating the coding of the protein and resulting in a truncated protein (Figure 2C). To examine the effect of tnfaip1 mutation on its mRNA and protein expression, we collected 72 hpf wild-type sibling and tnfaip1 mutant embryos and performed qPCR, in situ hybridization, and Western blotting. The results of qPCR and in situ hybridization showed a decrease in mRNA levels by approximately 40% after mutation (Figure 2E,F). Western blotting showed decreased expression of Tnfaip1 protein in mutants (Figure 2G). However, a small amount of the protein was still present in the tnfaip1 mutant, possibly due to residual maternal expression. These results indicated that we had successfully established a tnfaip1 mutant model and can use this mutant for the next step in our research.

Figure 2.
Generation of the tnfaip1 mutant in zebrafish using the CRISPR/Cas9 system. (A) Gene structure of tnfaip1 and location of CRISPR/Cas9 targets (red arrow), Target 1 and Target 2 are two knockout target sgRNAs, with the specific target sequence shown in black and the PAM (NGG) sequence in red. (B) The DNA sequencing peak and sequence of the tnfaip1 mutant. DNA sequencing of the corresponding genomic region of zebrafish tnfaip1 revealed a 160 bp deletion mutation. (C) The tnfaip1−160 mutation resulted in a gene frame shift and early termination of the encoded protein. This mutation resulted in a protein truncated to 30 aa. The asterisks mark the termination codon. aa: amino acids. (D) Identification of genotype by DNA electrophoresis. +/+, wild type; +/−, heterozygote; −/−, homozygote. (E) Relative expression of tnfaip1 mRNA at 72 hpf detected by qPCR and 18SrRNA as an internal control. “*” represents p < 0.05, analyzed with unpaired two-tailed t-tests. (F) In situ hybridization to detect expression of tnfaip1 in the mutants. (G) Decreased protein levels in tnfaip1−/− embryos at 72 hpf as measured by Western blot. Tubulin as an internal control.
3.3. Mutation of tnfaip1 Affects the Early Development of Zebrafish Embryos
We crossed tnfaip1 F1 heterozygous mutants to generate F2 embryos. Compared with wild-type sibling embryos at the same period, a small number of homozygous mutant embryos died during early development. This phenomenon may have been related to our early observation of pericardial edema and abnormal arrest of blood flow in homozygous mutant embryos. Subsequently, we observed the early development of these embryos under a stereomicroscope. We found that 48–96 hpf tnfaip1 mutant embryos exhibited significant morphological abnormalities, including microphthalmia, smaller heads, and shortened body lengths (Figure 3A). Subsequently, we performed statistical analyses of body length (Figure 3B) as well as eye size (Figure 3C) of tnfaip1−/− embryos at different developmental stages.

Figure 3.
Mutation of tnfaip1 in zebrafish affects early embryonic development. (A) Phenotypic analysis of 48–96 hpf wild-type sibling and tnfaip1−/− embryonic development. Scale bar: 750 μm. (B) Statistical analysis of 48–96 hpf wild-type sibling and tnfaip1−/− embryo body lengths. n = 30. “**” represents p < 0.01, analyzed with unpaired two-tailed t-tests. (C) Statistical analysis of 48–96 hpf wild-type sibling and tnfaip1−/− embryo eye sizes, n = 30. “**” represents p < 0.01, analyzed with unpaired two-tailed t-tests.
Early embryonic in situ hybridization showed that tnfaip1 was predominantly expressed in the anterior structures, and we found that tnfaip1 mutation resulted in microphthalmia and microcephaly. To identify the cause of this developmental malformation in tnfaip1−/− zebrafish, we collected 72 hpf wild-type sibling and tnfaip1−/− embryos for in situ hybridization assays for the neuronal marker genes ccnd1, tuba1b, and neurod1. The results showed that the neuronal marker genes ccnd1 (Figure 4A), tuba1b (Figure 4B), and neurod1 (Figure 4C) were significantly reduced in the mutants compared to the wild type. These results suggested that the reduction in tnfaip1 expression affected the expression of neuronal marker genes, leading to microphthalmia and microcephaly.

Figure 4.
Tnfaip1 mutation in zebrafish results in reduced expression of neuronal marker genes. (A) In situ hybridization assay for the 72 hpf wild-type sibling and tnfaip1−/− embryos neuronal marker gene ccnd1. Yellow arrows indicate the retinal; white arrows indicate the tectum proliferative zone; black arrows indicate the hindbrain. Scale bar: 250 μm. (B) In situ hybridization assay for the 72 hpf wild-type sibling and tnfaip1−/− embryos neuronal marker gene tuba1b. Red arrow indicates the retinal. Scale bar: 250 μm. (C) In situ hybridization assay for the 72 hpf wild-type sibling and tnfaip1−/− embryos neuronal marker gene neurod1. Red asterisks indicate the brain; yellow arrows indicate the retinal. Scale bar: 250 μm.
3.4. Transcriptome Levels Are Altered in tnfaip1 Mutants
To explore the downstream molecular mechanisms by which reduction in tnfaip1 affects the embryonic development of zebrafish, we collected 72 hpf embryos for transcriptome sequencing, with four independent biological replicates per group. Analysis of the sequencing results identified a total of 79 differentially expressed genes (DEGs), including 59 downregulated genes and 20 upregulated genes (Figure 5A). Altered expression of differentially expressed genes dhx40, hspa13, tnfrsf19, nppa, lrp2b, hspb9, clul1, zbtb47a, cryba1a, and adgrg4a in tnfaip1 mutants was associated with early developmental regulation (Figure 5B,C). To determine the function of the differentially expressed genes of sibling and tnfaip1−/−, we performed GO functional and KEGG pathway enrichment analyses. The results showed that DEGs were mainly enriched in processes such as “misfolded protein binding”, “phosphatase activity”, and “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen” (Figure 5D). KEGG pathway enrichment analysis showed that DEGs were mainly involved in the regulation of “steroid hormone biosynthesis”, “retinol metabolism”, “biosynthesis of cofactors”, and “nucleotide metabolism” (Figure 5E).
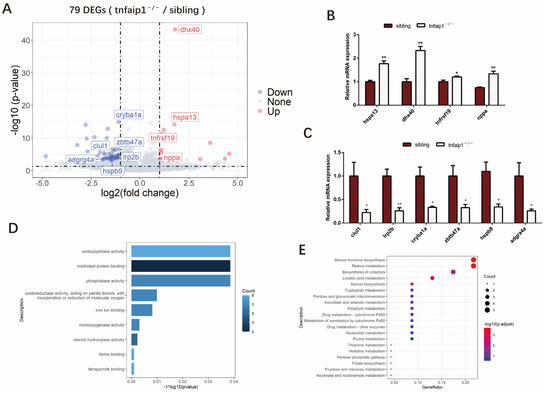
Figure 5.
Identification of differentially expressed genes between siblings and tnfaip1−/−. (A) Volcano plot showing the overall differences in upregulated and downregulated genes in the mutants. Some differentially expressed genes are highlighted in the figure. DEGs, differentially expressed genes. (B,C) qPCR validation of differentially expressed genes associated with embryonic development in siblings and tnfaip1−/−. “*” represents p < 0.05, “**” represents p < 0.01, analyzed with unpaired two-tailed t-tests. (D) Gene Ontology (GO) enrichment analysis of differentially expressed genes from siblings and tnfaip1 mutants. (E) KEGG pathway enrichment analysis of differentially expressed genes in siblings and tnfaip1 mutants.
4. Discussion
TNFAIP1 is thought to be associated with tumors and neurological disorders, and, to date, there have been no studies on the function of the TNFAIP1 gene during early development. In the past, the study of the function of TNFAIP1 have been limited to cellular models, and there is a lack of transgenic or knockout animal models of TNFAIP1, as zebrafish tnfaip1 is highly homologous to the human TNFAIP1 sequence. Therefore, this study focused on the expression pattern of tnfaip1 in zebrafish during early development and the establishment of a zebrafish tnfaip1 mutant model to lay the foundation for the elaboration of the biological functions of tnfaip1 during early development.
The results of whole mount in situ hybridization and qPCR showed that zebrafish tnfaip1 was a maternally expressed gene, and its expression level was relatively high in the early stage. With the depletion of maternal mRNA, its embryonic expression level began to increase gradually. Additionally, it was mainly expressed in the anterior structures of the embryo, including the head and eyes. This early developmental expression pattern of the zebrafish tnfaip1 gene revealed its possible involvement in the early development and functional maintenance of multiple tissues and organs in the anterior structure of the embryo. In this study, for the first time, we constructed a zebrafish tnfaip1 mutant model using CRISPR/Cas9 gene editing technology. Whole mount in situ hybridization and qPCR showed that the mRNA expression levels in the tnfaip1 mutant were reduced by approximately 40%. We speculated that the reason for the incomplete suppression of mRNA expression in the mutant may have been due to incomplete nonsense-mediated mRNA degradation [37,38]. In addition, Western blotting showed that a small amount of the protein was still present in the tnfaip1 mutant, possibly due to residual maternal expression. There was another possibility that the stop codon created by the mutation had some leakiness, which resulted in some expression of mRNA and protein remaining in the mutant. Due to our transcriptomic data showing no significant changes in the expression of the two orthologs of tnfaip1, kctd10, and kctd13, and no significant changes in the expression of other KCTD family paralogs were observed.
In this study, we crossed tnfaip1 F1 heterozygous mutants to generate F2 embryos. During early embryonic development from 48–96 hpf, we found that tnfaip1 homozygous mutants exhibited markedly shortened body lengths, small eyes, and small heads. Due to the expression pattern of tnfaip1 during early development and the observed microphthalmia and microcephaly in mutants, we hypothesized that tnfaip1 may be required to maintain normal head and eye morphologies in zebrafish embryos. To further confirm this conjecture, we collected 72 hpf wild-type and tnfaip1 mutant embryos for in situ hybridization of the neuronal marker genes ccnd1, tuba1b, and neurod1. The results showed that the expression of ccnd1 in the tectum proliferative zone, hindbrain, and retina was significantly lower in the mutants than the wild-type sibling. The expression of tuba1b in retinal ganglion cells was significantly reduced in the mutants. It was significantly reduced expression of neurod1 throughout the brain and retina of the mutants. These results suggested that microphthalmia and microcephaly caused by tnfaip1 mutations may have been due to affecting the expression of neuronal marker genes during early development.
To explore possible downstream mechanisms by which tnfaip1 mutations affect morphological abnormalities in early embryos, we collected 72 hpf wild-type sibling and tnfaip1 mutant embryos for transcriptome sequencing. Analysis of the sequencing data revealed that most of the differentially expressed genes in the tnfaip1 mutant were associated with early development. The altered expressions of clul1, hspa13, dhx40, cryba1a, tnfrsf19, zbtb47a, lrp2b, hspb9, and adgrg4a were closely associated with eye development, ocular diseases, formation of the nervous system, and embryonic development. Clul1 was highly expressed in the retina, suggesting that it had an important role in the retina [39,40]. Tnfaip1 mutation resulted in down-regulation of clul1 expression, indicating that the microphthalmia caused by tnfaip1 may have been due to the inhibition of clul1 expression. Hspa13 is a candidate gene for retinal degeneration, and its expression changes may lead to abnormal development of the retina [41]. GO functional enrichment analysis showed that differentially expressed genes were mainly involved in processes such as “misfolded protein binding”, “phosphatase activity”, and “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen”; KEGG Pathway enrichment analysis results showed that the differentially expressed genes were mainly involved in the regulation of pathways such as “Steroid hormone biosynthesis”, “Retinol metabolism”, “Biosynthesis of cofactors”, and “Nucleotide metabolism”. These results suggested that tnfaip1 mutations lead to alterations in the expression of a number of genes and associated signaling pathways that affected early embryonic development. However, studies on the function of tnfaip1 in embryonic development are still in the initial stages. Our previous results showed that tnfaip1 was highly expressed in multiple tissues and organs in the anterior structures of the embryo, and it was hypothesized that tnfaip1 was likely to be involved in the formation and functional maintenance of these tissues and organs. Therefore, the early developmental regulation of specific tissues and organs by tnfaip1 and details of its molecular mechanisms are worthy to further investigate in the future.
5. Conclusions
In this study, we found that tnfaip1 was highly expressed during the early development of zebrafish. Between 11 and 24 hpf, the expression began to localize in the anterior organ of the embryo, and the expression of tnfaip1 was still localized in the anterior embryo between 24 and 72 hpf. We generated a zebrafish tnfaip1 mutant model using CRISPR/Cas9 gene editing technology. Additionally, using this mutant model, we observed morphological abnormalities in zebrafish embryonic development, including shortened body length, microphthalmia, and microcephaly. In addition, we found reduced expression of neuronal marker genes in tnfaip1 mutants. RNA-seq data revealed altered expression of genes involved in the regulation of early development in tnfaip1 mutants. Taken together, our findings suggest an important function of tnfaip1 in the early development of zebrafish.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14051005/s1, Table S1: In situ hybridization primer sequences used in this study, Table S2: tnfaip1 sgRNA sequence and genotyping primer sequence used in this study, Table S3: qPCR primer sequences used in this study, Figure S1: Partial sequence of the tnfaip1 mutant related to Figure 2.
Author Contributions
Conceptualization, S.H., S.X. and X.H.; methodology, S.H.; software, H.Z. and W.C.; validation, S.H. and N.S.; formal analysis, S.H., H.Z. and W.C.; investigation, S.H.; resources, S.H., J.Z., S.X. and X.H.; data curation, S.H.; writing—original draft preparation, S.H.; writing—review and editing, S.H., H.Z., W.C., N.S., C.Y., J.Z., S.X. and X.H; visualization, S.H.; supervision, S.X. and X.H.; project administration, S.X. and X.H.; funding acquisition, X.H. and C.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81972642), the Hunan Provincial Natural Science Foundation of China (2021JJ30444), the Changsha City Natural Science Foundation of China (kq2007013), and the Innovation and entrepreneurship fund for college students of the Hunan Normal University (2021118).
Institutional Review Board Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC), Hunan Normal University.
Informed Consent Statement
Not applicable.
Data Availability Statement
All available raw data can be obtained from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wolfs, F.W.; Marks, R.M.; Sarma, V.; Byers, M.G.; Katz, R.W.; Shows, T.B.; Dixit, V.M. Characterization of a Novel Tumor Necrosis Factor-a-induced Endothelial Primary Response Gene. J. Biol. Chem. 1992, 267, 1317–1326. [Google Scholar] [CrossRef]
- Liu, Z.; Xiang, Y.; Sun, G. The KCTD family of proteins: Structure, function, disease relevance. Cell Biosci. 2013, 3, 5. [Google Scholar] [CrossRef]
- Sloan, D.C.; Cryan, C.E.; Muntean, B.S. Multiple potassium channel tetramerization domain (KCTD) family members interact with Gbetagamma, with effects on cAMP signaling. J. Biol. Chem. 2023, 299, 102924. [Google Scholar] [CrossRef]
- Muntean, B.S.; Marwari, S.; Li, X.; Sloan, D.C.; Young, B.D.; Wohlschlegel, J.A.; Martemyanov, K.A. Members of the KCTD family are major regulators of cAMP signaling. Proc. Natl. Acad. Sci. USA 2022, 119, e2119237119. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.N.; Corominas, R.; Lemmens, I.; Yang, X.; Tavernier, J.; Hill, D.E.; Vidal, M.; Sebat, J.; Iakoucheva, L.M. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron 2015, 85, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Golzio, C.; Willer, J.; Talkowski, M.E.; Oh, E.C.; Taniguchi, Y.; Jacquemont, S.; Reymond, A.; Sun, M.; Sawa, A.; Gusella, J.F.; et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature 2012, 485, 363–367. [Google Scholar] [CrossRef]
- Hu, X.; Gan, S.; Xie, G.; Li, L.; Chen, C.; Ding, X.; Han, M.; Xiang, S.; Zhang, J. KCTD10 is critical for heart and blood vessel development of zebrafish. Acta Biochim. Biophys. Sin. 2014, 46, 377–386. [Google Scholar] [CrossRef]
- Tong, X.; Zu, Y.; Li, Z.; Li, W.; Ying, L.; Yang, J.; Wang, X.; He, S.; Liu, D.; Zhu, Z.; et al. Kctd10 regulates heart morphogenesis by repressing the transcriptional activity of Tbx5a in zebrafish. Nat. Commun. 2014, 5, 3153. [Google Scholar] [CrossRef]
- Ren, K.; Yuan, J.; Yang, M.; Gao, X.; Ding, X.; Zhou, J.; Hu, X.; Cao, J.; Deng, X.; Xiang, S.; et al. KCTD10 is involved in the cardiovascular system and Notch signaling during early embryonic development. PLoS ONE 2014, 9, e112275. [Google Scholar] [CrossRef]
- Zhou, J.; Hu, X.; Xiong, X.; Liu, X.; Liu, Y.; Ren, K.; Jiang, T.; Hu, X.; Zhang, J. Cloning of two rat PDIP1 related genes and their interactions with proliferating cell nuclear antigen. J. Exp. Zool. Part A Comp. Exp. Biol. 2005, 303, 227–240. [Google Scholar] [CrossRef]
- Zhang, X.; Li, X.; Tan, Z.; Liu, X.; Yang, C.; Ding, X.; Hu, X.; Zhou, J.; Xiang, S.; Zhou, C.; et al. MicroRNA-373 is upregulated and targets TNFAIP1 in human gastric cancer, contributing to tumorigenesis. Oncol. Lett. 2013, 6, 1427–1434. [Google Scholar] [CrossRef]
- Kim, D.M.; Chung, K.S.; Choi, S.J.; Jung, Y.J.; Park, S.K.; Han, G.H.; Ha, J.S.; Song, K.B.; Choi, N.S.; Kim, H.M.; et al. RhoB induces apoptosis via direct interaction with TNFAIP1 in HeLa cells. Int. J. Cancer 2009, 125, 2520–2527. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Li, X.; Zhang, X.; Liu, X.; Tan, Z.; Yang, C.; Zhang, J. microRNA-372 maintains oncogene characteristics by targeting TNFAIP1 and affects NFkappaB signaling in human gastric carcinoma cells. Int. J. Oncol. 2013, 42, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.Y.; Kim, K.; Han, T.S.; Lee, M.O.; Lee, J.; Choi, J.; Jung, K.B.; Jeong, E.J.; An, D.M.; Jung, C.R.; et al. Human gut-microbiome-derived propionate coordinates proteasomal degradation via HECTD2 upregulation to target EHMT2 in colorectal cancer. ISME J. 2022, 16, 1205–1221. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.W.; Xie, S.; Hu, S.Y.; Liao, T.; Liu, P.; Peng, K.H.; Yang, X.Z.; He, Z.L.; Tang, H.Y.; Cui, Y.; et al. Caudatin targets TNFAIP1/NF-kappaB and cytochrome c/caspase signaling to suppress tumor progression in human uterine cancer. Int. J. Oncol. 2016, 49, 1638–1650. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Meng, W.; Sun, H.-L.; Kim, T.; Ye, Z.; Fassan, M.; Jeon, Y.-J.; Li, B.; Vincenti, C.; Peng, Y.; et al. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc. Natl. Acad. Sci. USA 2015, 112, E4288–E4297. [Google Scholar] [CrossRef]
- Zhang, M.; Gao, C.; Yang, Y.; Li, G.; Dong, J.; Ai, Y.; Ma, Q.; Li, W. MiR-424 Promotes Non-Small Cell Lung Cancer Progression and Metastasis through Regulating the Tumor Suppressor Gene TNFAIP1. Cell. Physiol. Biochem. 2017, 42, 211–221. [Google Scholar] [CrossRef]
- Xiao, Y.; Huang, S.; Qiu, F.; Ding, X.; Sun, Y.; Wei, C.; Hu, X.; Wei, K.; Long, S.; Xie, L.; et al. Tumor necrosis factor alpha-induced protein 1 as a novel tumor suppressor through selective downregulation of CSNK2B blocks nuclear factor-kappaB activation in hepatocellular carcinoma. eBioMedicine 2020, 51, 102603. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, W.; Wang, S.; Cai, L.; Jiang, Y.; Pan, Y.; Liang, Y.; Xian, J.; Jia, L.; Li, L.; et al. Cullin3-TNFAIP1 E3 Ligase Controls Inflammatory Response in Hepatocellular Carcinoma Cells via Ubiquitination of RhoB. Front. Cell Dev. Biol. 2021, 9, 617134. [Google Scholar] [CrossRef]
- Lin, M.C.; Lee, N.P.; Zheng, N.; Yang, P.-H.; Wong, O.G.; Kung, H.-F.; Hui, C.-K.; Luk, J.M.; Lau, G.K.-K. Tumor necrosis factor-α-induced protein 1 and immunity to hepatitis B virus. World J. Gastroenterol. 2005, 11, 7564–7568. [Google Scholar] [CrossRef]
- Link, C.D.; Taft, A.; Kapulkin, V.; Duke, K.; Kim, S.; Fei, Q.; Wood, D.E.; Sahagan, B.G. Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer's disease model. Neurobiol. Aging 2003, 24, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Yu, Z.; Xun, Y.; Li, M.; Peng, X.; Xiao, Y.; Hu, X.; Sun, Y.; Yang, M.; Gan, S.; et al. TNFAIP1 contributes to the neurotoxicity induced by Abeta25-35 in Neuro2a cells. BMC Neurosci. 2016, 17, 51. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Tan, J.; Zhang, J. miR-137 attenuates Abeta-induced neurotoxicity through inactivation of NF-kappaB pathway by targeting TNFAIP1 in Neuro2a cells. Biochem. Biophys. Res. Commun. 2017, 490, 941–947. [Google Scholar] [CrossRef]
- Li, Y.; Jin, L.; Wang, F.; Ren, L.; Pen, R.; Bo, G.; Wang, L. Epigenetic axis of SNHG19/miR-137/TNFAIP1 modulates amyloid beta peptide 25-35-induced SH-SY5Y cytotoxicity. Epigenomics 2022, 14, 187–198. [Google Scholar] [CrossRef]
- Xiong, L.; Zhang, J.; Shi, H.; Zhu, G.; Ji, X.; Li, M.; Zhu, P.; Luo, K. Downregulation of TNFAIP1 alleviates OGD/R-induced neuronal damage by suppressing Nrf2/GPX4-mediated ferroptosis. Exp. Ther. Med. 2023, 25, 25. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, Y.; Zhang, H.; Yang, L.; Jiang, Y.; Wei, C.; Feng, X.; Xun, Y.; Yuan, S.; Xiang, S.; et al. TNFAIP1 Is Upregulated in APP/PS1 Mice and Promotes Apoptosis in SH-SY5Y Cells by Binding to RhoB. J. Mol. Neurosci. MN 2021, 71, 1221–1233. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, S.; Xia, N.; Shi, Y.; Zhao, C.M. Effects of XIST/miR-137 axis on neuropathic pain by targeting TNFAIP1 in a rat model. J. Cell. Physiol. 2018, 233, 4307–4316. [Google Scholar] [CrossRef]
- Gladwyn-Ng, I.E.; Li, S.S.; Qu, Z.; Davis, J.M.; Ngo, L.; Haas, M.; Singer, J.; Heng, J.I. Bacurd2 is a novel interacting partner to Rnd2 which controls radial migration within the developing mammalian cerebral cortex. Neural Dev. 2015, 10, 9. [Google Scholar] [CrossRef]
- Gladwyn-Ng, I.; Huang, L.; Ngo, L.; Li, S.S.; Qu, Z.; Vanyai, H.K.; Cullen, H.D.; Davis, J.M.; Heng, J.I. Bacurd1/Kctd13 and Bacurd2/Tnfaip1 are interacting partners to Rnd proteins which influence the long-term positioning and dendritic maturation of cerebral cortical neurons. Neural Dev. 2016, 11, 7. [Google Scholar] [CrossRef]
- Varshney, G.K.; Carrington, B.; Pei, W.; Bishop, K.; Chen, Z.; Fan, C.; Xu, L.; Jones, M.; LaFave, M.C.; Ledin, J.; et al. A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 2016, 11, 2357–2375. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio); Univeristy of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Thisse, C.; Thisse, B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 2008, 3, 59–69. [Google Scholar] [CrossRef]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Ahkmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019, 20, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Nasif, S.; Contu, L.; Muhlemann, O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 2018, 75, 78–87. [Google Scholar] [CrossRef]
- Zhang, Q.; Ray, K.; Acland, G.M.; Czarnecki, J.M.; Aguirre, G.D. Molecular cloning, characterization and expression of a novel retinal clusterin-like protein cDNA. Gene 2000, 243, 151–160. [Google Scholar] [CrossRef]
- Zhang, Q.; Beltran, W.A.; Mao, Z.; Li, K.; Johnson, J.L.; Acland, G.M.; Aguirre, G.D. Comparative analysis and expression of CLUL1, a cone photoreceptor-specific gene. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4542–4549. [Google Scholar] [CrossRef]
- Appelbaum, T.; Murgiano, L.; Becker, D.; Santana, E.; Aguirre, G.D. Candidate Genetic Modifiers for RPGR Retinal Degeneration. Investig. Ophthalmol. Vis. Sci. 2020, 61, 20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).