Insight into the Natural History of Pathogenic Variant c.919-2A>G in the SLC26A4 Gene Involved in Hearing Loss: The Evidence for Its Common Origin in Southern Siberia (Russia)
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Ethics Statement
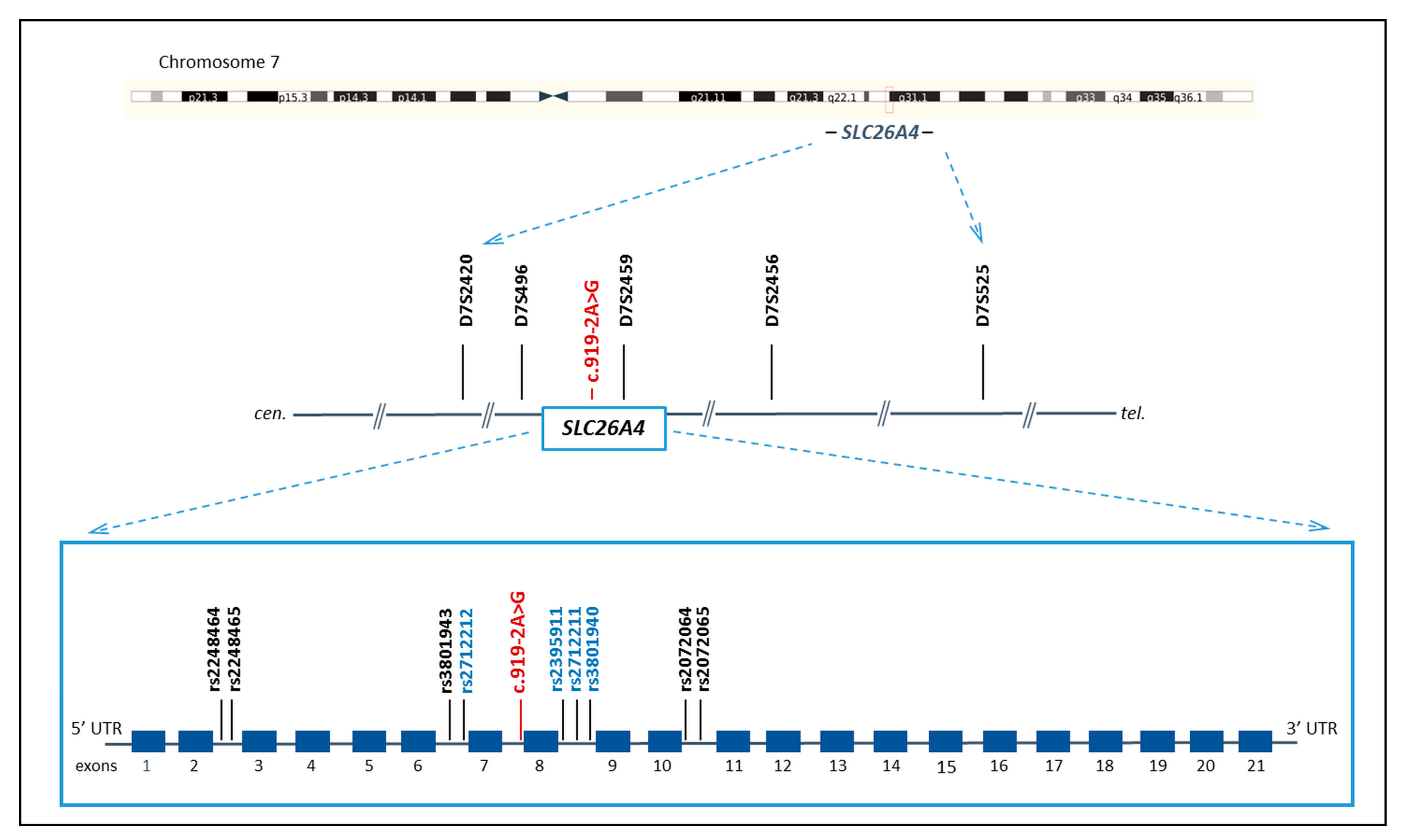
2.3. STRs and SNPs Genotyping
2.4. Reconstruction of STR and SNP Haplotypes
2.5. Estimation of c.919-2A>G Age
2.6. Statistical Analysis
3. Results
3.1. STR and SNP Haplotypes
3.2. Estimation of c.919-2A>G Age
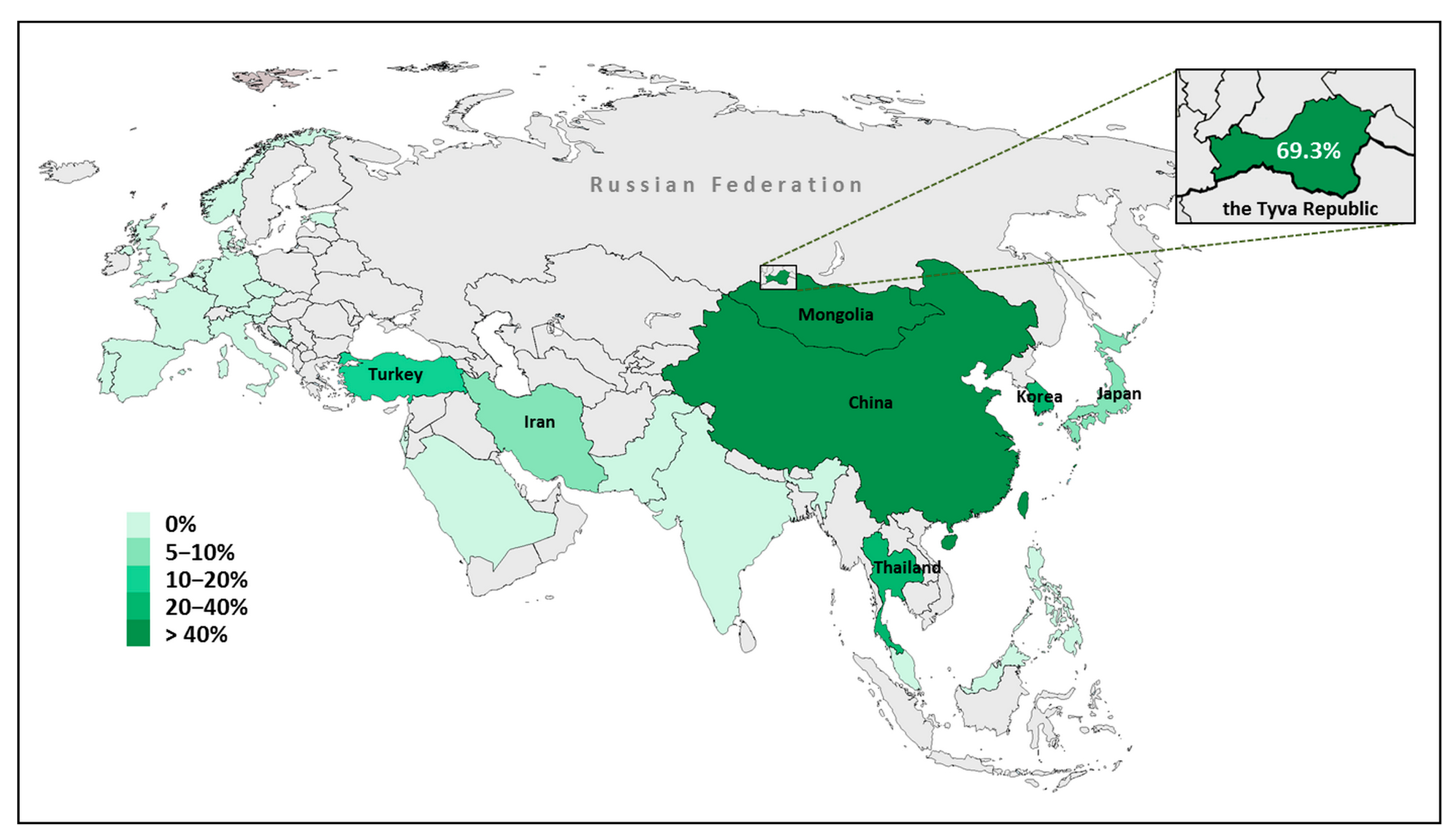
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Everett, L.A.; Glaser, B.; Beck, J.C.; Idol, J.R.; Buchs, A.; Heyman, M.; Adawi, F.; Hazani, E.; Nassir, E.; Baxevanis, A.D.; et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat. Genet. 1997, 17, 411–422. [Google Scholar] [CrossRef]
- Everett, L.A.; Morsli, H.; Wu, D.K.; Green, E.D. Expression pattern of the mouse ortholog of the Pendred’s syndrome gene (Pds) suggests a key role for pendrin in the inner ear. Proc. Natl. Acad. Sci. USA 1999, 96, 9727–9732. [Google Scholar] [CrossRef]
- Honda, K.; Griffith, A.J. Genetic architecture and phenotypic landscape of SLC26A4-related hearing loss. Hum. Genet. 2022, 141, 455–464. [Google Scholar] [CrossRef]
- Albert, S.; Blons, H.; Jonard, L.; Feldmann, D.; Chauvin, P.; Loundon, N.; Sergent-Allaoui, A.; Houang, M.; Joannard, A.; Schmerber, S.; et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. Eur. J. Hum. Genet. 2006, 14, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Guo, Y.; Wang, C.; Wang, Y.; Liu, X. A systematic review and meta-analysis of common mutations of SLC26A4 gene in Asian populations. Int. J. Pediatr. Otorhinolaryngol. 2013, 77, 1670–1676. [Google Scholar] [CrossRef] [PubMed]
- Miyagawa, M.; Nishio, S.Y.; Usami, S.; Deafness Gene Study Consortium. Mutation spectrum and genotype-phenotype correlation of hearing loss patients caused by SLC26A4 mutations in the Japanese: A large cohort study. J. Hum. Genet. 2014, 59, 262–268. [Google Scholar] [CrossRef]
- Lu, Y.J.; Yao, J.; Wei, Q.J.; Xing, G.Q.; Cao, X. Diagnostic Value of SLC26A4 Mutation Status in Hereditary Hearing Loss With EVA: A PRISMA-Compliant Meta-Analysis. Medicine 2015, 94, e2248. [Google Scholar] [CrossRef]
- Tsukada, K.; Nishio, S.Y.; Hattori, M.; Usami, S. Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: Their origin and a literature review. Ann. Otol. Rhinol. Laryngol. 2015, 124 (Suppl. S1), 61S–76S. [Google Scholar] [CrossRef]
- Koohiyan, M. A systematic review of SLC26A4 mutations causing hearing loss in the Iranian population. Int. J. Pediatr. Otorhinolaryngol. 2019, 125, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Naz, S. Molecular genetic landscape of hereditary hearing loss in Pakistan. Hum. Genet. 2022, 141, 633–648. [Google Scholar] [CrossRef]
- Park, H.J.; Shaukat, S.; Liu, X.Z.; Hahn, S.H.; Naz, S.; Ghosh, M.; Kim, H.N.; Moon, S.K.; Abe, S.; Tukamoto, K.; et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: Global implications for the epidemiology of deafness. J. Med. Genet. 2003, 40, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Yeh, T.H.; Chen, P.J.; Hsu, C.J. Prevalent SLC26A4 mutations in patients with enlarged vestibular aqueduct and/or Mondini dysplasia: A unique spectrum of mutations in Taiwan, including a frequent founder mutation. Laryngoscope 2005, 115, 1060–1064. [Google Scholar] [CrossRef]
- Borck, G.; Roth, C.; Martiné, U.; Wildhardt, G.; Pohlenz, J. Mutations in the PDS gene in German families with Pendred’s syndrome: V138F is a founder mutation. J. Clin. Endocrinol. Metab. 2003, 88, 2916–2921. [Google Scholar] [CrossRef]
- Pera, A.; Dossena, S.; Rodighiero, S.; Gandía, M.; Bottà, G.; Meyer, G.; Moreno, F.; Nofziger, C.; Hernández-Chico, C.; Paulmichl, M. Functional assessment of allelic variants in the SLC26A4 gene involved in Pendred syndrome and nonsyndromic EVA. Proc. Natl. Acad. Sci. USA 2008, 105, 18608–18613. [Google Scholar] [CrossRef] [PubMed]
- Mohseni, M.; Honarpour, A.; Mozafari, R.; Davarnia, B.; Najmabadi, H.; Kahrizi, K. Identification of a founder mutation for Pendred syndrome in families from northwest Iran. Int. J. Pediatr. Otorhinolaryngol. 2014, 78, 1828–1832. [Google Scholar] [CrossRef]
- Anwar, S.; Riazuddin, S.; Ahmed, Z.M.; Tasneem, S.; Ateeq-ul-Jaleel; Khan, S.Y.; Griffith, A.J.; Friedman, T.B.; Riazuddin, S. SLC26A4 mutation spectrum associated with DFNB4 deafness and Pendred’s syndrome in Pakistanis. J. Hum. Genet. 2009, 54, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Coucke, P.J.; van Hauwe, P.; Everett, L.A.; Demirhan, O.; Kabakkaya, Y.; Dietrich, N.L.; Smith, R.J.; Coyle, E.; Reardon, W.; Trembath, R.; et al. Identification of two different mutations in the PDS gene in an inbred family with Pendred syndrome. J. Med. Genet. 1999, 36, 475–477. [Google Scholar] [PubMed]
- Yang, J.J.; Tsai, C.C.; Hsu, H.M.; Shiao, J.Y.; Su, C.C.; Li, S.Y. Hearing loss associated with enlarged vestibular aqueduct and Mondini dysplasia is caused by splice-site mutation in the PDS gene. Hear Res. 2005, 199, 22–30. [Google Scholar] [CrossRef]
- Tsukamoto, K.; Suzuki, H.; Harada, D.; Namba, A.; Abe, S.; Usami, S. Distribution and frequencies of PDS (SLC26A4) mutations in Pendred syndrome and nonsyndromic hearing loss associated with enlarged vestibular aqueduct: A unique spectrum of mutations in Japanese. Eur. J. Hum. Genet. 2003, 11, 916–922. [Google Scholar] [CrossRef]
- Wang, Q.J.; Zhao, Y.L.; Rao, S.Q.; Guo, Y.F.; Yuan, H.; Zong, L.; Guan, J.; Xu, B.C.; Wang, D.Y.; Han, M.K.; et al. A distinct spectrum of SLC26A4 mutations in patients with enlarged vestibular aqueduct in China. Clin. Genet. 2007, 72, 245–254. [Google Scholar] [CrossRef]
- Dai, P.; Li, Q.; Huang, D.; Yuan, Y.; Kang, D.; Miller, D.T.; Shao, H.; Zhu, Q.; He, J.; Yu, F.; et al. SLC26A4 c.919-2A>G varies among Chinese ethnic groups as a cause of hearing loss. Genet. Med. 2008, 10, 586–592. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Bai-Cheng, X.; Chen, X.J.; Pan-Pan, B.; Jian-Li, M.; Xiao-Wen, L.; Zhang, Z.W.; Wan, D.; Zhu, Y.M.; Guo, Y.F. Common molecular etiology of patients with nonsyndromic hearing loss in Tibetan, Tu nationality, and Mongolian patients in the northwest of China. Acta Otolaryngol. 2013, 133, 930–934. [Google Scholar] [CrossRef]
- Lee, H.J.; Jung, J.; Shin, J.W.; Song, M.H.; Kim, S.H.; Lee, J.H.; Lee, K.A.; Shin, S.; Kim, U.K.; Bok, J.; et al. Correlation between genotype and phenotype in patients with bi-allelic SLC26A4 mutations. Clin. Genet. 2014, 86, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ao, L.; Ding, H.; Zhang, D. Genetic frequencies related to severe or profound sensorineural hearing loss in Inner Mongolia Autonomous Region. Genet. Mol. Biol. 2016, 39, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Erdenechuluun, J.; Lin, Y.-H.; Ganbat, K.; Bataakhuu, D.; Makhbal, Z.; Tsai, C.-Y.; Lin, Y.-H.; Chan, Y.-H.; Hsu, C.-J.; Hsu, W.-C.; et al. Unique spectra of deafness-associated mutations in Mongolians provide insights into the genetic relationships among Eurasian populations. PLoS ONE 2018, 13, e0209797. [Google Scholar] [CrossRef]
- Wu, C.C.; Tsai, C.Y.; Lin, Y.H.; Chen, P.Y.; Lin, P.H.; Cheng, Y.F.; Wu, C.M.; Lin, Y.H.; Lee, C.Y.; Erdenechuluun, J.; et al. Genetic Epidemiology and Clinical Features of Hereditary Hearing Impairment in the Taiwanese Population. Genes 2019, 10, 772. [Google Scholar] [CrossRef]
- Danilchenko, V.Y.; Zytsar, M.V.; Maslova, E.A.; Bady-Khoo, M.S.; Barashkov, N.A.; Morozov, I.V.; Bondar, A.A.; Posukh, O.L. Different Rates of the SLC26A4-Related Hearing Loss in Two Indigenous Peoples of Southern Siberia (Russia). Diagnostics 2021, 11, 2378. [Google Scholar] [CrossRef]
- Baldwin, C.T.; Weiss, S.; Farrer, L.A.; De Stefano, A.L.; Adair, R.; Franklyn, B.; Kidd, K.K.; Korostishevsky, M.; Bonné-Tamir, B. Linkage of congenital, recessive deafness (DFNB4) to chromosome 7q31 and evidence for genetic heterogeneity in the Middle Eastern Druze population. Hum. Mol. Genet. 1995, 4, 1637–1642. [Google Scholar] [CrossRef]
- Coucke, P.; Van Camp, G.; Demirhan, O.; Kabakkaya, Y.; Balemans, W.; Van Hauwe, P.; Van Agtmael, T.; Smith, R.J.; Parving, A.; Bolder, C.H.; et al. The gene for Pendred syndrome is located between D7S501 and D7S692 in a 1.7-cM region on chromosome 7q. Genomics 1997, 40, 48–54. [Google Scholar] [CrossRef]
- Gausden, E.; Coyle, B.; Armour, J.A.; Coffey, R.; Grossman, A.; Fraser, G.R.; Winter, R.M.; Pembrey, M.E.; Kendall-Taylor, P.; Stephens, D.; et al. Pendred syndrome: Evidence for genetic homogeneity and further refinement of linkage. J. Med. Genet. 1997, 34, 126–129. [Google Scholar] [CrossRef]
- López-Bigas, N.; Rabionet, R.; de Cid, R.; Govea, N.; Gasparini, P.; Zelante, L.; Arbonés, M.L.; Estivill, X. Splice-site mutation in the PDS gene may result in intrafamilial variability for deafness in Pendred syndrome. Hum. Mutat. 1999, 14, 520–526. [Google Scholar] [CrossRef]
- Gonzalez Trevino, O.; Karamanoglu Arseven, O.; Ceballos, C.J.; Vives, V.I.; Ramirez, R.C.; Gomez, V.V.; Medeiros-Neto, G.; Kopp, P. Clinical and molecular analysis of three Mexican families with Pendred’s syndrome. Eur. J. Endocrinol. 2001, 144, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Yazdanpanahi, N.; Tabatabaiefar, M.A.; Bagheri, N.; Azadegan Dehkordi, F.; Farrokhi, E.; Hashemzadeh Chaleshtori, M. The role and spectrum of SLC26A4 mutations in Iranian patients with autosomal recessive hereditary deafness. Int. J. Audiol. 2015, 54, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Bengtsson, B.O.; Thomson, G. Measuring the strength of associations between HLA antigens and diseases. Tissue Antigens 1981, 18, 356–363. [Google Scholar] [CrossRef]
- Rannala, B.; Bertorelle, G. Using linked markers to infer the age of a mutation. Hum. Mutat. 2001, 18, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Risch, N.; de Leon, D.; Ozelius, L.; Kramer, P.; Almasy, L.; Singer, B.; Fahn, S.; Breakefield, X.; Bressman, S. Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat. Genet. 1995, 9, 152–159. [Google Scholar] [CrossRef]
- Nonose, R.W.; Lezirovitz, K.; de Mello Auricchio, M.T.B.; Batissoco, A.C.; Yamamoto, G.L.; Mingroni-Netto, R.C. Mutation analysis of SLC26A4 (Pendrin) gene in a Brazilian sample of hearing-impaired subjects. BMC Med. Genet. 2018, 19, 73. [Google Scholar] [CrossRef]
- Pryor, S.P.; Madeo, A.C.; Reynolds, J.C.; Sarlis, N.J.; Arnos, K.S.; Nance, W.E.; Yang, Y.; Zalewski, C.K.; Brewer, C.C.; Butman, J.A.; et al. SLC26A4/PDS genotype-phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): Evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J. Med. Genet. 2005, 42, 159–165. [Google Scholar] [CrossRef]
- Dai, P.; Stewart, A.K.; Chebib, F.; Hsu, A.; Rozenfeld, J.; Huang, D.; Kang, D.; Lip, V.; Fang, H.; Shao, H.; et al. Distinct and novel SLC26A4/Pendrin mutations in Chinese and U.S. patients with nonsyndromic hearing loss. Physiol. Genom. 2009, 38, 281–290. [Google Scholar] [CrossRef]
- Dahl, H.H.; Ching, T.Y.; Hutchison, W.; Hou, S.; Seeto, M.; Sjahalam-King, J. Etiology and audiological outcomes at 3 years for 364 children in Australia. PLoS ONE 2013, 8, e59624. [Google Scholar] [CrossRef] [PubMed]
- de Moraes, V.C.; dos Santos, N.Z.; Ramos, P.Z.; Svidnicki, M.C.; Castilho, A.M.; Sartorato, E.L. Molecular analysis of SLC26A4 gene in patients with nonsyndromic hearing loss and EVA: Identification of two novel mutations in Brazilian patients. Int. J. Pediatr. Otorhinolaryngol. 2013, 77, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, S.D.C.E.S.; Grangeiro, C.H.P.; Picanço-Albuquerque, C.G.; Dos Anjos, T.O.; De Molfetta, G.A.; Silva, W.A., Jr.; Ferraz, V.E.F. Contribution of SLC26A4 to the molecular diagnosis of nonsyndromic prelingual sensorineural hearing loss in a Brazilian cohort. BMC Res. Notes 2018, 11, 546. [Google Scholar] [CrossRef] [PubMed]
- Talbi, S.; Bonnet, C.; Riahi, Z.; Boudjenah, F.; Dahmani, M.; Hardelin, J.P.; Wong Jun Tai, F.; Louha, M.; Ammar-Khodja, F.; Petit, C. Genetic heterogeneity of congenital hearing impairment in Algerians from the Ghardaïa province. Int. J. Pediatr. Otorhinolaryngol. 2018, 112, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Torre-González, C.; Villanueva-García, D.; García-Delgado, C.; Castillo-Castillo, S.; Huante-Guido, M.; Chichitz-Madrigal, J.; Juárez-Torres, M.E.; Sánchez-Sandoval, A.L.; Barrón-Palma, E.V.; Morán-Barroso, V.F. Congenital hearing loss: A literature review of the genetic etiology in a Mexican population. Bol. Med. Hosp. Infant. Mex. 2022, 79, 206–214. [Google Scholar] [CrossRef]
- Wonkam, A.; Adadey, S.M.; Schrauwen, I.; Aboagye, E.T.; Wonkam-Tingang, E.; Esoh, K.; Popel, K.; Manyisa, N.; Jonas, M.; deKock, C.; et al. Exome sequencing of families from Ghana reveals known and candidate hearing impairment genes. Commun. Biol. 2022, 5, 369. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Guo, W.; Tang, J.; Zhang, G.; Wang, G.; Han, M.; Zhang, X.; Yang, S.; He, D.Z.; Dai, P. Molecular epidemiology and functional assessment of novel allelic variants of SLC26A4 in non-syndromic hearing loss patients with enlarged vestibular aqueduct in China. PLoS ONE 2012, 7, e49984. [Google Scholar] [CrossRef] [PubMed]
- Dai, P.; Yuan, Y.; Huang, D.; Zhu, X.; Yu, F.; Kang, D.; Yuan, H.; Wu, B.; Han, D.; Wong, L.J. Molecular etiology of hearing impairment in Inner Mongolia: Mutations in SLC26A4 gene and relevant phenotype analysis. J. Transl. Med. 2008, 6, 74. [Google Scholar] [CrossRef]
- Chai, Y.; Huang, Z.; Tao, Z.; Li, X.; Li, L.; Li, Y.; Wu, H.; Yang, T. Molecular etiology of hearing impairment associated with nonsyndromic enlarged vestibular aqueduct in East China. Am. J. Med. Genet. A 2013, 161A, 2226–2233. [Google Scholar] [CrossRef]
- Xin, F.; Yuan, Y.; Deng, X.; Han, M.; Wang, G.; Zhao, J.; Gao, X.; Liu, J.; Yu, F.; Han, D.; et al. Genetic mutations in nonsyndromic deafness patients of Chinese minority and Han ethnicities in Yunnan, China. J. Transl. Med. 2013, 11, 312. [Google Scholar] [CrossRef]
- Chen, K.; Zong, L.; Liu, M.; Wang, X.; Zhou, W.; Zhan, Y.; Cao, H.; Dong, C.; Tang, H.; Jiang, H. Developing regional genetic counseling for southern Chinese with nonsyndromic hearing impairment: A unique mutational spectrum. J. Transl. Med. 2014, 12, 64. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Wang, Q.; Zhu, Y.; Wang, Y.; Guo, Y. Associations between GJB2, mitochondrial 12S rRNA, SLC26A4 mutations, and hearing loss among three ethnicities. Biomed. Res. Int. 2014, 2014, 746838. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.H.; Zhu, Y.M.; Wang, Y.L.; Guo, Y.F. Common molecular etiology of nonsyndromic hearing loss in 484 patients of 3 ethnicities in northwest China. Acta Otolaryngol. 2015, 135, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Han, M.; Wang, G.; Huang, S.; Zeng, J.; Yuan, Y.; Dai, P. Genetic mutations in non-syndromic deafness patients in Hainan Province have a different mutational spectrum compared to patients from Mainland China. Int. J. Pediatr. Otorhinolaryngol. 2018, 108, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, Y.; Wu, J.; Chen, S.; Tang, S.; Jiang, Y.; Dai, P. Study on the relationship between the pathogenic mutations of SLC26A4 and CT phenotypes of inner ear in patient with sensorineural hearing loss. Biosci. Rep. 2019, 39, BSR20182241. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, S.J.; Jin, H.S.; Lee, J.O.; Go, S.H.; Jang, H.S.; Moon, S.K.; Lee, S.C.; Chun, Y.M.; Lee, H.K.; et al. Genetic basis of hearing loss associated with enlarged vestibular aqueducts in Koreans. Clin. Genet. 2005, 67, 160–165. [Google Scholar] [CrossRef]
- Rah, Y.C.; Kim, A.R.; Koo, J.W.; Lee, J.H.; Oh, S.H.; Choi, B.Y. Audiologic presentation of enlargement of the vestibular aqueduct according to the SLC26A4 genotypes. Laryngoscope 2015, 125, E216–E222. [Google Scholar] [CrossRef]
- Snabboon, T.; Plengpanich, W.; Saengpanich, S.; Sirisalipoch, S.; Keelawat, S.; Sunthornyothin, S.; Khovidhunkit, W.; Suwanwalaikorn, S.; Sridama, V.; Shotelersuk, V. Two common and three novel PDS mutations in Thai patients with Pendred syndrome. J. Endocrinol. Investig. 2007, 30, 907–913. [Google Scholar] [CrossRef]
- Cengiz, F.B.; Yilmazer, R.; Olgun, L.; Sennaroglu, L.; Kirazli, T.; Alper, H.; Olgun, Y.; Incesulu, A.; Atik, T.; Huesca-Hernandez, F.; et al. Novel pathogenic variants underlie SLC26A4-related hearing loss in a multiethnic cohort. Int. J. Pediatr. Otorhinolaryngol. 2017, 101, 167–171. [Google Scholar] [CrossRef]
- Reiisi, S.; Sanati, M.H.; Tabatabaiefar, M.A.; Ahmadian, S.; Reiisi, S.; Parchami, S.; Porjafari, H.; Shahi, H.; Shavarzi, A.; Hashemzade Chaleshtori, M. The Study of SLC26A4 Gene Causing Autosomal Recessive Hearing Loss by Linkage Analysis in a Cohort of Iranian Populations. Int. J. Mol. Cell. Med. 2014, 3, 176–182. [Google Scholar]
- van Hauwe, P.; Everett, L.A.; Coucke, P.; Scott, D.A.; Kraft, M.L.; Ris-Stalpers, C.; Bolder, C.; Otten, B.; de Vijlder, J.J.; Dietrich, N.L.; et al. Two frequent missense mutations in Pendred syndrome. Hum. Mol. Genet. 1998, 7, 1099–1104. [Google Scholar] [CrossRef] [PubMed]
- Rendtorff, N.D.; Schrijver, I.; Lodahl, M.; Rodriguez-Paris, J.; Johnsen, T.; Hansén, E.C.; Nickelsen, L.A.; Tümer, Z.; Fagerheim, T.; Wetke, R.; et al. SLC26A4 mutation frequency and spectrum in 109 Danish Pendred syndrome/DFNB4 probands and a report of nine novel mutations. Clin. Genet. 2013, 84, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Azadegan-Dehkordi, F.; Ahmadi, R.; Bahrami, T.; Yazdanpanahi, N.; Farrokhi, E.; Tabatabaiefar, M.A.; Hashemzadeh-Chaleshtori, M. A novel variant of SLC26A4 and first report of the c.716T>A variant in Iranian pedigrees with non-syndromic sensorineural hearing loss. Am. J. Otolaryngol. 2018, 39, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Mojtabavi Naeini, M.; Mesrian Tanha, H.; Hashemzadeh Chaleshtori, M.; Vallian, S. Genotyping data and novel haplotype diversity of STR markers in the SLC26A4 gene region in five ethnic groups of the Iranian population. Genet. Test. Mol. Biomarkers 2014, 18, 820–825. [Google Scholar] [CrossRef]
- Slatkin, M.; Rannala, B. Estimating allele age. Annu. Rev. Genom. Hum. Genet. 2000, 1, 225–249. [Google Scholar] [CrossRef]
- Labuda, D.; Zietkiewicz, E.; Labuda, M. The genetic clock and the age of the founder effect in growing populations: A lesson from French Canadians and Ashkenazim. Am. J. Hum. Genet. 1997, 61, 768–771. [Google Scholar] [CrossRef]
- Colombo, R. Age estimate of the N370S mutation causing Gaucher disease in Ashkenazi Jews and European populations: A reappraisal of haplotype data. Am. J. Hum. Genet. 2000, 66, 692–697. [Google Scholar] [CrossRef]
- Mongush, M.V. Tuvans of Mongolia and China. Int. J. Cent. Asian Stud. 1996, 1, 225–243. [Google Scholar]
- Chen, Z.; Zhang, Y.; Fan, A.; Zhang, Y.; Wu, Y.; Zhao, Q.; Zhou, Y.; Zhou, C.; Bawudong, M.; Mao, X.; et al. Brief communication: Y-chromosome haplogroup analysis indicates that Chinese Tuvans share distinctive affinity with Siberian Tuvans. Am. J. Phys. Anthropol. 2011, 144, 492–497. [Google Scholar] [CrossRef]
- Vainshtein, S.I.; Mannay-Ool, M.H. History of Tyva, 2nd ed.; Science: Novosibirsk, Russia, 2001. (In Russian) [Google Scholar]
- Mannai-ool, M.K. Tuvan People. The Origin and Formation of the Ethnos; Nauka Publ.: Novosibirsk, Russia, 2004; pp. 99–166. (In Russian) [Google Scholar]
| STR Haplotypes D7S2420-D7S496-/c.919-2A>G/-D7S2459-D7S2456-D7S525 (~2.8 Mb) | Frequency of Haplotypes | X2 | p | |
|---|---|---|---|---|
| Mutant Chromosomes | Normal Chromosomes | |||
| 278-120-147-244-227 | 0.9130 | 0.0 | 150 | <10−35 |
| 278-120-147-244-229 | 0.0435 | 0.0 | 2.4 | 0.0704 |
| 278-120-147-244-221 | 0.0217 | 0.0 | 0.28 | 0.2674 |
| 278-120-147-244-225 | 0.0217 | 0.0 | 0.28 | 0.2674 |
| Other haplotypes | 0.0 | 1.0 | - | - |
| SNP Haplotypes rs2248464-rs2248465-rs3801943-rs2712212*-/c.919-2A>G/-rs2395911*-rs2712211*-rs3801940*-rs2072064-rs2072065 (31.039 kb) | Frequency of Haplotypes | X2 | p | |
| Mutant Chromosomes | Normal Chromosomes | |||
| A-C-T-A-G-G-C-A-C | 1.0 | 0.0280 | 150 | <10−36 |
| Other haplotypes | 0.0 | 0.9720 | - | - |
| Genetic Markers Used for Calculations | d | The Single-Marker Method | The DMLE + Calculation | ||
|---|---|---|---|---|---|
| g | Age | g (95% CI) | Age (95% CI) | ||
| STR markers * | 0.05 | 22 | 550 years | 103–198 | 2575–4950 years |
| 0.1 | 21 | 525 years | 63–107 | 1575–2675 years | |
| 0.2 | 17 | 425 years | 35–59 | 875–1475 years | |
| SNP markers | 0.05 | - | - | 91–191 | 2275–4775 years |
| 0.1 | 53–103 | 1325–2575 years | |||
| 0.2 | 29–54 | 725–1350 years | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilchenko, V.Y.; Zytsar, M.V.; Maslova, E.A.; Orishchenko, K.E.; Posukh, O.L. Insight into the Natural History of Pathogenic Variant c.919-2A>G in the SLC26A4 Gene Involved in Hearing Loss: The Evidence for Its Common Origin in Southern Siberia (Russia). Genes 2023, 14, 928. https://doi.org/10.3390/genes14040928
Danilchenko VY, Zytsar MV, Maslova EA, Orishchenko KE, Posukh OL. Insight into the Natural History of Pathogenic Variant c.919-2A>G in the SLC26A4 Gene Involved in Hearing Loss: The Evidence for Its Common Origin in Southern Siberia (Russia). Genes. 2023; 14(4):928. https://doi.org/10.3390/genes14040928
Chicago/Turabian StyleDanilchenko, Valeriia Yu., Marina V. Zytsar, Ekaterina A. Maslova, Konstantin E. Orishchenko, and Olga L. Posukh. 2023. "Insight into the Natural History of Pathogenic Variant c.919-2A>G in the SLC26A4 Gene Involved in Hearing Loss: The Evidence for Its Common Origin in Southern Siberia (Russia)" Genes 14, no. 4: 928. https://doi.org/10.3390/genes14040928
APA StyleDanilchenko, V. Y., Zytsar, M. V., Maslova, E. A., Orishchenko, K. E., & Posukh, O. L. (2023). Insight into the Natural History of Pathogenic Variant c.919-2A>G in the SLC26A4 Gene Involved in Hearing Loss: The Evidence for Its Common Origin in Southern Siberia (Russia). Genes, 14(4), 928. https://doi.org/10.3390/genes14040928






