Genome–Transcriptome Transition Approaches to Characterize Anthocyanin Biosynthesis Pathway Genes in Blue, Black and Purple Wheat
Abstract
1. Introduction
2. Results
2.1. In Silico Identification of Anthocyanin Biosynthesis Pathway Genes in Wheat
2.2. Physiochemical Properties and Domain Analysis of Putative Anthocyanin Biosynthetic Genes in Wheat
2.3. Comparative Transcriptomic Analysis of the Putative Anthocyanin Biosynthetic Pathway Genes in Colored Wheat
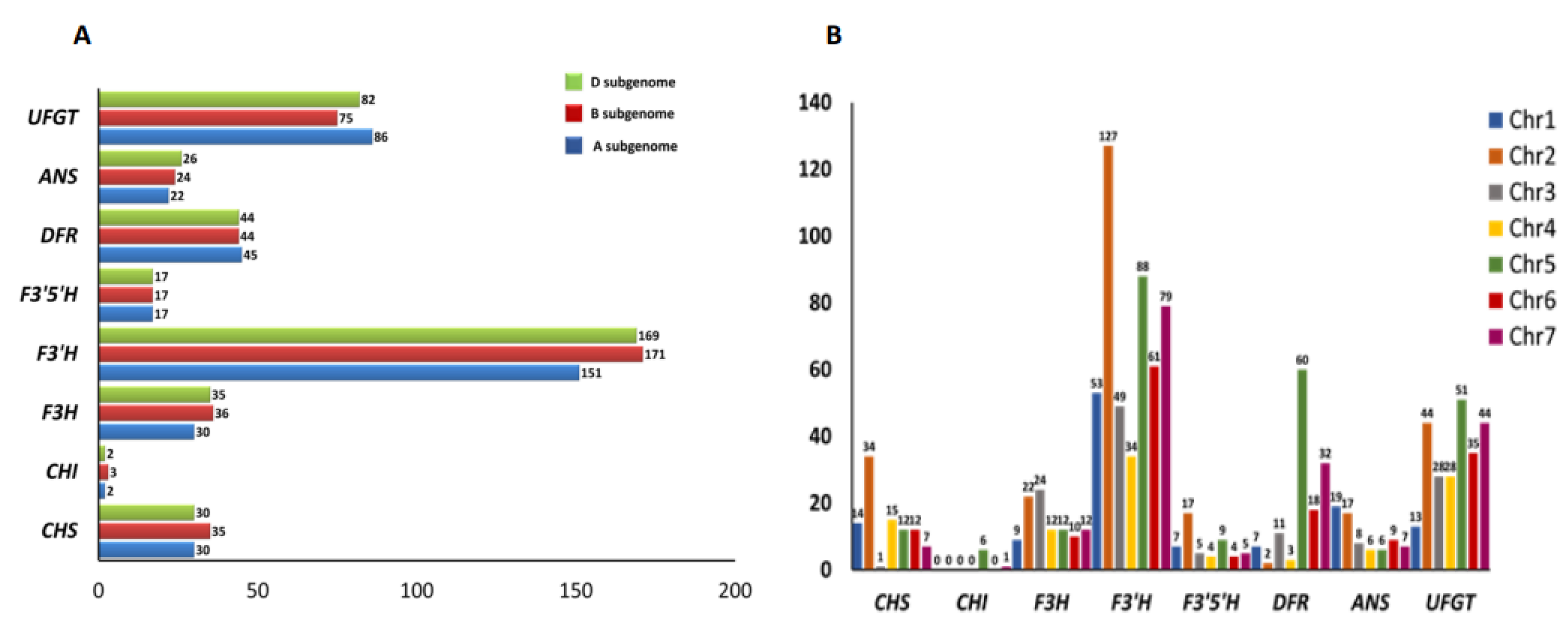
2.4. Genomic Distribution and Chromosomal Localization of the Putative Anthocyanin Biosynthesis Pathway Genes in Wheat
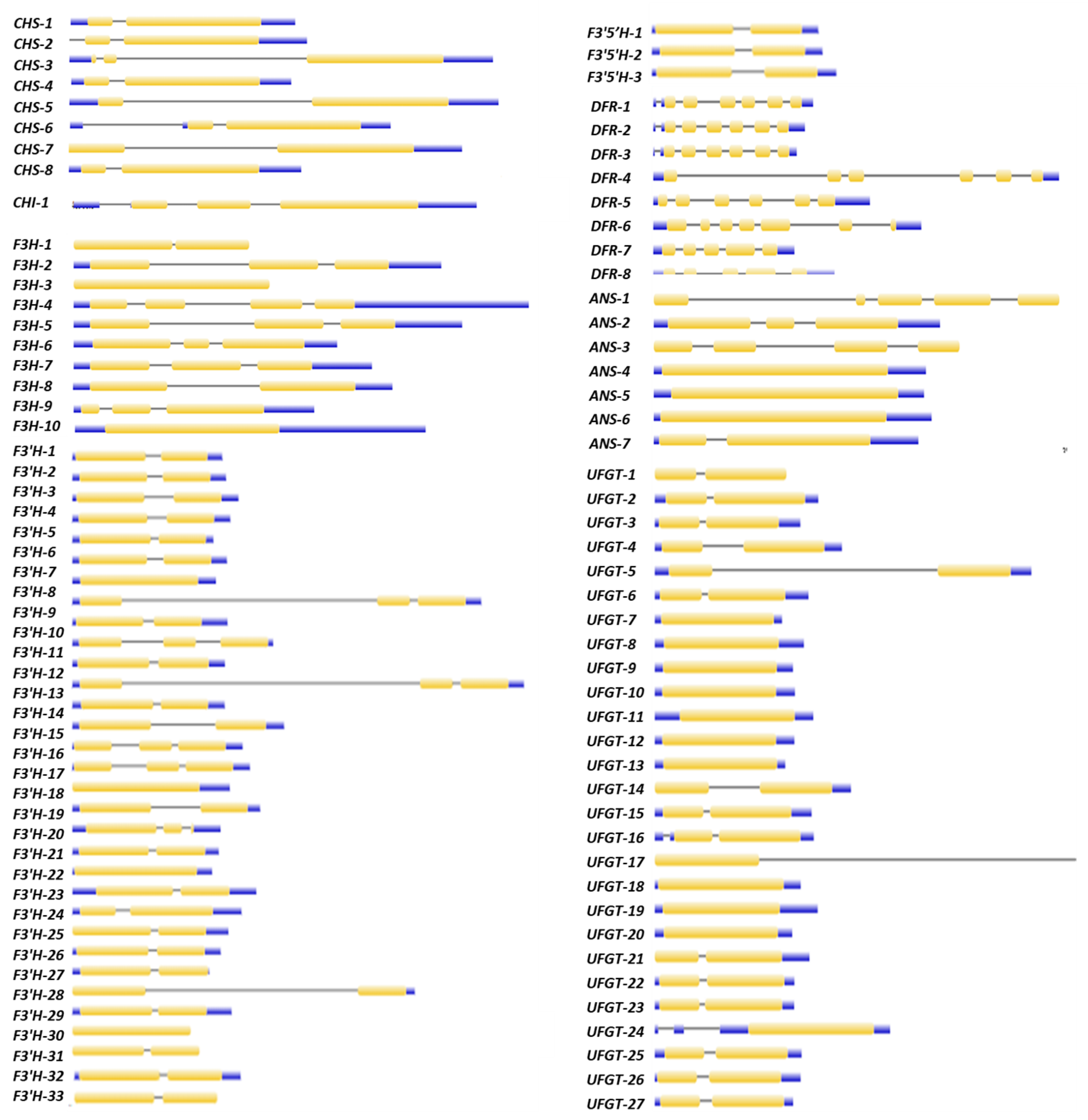
2.5. Gene Structure and Motif Analysis of Differentially Expressed Putative Anthocyanin Biosynthesis Pathway Genes
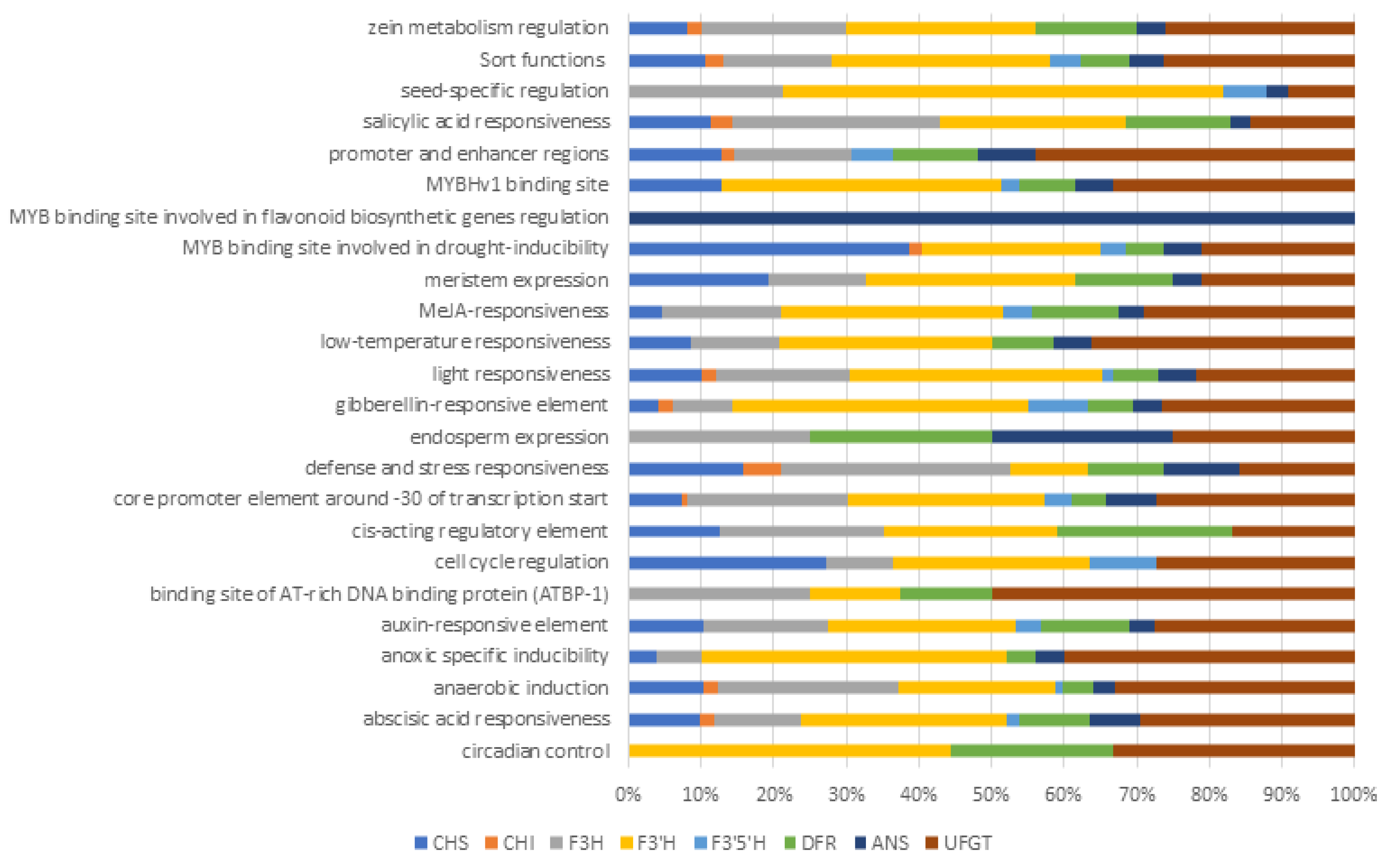
2.6. Cis-Acting Regulatory Elements (CAREs)
2.7. Phylogenetic and Homolog Analysis
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification and Distribution of Anthocyanin Biosynthetic Genes in Wheat
4.2. Comparative Transcriptome Data Analysis of Anthocyanin Biosynthesis Related Genes in Color Wheat
4.2.1. Plant Material
4.2.2. RNA Isolation and RNA-Seq Analysis
4.2.3. Characterization of Identified Anthocyanin Biosynthetic Genes
4.2.4. Gene Motif, and Cis-Acting Regulatory Elements (CAREs) Identification
4.2.5. Phylogenetic and Homology Analysis
4.2.6. qRT-PCR Validation of Anthocyanin Biosynthetic Genes during Seed Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samanta, A.; Das, G.; Das, S.K. Roles of flavonoids in plants. Carbon N. Y. 2011, 100, 12–35. [Google Scholar]
- Winkel-Shirley, B. Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef]
- Yadav, V.; Wang, Z.; Wei, C.; Amo, A.; Ahmed, B.; Yang, X.; Zhang, X. Phenylpropanoid Pathway Engineering: An Emerging Approach towards Plant Defense. Pathogens 2020, 9, 312. [Google Scholar] [CrossRef]
- Jaakola, L.; Maatta, K.; Pirttila, A.M.; Torronen, R.; Karenlampi, S.; Hohtola, A. Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Physiol. 2002, 130, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Wang, Y.; Yang, S.; Xu, Y.; Chen, X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 2010, 232, 245–255. [Google Scholar] [CrossRef]
- Chen, S.M.; Li, C.H.; Zhu, X.R.; Deng, Y.M.; Sun, W.; Wang, L.S.; Chen, F.-D.; Zhang, Z. The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol. Plant. 2012, 56, 458–464. [Google Scholar] [CrossRef]
- Tanaka, Y.; Brugliera, F. Flower colour and cytochromes P450. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120432. [Google Scholar] [CrossRef] [PubMed]
- Havrlentová, M.; Pšenáková, I.; Žofajová, A.; Rückschloss, L.; Kraic, J. Anthocyanins in wheat seed—A mini review. Nova Biotechnol. Chim. 2014, 13, 1–12. [Google Scholar] [CrossRef]
- Jeewani, D.C.; Hua, W.Z. Recent advances in anthocyanin biosynthesis in colored wheat. Res. J. Biotechnol. 2017, 12, 6. [Google Scholar]
- Wu, S.; O’Leary, S.; Gleddie, S.; Eudes, F.; Laroche, A.; Robert, L. A chalcone synthase-like gene is highly expressed in the tapetum of both wheat (Triticum aestivum L.) and triticale (×Triticosecale Wittmack). Plant Cell Rep. 2008, 27, 1441–1449. [Google Scholar] [CrossRef]
- Li, W.L.; Faris, J.D.; Chittoor, J.M.; Leach, J.E.; Hulbert, S.H.; Liu, D.J.; Chen, P.D.; Gill, B.S. Genomic mapping of defense response genes in wheat. Theor. Appl. Genet. 1999, 98, 226–233. [Google Scholar] [CrossRef]
- Himi, E.; Maekawa, M.; Noda, K. Differential Expression of Three Flavanone 3-Hydroxylase Genes in Grains and Coleoptiles of Wheat. Int. J. Plant Genom. 2011, 2011, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Li, S.; Chen, W.; Zhang, B.; Liu, D.; Liu, B.; Zhang, H. Transcriptome analysis of purple pericarps in common wheat (Triticum aestivum L.). PLoS ONE 2016, 11, e0155428. [Google Scholar] [CrossRef]
- Himi, E.; Noda, K. Isolation and location of three homoeologous dihydroflavonol-4-reductase (DFR) genes of wheat and their tissue-dependent expression. J. Exp. Bot. 2004, 55, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.-S.; Wang, F.; Dong, Y.-X.; Zhang, X.-S. Expression Analysis of Dihydroflavonol 4-Reductase Genes Involved in Anthocyanin Biosynthesis in Purple Grains of Wheat. J. Integr. Plant Biol. 2005, 47, 1107–1114. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Gordeeva, E.I.; Khlestkina, E.K. The Regulation of Anthocyanin Synthesis in the Wheat Pericarp. Molecules 2014, 19, 20266–20279. [Google Scholar] [CrossRef]
- Yang, G.; Li, B.; Gao, J.; Liu, J.; Zhao, X.; Zheng, Q.; Tong, Y.; Li, Z. Cloning and expression of two chalcone synthase and a flavonoid 3’5’-Hydroxylase 3’-end cDNAs from developing seeds of blue-grained wheat involved in anthocyanin biosynthetic pathway. Acta Bot. Sin. Engl. Ed. 2004, 46, 588–594. [Google Scholar]
- Shih, C.H.; Chu, H.; Tang, L.K.; Sakamoto, W.; Maekawa, M.; Chu, I.K.; Wang, M.; Lo, C. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 2008, 228, 1043–1054. [Google Scholar] [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef]
- Zhang, D.-W.; Liu, L.-L.; Zhou, D.-G.; Liu, X.-J.; Liu, Z.-S.; Yan, M.-L. Genome-wide identification and expression analysis of anthocyanin biosynthetic genes in Brassica juncea. J. Integr. Agric. 2020, 19, 1250–1260. [Google Scholar] [CrossRef]
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Stein, N.; Choulet, F. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef]
- Dhaliwal, H.; Chhuneja, P.; Singh, I.; Ghai, M.; Goel, R.; Garg, M.; Keller, B.; Röder, M.; Singh, K. Triticum monococcum—A novel source for transfer and exploitation of disease resistance in wheat. In Proceedings of the 10th International Wheat Genetics Symposium, Paestum, Italy, 1–6 September 2003; pp. 346–349. [Google Scholar]
- Garg, M.; Rao, Y.; Goyal, A.; Singh, B. Variations in Seed Storage Protein-Triticin among Diploid Triticum and Aegilops Species. Biotechnology 2007, 6, 444–446. [Google Scholar] [CrossRef]
- Abdel-Aal, E.-S.M.; Young, J.C.; Rabalski, I. Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food Chem. 2006, 54, 4696–4704. [Google Scholar] [CrossRef]
- Garg, M.; Chawla, M.; Chunduri, V.; Kumar, R.; Sharma, S.; Sharma, N.K.; Kaur, N.; Kumar, A.; Mundey, J.K.; Saini, M.K.; et al. Transfer of grain colors to elite wheat cultivars and their characterization. J. Cereal Sci. 2016, 71, 138–144. [Google Scholar] [CrossRef]
- Tikhonova, M.; Shoeva, O.; Tenditnik, M.; Ovsyukova, M.; Akopyan, A.; Dubrovina, N.; Amstislavskaya, T.; Khlestkina, E. Evaluating the Effects of Grain of Isogenic Wheat Lines Differing in the Content of Anthocyanins in Mouse Models of Neurodegenerative Disorders. Nutrients 2020, 12, 3877. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Kumar, N.; Kumar, S.; Mwaurah, P.W.; Panghal, A.; Attkan, A.K.; Singh, V.K.; Garg, M.K. Bioactive compounds, nutritional benefits and food applications of colored wheat: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2021, 61, 3197–3210. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, P.; Kumari, A.; Sheoran, B.; Sharma, S.; Kaur, S.; Bhunia, R.K.; Rajarammohan, S.; Bishnoi, M.; Kondepudi, K.K.; Garg, M. Anthocyanin biofortified colored wheat modifies gut microbiota in mice. J. Cereal Sci. 2022, 104, 103433. [Google Scholar] [CrossRef]
- Lepiniec, L.; Debeaujon, I.; Routaboul, J.-M.; Baudry, A.; Pourcel, L.; Nesi, N.; Caboche, M. Genetics and Biochemistry of Seed Flavonoids. Annu. Rev. Plant Biol. 2006, 57, 405–430. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome Evolution Due to Allopolyploidization in Wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10–21. [Google Scholar] [CrossRef]
- Deng, X.; Bashandy, H.; Ainasoja, M.; Kontturi, J.; Pietiäinen, M.; Laitinen, R.A.E.; Albert, V.A.; Valkonen, J.P.T.; Elomaa, P.; Teeri, T.H. Functional diversification of duplicated chalcone synthase genes in anthocyanin biosynthesis of Gerbera hybrida. New Phytol. 2014, 201, 1469–1483. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Ding, T.; Su, B.; Jiang, H. Genome-Wide Identification, Characterization and Expression Analysis of the Chalcone Synthase Family in Maize. Int. J. Mol. Sci. 2016, 17, 161. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; He, H.; Zhu, C.; Peng, X.; Fu, J.; He, X.; Chen, X.; Ouyang, L.; Bian, J.; Liu, S. Genome-wide identification and phylogenetic analysis of the chalcone synthase gene family in rice. J. Plant Res. 2016, 130, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Vadivel, A.K.A.; Krysiak, K.; Tian, G.; Dhaubhadel, S. Genome-wide identification and localization of chalcone synthase family in soybean (Glycine max [L]Merr). BMC Plant Biol. 2018, 18, 1–13. [Google Scholar] [CrossRef]
- Tsai, C.; Harding, S.A.; Tschaplinski, T.J.; Lindroth, R.L.; Yuan, Y. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol. 2006, 172, 47–62. [Google Scholar] [CrossRef]
- Sun, W.; Meng, X.; Liang, L.; Jiang, W.; Huang, Y.; He, J.; Hu, H.; Almqvist, J.; Gao, X.; Wang, L. Molecular and Biochemical Analysis of Chalcone Synthase from Freesia hybrid in Flavonoid Biosynthetic Pathway. PLoS ONE 2015, 10, e0119054. [Google Scholar] [CrossRef]
- Deng, Y.; Li, C.; Li, H.; Lu, S. Identification and Characterization of Flavonoid Biosynthetic Enzyme Genes in Salvia miltiorrhiza (Lamiaceae). Molecules 2018, 23, 1467. [Google Scholar] [CrossRef]
- Abe, I.; Morita, H. Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases. Nat. Prod. Rep. 2010, 27, 809–838. [Google Scholar] [CrossRef]
- Cheng, A.-X.; Han, X.-J.; Wu, Y.-F.; Lou, H.-X. The Function and Catalysis of 2-Oxoglutarate-Dependent Oxygenases Involved in Plant Flavonoid Biosynthesis. Int. J. Mol. Sci. 2014, 15, 1080–1095. [Google Scholar] [CrossRef]
- Vikhorev, A.V.; Strygina, K.V.; Khlestkina, E.K. Duplicated flavonoid 3’-hydroxylase and flavonoid 3’, 5’-hydroxylase genes in barley genome. PeerJ 2019, 7, e6266. [Google Scholar] [CrossRef]
- Johnson, E.; Ryu, S.; Yi, H.; Shin, B.; Cheong, H.; Choi, G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J. 2001, 25, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Petit, P.; Granier, T.; d’Estaintot, B.L.; Manigand, C.; Bathany, K.; Schmitter, J.M.; Lauvergeat, V.; Hamdi, S.; Gallois, B. Crystal structure of grape dihydroflavonol 4-reductase, a key enzyme in flavonoid biosynthesis. J. Mol. Biol. 2007, 368, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, X.; Wilson, I.W.; Shao, F.; Qiu, D. Identification of the Genes Involved in Anthocyanin Biosynthesis and Accumulation in Taxus chinensis. Genes 2019, 10, 982. [Google Scholar] [CrossRef]
- He, Y.; Ahmad, D.; Zhang, X.; Zhang, Y.; Wu, L.; Jiang, P.; Ma, H. Genome-wide analysis of family-1 UDP glycosyltransferases (UGT) and identification of UGT genes for FHB resistance in wheat (Triticum aestivum L.). BMC Plant Biol. 2018, 18, 1–20. [Google Scholar] [CrossRef]
- Sundaramoorthy, J.; Park, G.T.; Lee, J.-D.; Kim, J.H.; Seo, H.S.; Song, J.T. Genetic and molecular regulation of flower pigmentation in soybean. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 555–562. [Google Scholar] [CrossRef]
- Yang, K.; Jeong, N.; Moon, J.-K.; Lee, Y.-H.; Lee, S.-H.; Kim, H.M.; Hwang, C.H.; Back, K.; Palmer, R.G.; Jeong, S.-C. Genetic Analysis of Genes Controlling Natural Variation of Seed Coat and Flower Colors in Soybean. J. Hered. 2010, 101, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, P.; Wang, Y.; Dong, R.; Yu, H.; Hou, B. Genome-wide identification and phylogenetic analysis of Family-1 UDP glycosyltransferases in maize (Zea mays). Planta 2014, 239, 1265–1279. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.-Z.; Zhou, D.-R.; Ye, X.-F.; Jiang, C.-C.; Pan, S.-L. Identification of candidate anthocyanin-related genes by transcriptomic analysis of ‘Furongli’plum (Prunus salicina L.) during fruit ripening using RNA-seq. Front. Plant Sci. 2016, 7, 1338. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, S.; Cheng, Y.; Peng, Z.; Han, J. Transcriptome profiling of anthocyanin-related genes reveals effects of light intensity on anthocyanin biosynthesis in red leaf lettuce. PeerJ 2018, 6, e4607. [Google Scholar] [CrossRef]
- Otto, S.P.; Whitton, J. Polyploid incidence and evolution. Annu. Rev. Genet. 2000, 34, 401–437. [Google Scholar] [CrossRef]
- Heidari, P.; Puresmaeli, F.; Mora-Poblete, F. Genome-Wide Identification and Molecular Evolution of the Magnesium Transporter (MGT) Gene Family in Citrullus lanatus and Cucumis sativus. Agronomy 2022, 12, 2253. [Google Scholar] [CrossRef]
- Puresmaeli, F.; Heidari, P.; Lawson, S. Insights into the Sulfate Transporter Gene Family and Its Expression Patterns in Durum Wheat Seedlings under Salinity. Genes 2023, 14, 333. [Google Scholar] [CrossRef] [PubMed]
- Glagoleva, A.Y.; Ivanisenko, N.V.; Khlestkina, E.K. Organization and evolution of the chalcone synthase gene family in bread wheat and relative species. BMC Genet. 2019, 20, 30. [Google Scholar] [CrossRef]
- Richard, S.; Lapointe, G.; Rutledge, R.G.; Séguin, A. Induction of Chalcone Synthase Expression in White Spruce by Wounding and Jasmonate. Plant Cell Physiol. 2000, 41, 982–987. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Strid, A. Anthocyanin accumulation and changes in CHS and PR-5 gene expression in Arabidopsis thaliana after removal of the inflorescence stem (decapitation). Plant Physiol. Biochem. 2005, 43, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Shoeva, O.Y.; Khlestkina, E.K. The specific features of anthocyanin biosynthesis regulation in wheat. In Advances in Wheat Genetics: From Genome to Field: Proceedings of the 12th International Wheat Genetics Symposium; Springer: Berlin/Heidelberg, Germany, 2015; pp. 147–157. [Google Scholar]
- Khlestkina, E.K.; Salina, E.A.; Matthies, I.E.; Leonova, I.N.; Börner, A.; Röder, M.S. Comparative molecular marker-based genetic mapping of flavanone 3-hydroxylase genes in wheat, rye and barley. Euphytica 2010, 179, 333–341. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, A.; Singh, D.; Kumari, A.; Kapoor, P.; Kaur, S.; Shreon, B.; Garg, M. Integrated transcriptional and metabolomics signature pattern of pigmented wheat to insight the seed pigmentation and other associated features. Plant Physiol. Biochem. 2022, 189, 59–70. [Google Scholar] [CrossRef]
- Tanaka, Y.; Ohmiya, A. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Trainin, T.; Harel-Beja, R.; Bar-Ya’Akov, I.; Borochov-Neori, H.; Holland, D. A “White” Anthocyanin-less Pomegranate (Punica granatum L.) Caused by an Insertion in the Coding Region of the Leucoanthocyanidin Dioxygenase (LDOX; ANS) Gene. PLoS ONE 2015, 10, e0142777. [Google Scholar] [CrossRef]
- Rafique, M.Z.; Carvalho, E.; Stracke, R.; Palmieri, L.; Herrera, L.; Feller, A.; Malnoy, M.; Martens, S. Nonsense Mutation Inside Anthocyanidin Synthase Gene Controls Pigmentation in Yellow Raspberry (Rubus idaeus L.). Front. Plant Sci. 2016, 7, 1892. [Google Scholar] [CrossRef] [PubMed]
- Gonda, I.; Abu-Abied, M.; Adler, C.; Milavsky, R.; Tal, O.; Davidovich-Rikanati, R.; Faigenboim, A.; Kahane-Achinoam, T.; Shachter, A.; Chaimovitsh, D. Two independent loss-of-function mutations in anthocyanidin synthase homeologous genes make sweet basil all green. BioRxiv 2022, 2005–2022. [Google Scholar] [CrossRef]
- Lin, C.; Xing, P.; Jin, H.; Zhou, C.; Li, X.; Song, Z. Loss of anthocyanidin synthase gene is associated with white flowers of Salvia miltiorrhiza Bge. f. alba, a natural variant of S. miltiorrhiza. Planta 2022, 256, 15. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Maekawa, M.; Noda, K. Anthocyanin accumulation and expression pattern of anthocyanin biosynthesis genes in developing wheat coleoptiles. Biol. Plant. 2009, 53, 223–228. [Google Scholar] [CrossRef]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.; Liang, J.; Wang, X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014, 15, 1–11. [Google Scholar] [CrossRef]
- Kaur, S.; Tiwari, V.; Kumari, A.; Chaudhary, E.; Sharma, A.; Ali, U.; Garg, M. Protective and defensive role of anthocyanins under plant abiotic and biotic stresses: An emerging application in sustainable agriculture. J. Biotechnol. 2023, 361, 12–29. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhou, Y.; Yao, D.; Iqbal, S.; Gao, Z.; Zhang, Z. DNA methylation of LDOX gene contributes to the floral colour variegation in peach. J. Plant Physiol. 2020, 246–247, 153116. [Google Scholar] [CrossRef]
- Zhu, Q.; Yu, S.; Zeng, D.; Liu, H.; Wang, H.; Yang, Z.; Xie, X.; Shen, R.; Tan, J.; Li, H.; et al. Development of “Purple Endosperm Rice” by Engineering Anthocyanin Biosynthesis in the Endosperm with a High-Efficiency Transgene Stacking System. Mol. Plant 2017, 10, 918–929. [Google Scholar] [CrossRef]
- Garg, M. NABIMG-11-Black (BW/2* PBW621)(IC0620916; INGR17003), a Wheat (Triticum aestivum) Germplasm with Black grain colour; (purple pericarp + blue aleuron). Indian J. Plant Genet. Resour. 2018, 31, 334–335. [Google Scholar]
- Garg, M. NABIMG-10-Purple (BW/2* PBW621)(IC0620915; INGR17002), a Wheat (Triticum aestivum) Germplasm with purple grain (pericarp) Color. Indian J. Plant Genet. Resour. 2018, 31, 333–334. [Google Scholar]
- Garg, M. NABIMG-9-Blue; BW/2*/PBW621 (IC0620914; INGR17001), a Wheat (Triticum aestivum) Germplasm with blue grain (aleurone) Color. Indian J. Plant Genet. Resour. 2018, 31, 332–333. [Google Scholar]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.-J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34 (Suppl. S2), W369–W373. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]





| S.No. | Gene Name | Abbreviation | Gene Superfamily | Number of Genes in Triticum aestivum (% Identity) |
|---|---|---|---|---|
| 1 | Chalcone synthase | CHS | Polyketide synthase enzymes (PKS) | 95 (37–92) |
| 2 | Chalcone isomerase | CHI | Isomerases | 7 (50–61) |
| 3 | Flavanone hydroxylase | F3H | 2OG-Fe (II) oxygenase superfamily | 101 (37–58) |
| 4 | Flavonoid 3′-hydroxylase | F3′H | P450-dependent monooxygenase (P450) | 491 (37–81) |
| 5 | Flavonoid 3′,5′-hydroxylase | F3′5′H | 51 (39–40) | |
| 6 | Dihydroflavanol reductase | DFR | NAD dependent epimerase/dehydratase | 133 (37–66) |
| 7 | Anthocyanidin synthase | ANS | 2OG-Fe (II) oxygenase superfamily | 73 (27–68) |
| 8 | UDP glycosyltransferases | UGFT | UDP-glucosyl transferases | 242 (38–76) |
| Gene | Subcellular Localization | Predicted Molecular Mass KD (Min–Max) | Isoelectric Point (Min–Max) | Domain |
|---|---|---|---|---|
| CHS | Cytoplasm | 23.6–52.04 | 5.06–9.21 | Chal_sti_synt |
| CHI | Cytoplasm | 23.54–24.18 | 4.54–5.09 | Chalcone |
| F3H | Cytoplasm | 30.1–48.13 | 4.85–8.04 | 2-Oxoglutatate dependent dioxygense |
| F3′H | Chloroplast | 21.20–63.69 | 5.76–9.95 | Cytochrome p450(P450-dependent monooxygenase) |
| F3′5′H | Chloroplast | 39.27–66.59 | 5.63–9.91 | Cytochrome p450(P450-dependent monooxygenase) |
| DFR | Cytoplasm | 16.06–46.68 | 4.74–9.02 | Epimerase |
| ANS | Cytoplasm | 29.45–47.62 | 4.61–8.95 | 2OG-Fe (II) oxygenase superfamily |
| UFGT | Chloroplast | 28.05–76.67 | 4.60–8.41 | Glycosyltransferase_GTB-type superfamily |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kapoor, P.; Sharma, S.; Tiwari, A.; Kaur, S.; Kumari, A.; Sonah, H.; Goyal, A.; Krishania, M.; Garg, M. Genome–Transcriptome Transition Approaches to Characterize Anthocyanin Biosynthesis Pathway Genes in Blue, Black and Purple Wheat. Genes 2023, 14, 809. https://doi.org/10.3390/genes14040809
Kapoor P, Sharma S, Tiwari A, Kaur S, Kumari A, Sonah H, Goyal A, Krishania M, Garg M. Genome–Transcriptome Transition Approaches to Characterize Anthocyanin Biosynthesis Pathway Genes in Blue, Black and Purple Wheat. Genes. 2023; 14(4):809. https://doi.org/10.3390/genes14040809
Chicago/Turabian StyleKapoor, Payal, Saloni Sharma, Apoorv Tiwari, Satveer Kaur, Anita Kumari, Humira Sonah, Ajay Goyal, Meena Krishania, and Monika Garg. 2023. "Genome–Transcriptome Transition Approaches to Characterize Anthocyanin Biosynthesis Pathway Genes in Blue, Black and Purple Wheat" Genes 14, no. 4: 809. https://doi.org/10.3390/genes14040809
APA StyleKapoor, P., Sharma, S., Tiwari, A., Kaur, S., Kumari, A., Sonah, H., Goyal, A., Krishania, M., & Garg, M. (2023). Genome–Transcriptome Transition Approaches to Characterize Anthocyanin Biosynthesis Pathway Genes in Blue, Black and Purple Wheat. Genes, 14(4), 809. https://doi.org/10.3390/genes14040809








