NGS Sequencing Reveals New UCP1 Gene Variants Potentially Associated with MetS and/or T2DM Risk in the Polish Population—A Preliminary Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Controls
2.3. DNA Isolation and Genotyping
2.4. Library Preparation and Next Generation Sequencing
2.5. NGS Data Analysis
3. Results
3.1. UCP1 Gene Variants Identified by NGS Sequencing
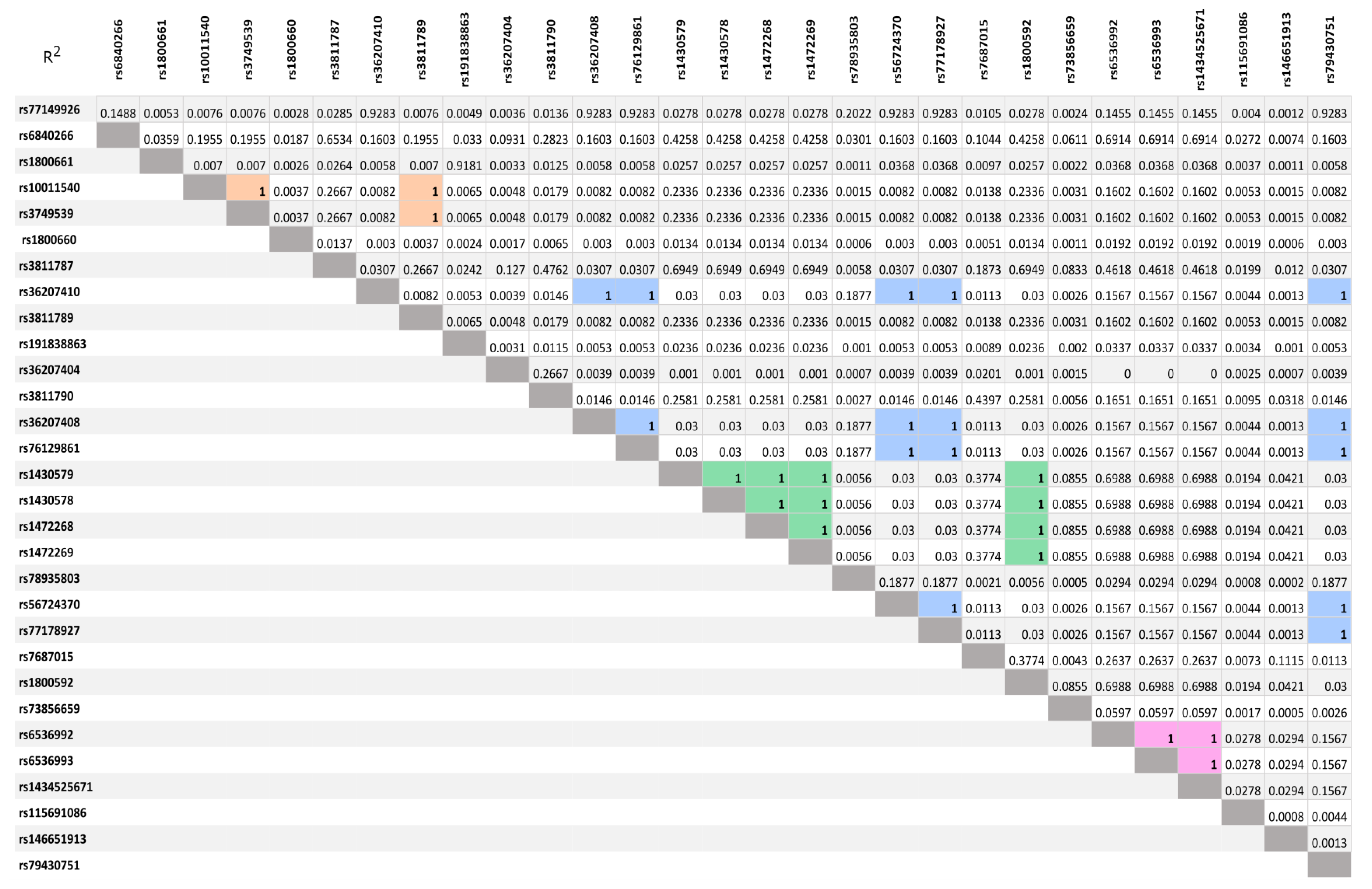
3.2. LD Analysis of SNVs Idetyfied by NGS Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, V.; Hsueh, W.A.; Raman, S.V. Multiorgan, multimodality imaging in cardiometabolic disease. Circ. Cardiovasc. Imaging 2017, 10, e005447. [Google Scholar] [CrossRef] [PubMed]
- Pawłowska, J.; Witkowski, J.; Bryl, E. Zespół metaboliczny—Rys historyczny i współczesność. Forum Med. Rodz. 2009, 3, 222–228. [Google Scholar]
- Rajca, A.; Wojciechowska, A.; Śmigielski, W.; Drygas, W.; Piwońska, A.; Pająk, A.; Tykarski, A.; Kozakiewicz, K.; Kwaśniewska, M.; Zdrojewski, T. Increase in the prevalence of the metabolic syndrome in Poland. comparison of the results of the WOBASZ (2003–2005) and Wobasz II (2013–2014) studies. Pol. Arch. Intern. Med. 2021, 131, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Raposo, L. Metabolic syndrome in Poland: The WOBASZ II Study. Pol. Arch. Intern. Med. 2021, 131, 501–502. [Google Scholar] [CrossRef]
- Saklayen, M.G. The global epidemic of the metabolic syndrome. Curr. Hypertens. Rep. 2018, 20, 12. [Google Scholar] [CrossRef]
- Cardiovascular Diseases (cvds) [Internet]. World Health Organization. Available online: https://www.who.int/en/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 10 October 2022).
- Towpik, I.; Walicka, M.; Marcinkowska, K.; Lisicka, I.; Raczyńska, M.; Wierzba, W.; Strojek, K.; Ryś, P.; Wajda-Cuszlag, M.; Franek, E. Epidemiology of diabetes in Poland in 2014–2017. Clin. Diabetol. 2020, 9, 279–285. [Google Scholar] [CrossRef]
- Lowell, B.B.; Spiegelman, B.M. Towards a molecular understanding of adaptive thermogenesis. Nature 2000, 404, 652–660. [Google Scholar] [CrossRef]
- Virtanen, K.A.; Nuutila, P. The importance of Human Brown Adipose Tissue Volume. Nat. Rev. Endocrinol. 2021, 17, 453–454. [Google Scholar] [CrossRef]
- Saito, M.; Matsushita, M.; Yoneshiro, T.; Okamatsu-Ogura, Y. Brown adipose tissue, diet-induced thermogenesis, and thermogenic food ingredients: From mice to men. Front. Endocrinol. 2020, 11, 222. [Google Scholar] [CrossRef]
- Trayhurn, P. Brown adipose tissue—A therapeutic target in obesity? Front. Physiol. 2018, 9, 1672. [Google Scholar] [CrossRef]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.-H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef]
- Vijgen, G.H.; Bouvy, N.D.; Teule, G.J.; Brans, B.; Schrauwen, P.; van MarkenLichtenbelt, W.D. Brown adipose tissue in morbidly obese subjects. PLoS ONE 2011, 6, e17247. [Google Scholar] [CrossRef]
- Fedorenko, A.; Lishko, P.V.; Kirichok, Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012, 151, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Bertholet, A.M.; Kirichok, Y. The mechanism FA-dependent H+ transport by UCP1. Handb. Exp. Pharmacol. 2019, 251, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Villarroya, F.; Peyrou, M.; Giralt, M. Transcriptional regulation of the uncoupling protein-1 gene. Biochimie 2017, 134, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Vimaleswaran, K.S.; Loos, R.J.F. Progress in the genetics of common obesity and type 2 diabetes. Expert Rev. Mol. Med. 2010, 12, e7. [Google Scholar] [CrossRef] [PubMed]
- Jia, J.-J.; Tian, Y.-B.; Cao, Z.-H.; Tao, L.-L.; Zhang, X.; Gao, S.-Z.; Ge, C.-R.; Lin, Q.-Y.; Jois, M. The polymorphisms of UCP1 genes associated with fat metabolism, obesity and diabetes. Mol. Biol. Rep. 2009, 37, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- BrondaniLde Assmann, T.S.; Duarte, G.C.; Gross, J.L.; Canani, L.H.; Crispim, D. The role of the uncoupling protein 1 (UCP1) on the development of obesity and type 2 diabetes mellitus. Arq. De Endocrinol. E Metabol. 2012, 56, 215–225. [Google Scholar] [CrossRef]
- Brondani, L.A.; Assmann, T.S.; de Souza, B.M.; Bouças, A.P.; Canani, L.H.; Crispim, D. Meta-analysis reveals the association of common variants in the uncoupling protein (UCP) 1–3 genes with body mass index variability. PLoS ONE 2014, 9, e96411. [Google Scholar] [CrossRef]
- Stosio, M.; Witkowicz, A.; Kowalska, A.; Karabon, L. Genetic background of aberrant thermogenin expression (UCP-1) in obesity leading to metabolic syndrome. Postępy Hig. Med. Doświadczalnej 2016, 70, 1389–1403. [Google Scholar] [CrossRef]
- Dinas, P.C.; Nintou, E.; Vliora, M.; Pravednikova, A.E.; Sakellariou, P.; Witkowicz, A.; Kachaev, Z.M.; Kerchev, V.V.; Larina, S.N.; Cotton, J.; et al. Prevalence of uncoupling protein one genetic polymorphisms and their relationship with cardiovascular and Metabolic Health. PLoS ONE 2022, 17, e0266386. [Google Scholar] [CrossRef] [PubMed]
- Flouris, A.D.; Shidlovskii, Y.V.; Shaposhnikov, A.V.; Yepiskoposyan, L.; Nadolnik, L.; Karabon, L.; Kowalska, A.; Carrillo, A.E.; Metsios, G.S.; Sakellariou, P. Role of UCP1 gene variants in interethnic differences in the development of cardio-metabolic diseases. Front. Genet. 2017, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FASTQ SCREEN: A tool for multi-genome mapping and Quality Control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms. SnpEff Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.Y.; He, L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005, 15, 97–98. [Google Scholar] [CrossRef]
- Ho Cha, M.; Soo Kim, K.; Suh, D.; Chung, S.-I.; Yoon, Y. A UCP1-412A>C polymorphism is associated with abdominal fat area in Korean women. Hereditas 2008, 145, 231–237. [Google Scholar] [CrossRef]
- Jin, P.; Li, Z.; Xu, X.; He, J.; Chen, J.; Xu, X.; Du, X.; Bai, X.; Zhang, B.; He, X.; et al. Analysis of association between common variants of uncoupling proteins genes and diabetic retinopathy in a Chinese population. BMC Med. Genet. 2020, 21, 25. [Google Scholar] [CrossRef]
- Nikanorova, A.A.; Barashkov, N.A.; Pshennikova, V.G.; Nakhodkin, S.S.; Gotovtsev, N.N.; Romanov, G.P.; Solovyev, A.; Kuzmina, S.; Sazonov, N.; Fedorova, S. The role of nonshivering thermogenesis genes on leptin levels regulation in residents of the coldest region of Siberia. Int. J. Mol. Sci. 2021, 22, 4657. [Google Scholar] [CrossRef] [PubMed]
- Bovolini, A.; Garcia, J.; Andrade, M.A.; Duarte, J.A. Metabolic syndrome pathophysiology and predisposing factors. Int. J. Sport. Med. 2020, 42, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Giglio, R.V.; Stoian, A.P.; Patti, A.M.; Rizvi, A.A.; Sukhorukov, V.; Ciaccio, M.; Orekhov, A.; Rizzo, M. Genetic and epigenetic biomarkers for diagnosis, prognosis and treatment of metabolic syndrome. Curr. Pharm. Des. 2021, 27, 3729–3740. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Zhou, H.; Shen, C.; Yu, L.-G.; Ding, Y.; Zhang, Y.-H.; Guo, Z.-R. Role of peroxisome proliferator-activated receptors gene polymorphisms in type 2 diabetes and metabolic syndrome. World J. Diabetes 2015, 6, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zou, Y.; Shen, Z.; Xiong, Y.; Zhang, W.; Liu, C.; Chen, S. Trace elements, PPARs, and metabolic syndrome. Int. J. Mol. Sci. 2020, 21, 2612. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.W.; Willershäuser, M.; Jastroch, M.; Rourke, B.C.; Fromme, T.; Oelkrug, R.; Heldmaier, G.; Klingenspor, M. Adaptive thermogenesis and thermal conductance in wild-type and UCP1-Ko Mice. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2010, 99, R1396–R1406. [Google Scholar] [CrossRef]
- Ukropec, J.; Anunciado, R.P.; Ravussin, Y.; Hulver, M.W.; Kozak, L.P. UCP1-independent thermogenesis in white adipose tissue of cold-acclimated Ucp1−/− mice. J. Biol. Chem. 2006, 281, 31894–31908. [Google Scholar] [CrossRef]
- Esterbauer, H.; Oberkofler, H.; Liu, Y.M.; Breban, D.; Hell, E.; Krempler, F.; Patsch, W. Uncoupling protein-1 mRNA expression in obese human subjects: The role of sequence variations at the uncoupling protein-1 gene locus. J. Lipid Res. 1998, 39, 834–844. [Google Scholar] [CrossRef]
- Rose, G.; Crocco, P.; D’Aquila, P.; Montesanto, A.; Bellizzi, D.; Passarino, G. Two variants located in the upstream enhancer region of human UCP1 gene affect gene expression and are correlated with human longevity. Exp. Gerontol. 2011, 46, 897–904. [Google Scholar] [CrossRef]
- Hamann, A.; Tafel, J.; Büsing, B.; Münzberg, H.; Hinney, A.; Mayer, H.; Siegfried, W.; Ricquier, D.; Greten, H.; Hebebrand, J.; et al. Analysis of the uncoupling protein-1 (UCP1) gene in obese and lean subjects: Identification of four amino acid variants. Int. J. Obes. 1998, 22, 939–941. [Google Scholar] [CrossRef]
- Heilbronn, L.K.; Kind, K.L.; Pancewicz, E.; Morris, A.M.; Noakes, M.; Clifton, P.M. Association of −3826 G variant in uncoupling protein-1 with increased BMI in overweight Australian women. Diabetologia 2000, 43, 242–244. [Google Scholar] [CrossRef] [PubMed]
- Iwata, K.; Iwasa, M.; Nakatani, T.; Yano, Y.; Mifuji-Moroka, R.; Hara, N.; Akamatsu, M.; Ishidome, M.; Takei, Y. Seasonal variation in visceral fat and blood hba1c in people with type 2 diabetes. Diabetes Res. Clin. Pract. 2012, 96, e53–e54. [Google Scholar] [CrossRef] [PubMed]

| Controls (n = 36) | MetS Patients | |||
|---|---|---|---|---|
| All (n = 59) | with T2DM (n = 29) | w/o T2DM (n = 30) | ||
| Sex (females/males) | 18/18 | 32/27 | 17/12 | 15/15 |
| Age | 36.33 ± 13.03 | 61.5 ± 12.91 | 63 ± 14.06 | 60 ± 11.49 |
| Total cholesterol [mg/dL] | - | 192.82 ± 51.52 | 186.94 ± 55.45 | 198.69 ± 46.34 |
| LDL [mg/dL] | - | 102.36 ± 41.88 | 101.16 ± 40.18 | 103.69 ± 43.73 |
| HDL [mg/dL] | ||||
| - | 53.53 ± 17.51 | 51.72 ± 21.58 | 55.71 ± 11.17 |
| - | 45.81 ± 12.35 | 43.65 ± 15.75 | 47.53 ± 9.01 |
| Triglycerides [mg/dL] | - | 242.46 ± 188.70 | 254.25 ± 246.92 | 231.07 ± 110.24 |
| Fasting blood glucose [mg/dL] | - | 128.13 ± 51.01 | 151.80 ± 57.17 | 106.03 ± 32.01 |
| Body fat percentage [%] | 23.89 ± 6.12 | - | - | - |
| BMI | 22.77 ± 1.86 | 34.29 ± 7.55 | 33.96 ± 9.61 | 34.60 ± 4.76 |
| Waist circumference [cm] | ||||
| 75.24 ± 5.82 | 111.30 ± 19.52 | 111.82 ± 23.49 | 110.70 ± 14.59 |
| 84.11 ± 5.53 | 110.29 ± 12.54 | 105.04 ± 12.81 | 114.48 ± 10.99 |
| WHR | ||||
| 0.78 ± 0.03 | 0.97 ± 0.13 | 0.97 ± 0.13 | 0.98 ± 0.12 |
| 0.85 ± 0.05 | 1.07 ± 0.09 | 1.06 ± 0.05 | 1.08 ± 0.12 |
| Systolic blood pressure [mmHg] | 120.31 ± 7.26 | 142.73 ± 19.86 | 134.86 ± 18.90 | 150.33 ± 17.95 |
| Diastolic blood pressure [mmHg] | 76.77 ± 6.07 | 83.41 ± 12.61 | 77.34 ± 11.73 | 89.27 ± 10.62 |
| SNV Position | Control | MetS | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GRCh38/hg38 | rsID | Variable | N | % | N | % | OR * | CI95% | p Value | |
| 140558375 | rs77149926 | C | 55 | 88.71 | 100 | 98.04 | 1 | 0.011 | ||
| T | 7 | 11.29 | 2 | 1.96 | 0.184 | 0.04 | 0.80 | |||
| CC | 25 | 80.65 | 49 | 96.08 | 1 | 0.023 | ||||
| CT+TT | 6 | 19.35 | 2 | 3.92 | 0.198 | 0.04 | 0.92 | |||
| 140560701 | unknown | T | 68 | 97.14 | 75 | 87.21 | 1 | 0.026 | ||
| C | 2 | 2.857 | 11 | 12.79 | 4.174 | 1.02 | 17.03 | |||
| TT | 33 | 94.29 | 32 | 74.42 | 1 | 0.020 | ||||
| CT+CC | 2 | 5.71 | 11 | 25.58 | 4.742 | 1.11 | 20.22 | |||
| 140561713 | rs183105785 | A | 69 | 98.57 | 77 | 91.67 | 1 | 0.055 | ||
| C | 1 | 1.43 | 7 | 8.33 | 4.484 | 0.75 | 26.66 | |||
| AA | 34 | 97.14 | 35 | 83.33 | 1 | 0.049 | ||||
| AC+CC | 1 | 2.86 | 7 | 16.67 | 4.859 | 0.79 | 29.83 | |||
| 140563477 | unknown | T | 69 | 98.57 | 79 | 91.86 | 1 | 0.060 | ||
| C | 1 | 1.43 | 7 | 8.14 | 4.371 | 0.74 | 25.98 | |||
| TT | 34 | 97.14 | 36 | 83.72 | 1 | 0.054 | ||||
| CT+CC | 1 | 2.86 | 7 | 16.28 | 4.726 | 0.77 | 28.99 | |||
| 140569304 | rs36207410 | G | 55 | 88.71 | 100 | 98.04 | 1 | 0.011 | ||
| A | 7 | 11.29 | 2 | 1.96 | 0.184 | 0.04 | 0.80 | |||
| GG | 25 | 80.65 | 49 | 96.08 | 1 | 0.023 | ||||
| AG+AA | 6 | 19.35 | 2 | 3.92 | 0.198 | 0.04 | 0.92 | |||
| 140570776 | rs36207408 | G | 55 | 88.71 | 99 | 97.06 | 1 | 0.031 | ||
| A | 7 | 11.29 | 3 | 2.94 | 0.260 | 0.07 | 0.97 | |||
| GG | 25 | 80.65 | 48 | 94.12 | 1 | 0.060 | ||||
| AG+AA | 6 | 19.35 | 3 | 5.88 | 0.283 | 0.07 | 1.13 | |||
| 140571476 | rs76129861 | G | 55 | 88.71 | 99 | 97.06 | 1 | 0.031 | ||
| A | 7 | 11.29 | 3 | 2.94 | 0.260 | 0.07 | 0.97 | |||
| GG | 25 | 80.65 | 48 | 94.12 | 1 | 0.060 | ||||
| AG+AA | 6 | 19.35 | 3 | 5.88 | 0.283 | 0.07 | 1.13 | |||
| 140572036 | rs56724370 | A | 55 | 88.71 | 99 | 97.06 | 1 | 0.031 | ||
| C | 7 | 11.29 | 3 | 2.94 | 0.260 | 0.07 | 0.97 | |||
| AA | 25 | 80.65 | 48 | 94.12 | 1 | 0.060 | ||||
| AC+CC | 6 | 19.35 | 3 | 5.88 | 0.283 | 0.07 | 1.13 | |||
| 140572113 | rs77178927 | T | 55 | 88.71 | 99 | 97.06 | 1 | 0.031 | ||
| G | 7 | 11.29 | 3 | 2.94 | 0.260 | 0.07 | 0.97 | |||
| TT | 25 | 80.65 | 48 | 94.12 | 1 | 0.060 | ||||
| GT+GG | 6 | 19.35 | 3 | 5.88 | 0.283 | 0.07 | 1.13 | |||
| SNV Position | Control | T2DM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| GRCh38/hg38 | rsID | Variation | N | % | N | % | OR * | CI95% | p Value | |
| 140558375 | rs77149926 | C | 55 | 88.71 | 53 | 98.15 | 1 | 0.046 | ||
| T | 7 | 11.29 | 1 | 1.85 | 0.207 | 0.03 | 1.25 | |||
| CC | 25 | 80.65 | 26 | 96.30 | 1 | 0.070 | ||||
| CT+TT | 6 | 19.35 | 1 | 3.70 | 0.222 | 0.03 | 1.42 | |||
| 140560701 | unknown | T | 68 | 97.14 | 21 | 75.00 | 1 | 0.001 | ||
| C | 2 | 2.86 | 7 | 25.00 | 9.558 | 2.11 | 43.32 | |||
| TT | 33 | 94.29 | 7 | 50.00 | 1 | 0.001 | ||||
| CT+CC | 2 | 5.71 | 7 | 50.00 | 13.4 | 2.61 | 68.79 | |||
| 140561203 | unknown | G | 64 | 91.43 | 20 | 71.43 | 1 | 0.011 | ||
| A | 6 | 8.57 | 8 | 28.57 | 4.114 | 1.32 | 12.81 | |||
| GG | 29 | 82.86 | 6 | 42.86 | 1 | 0.006 | ||||
| AG+AA | 6 | 17.14 | 8 | 57.14 | 5.935 | 1.57 | 22.40 | |||
| 140561340 | unknown | T | 58 | 82.86 | 16 | 57.14 | 1 | 0.008 | ||
| A | 12 | 17.14 | 12 | 42.86 | 3.545 | 1.36 | 9.22 | |||
| TT | 23 | 65.71 | 2 | 14.29 | 1 | 0.001 | ||||
| AT+AA | 12 | 34.29 | 12 | 85.71 | 9.400 | 2.05 | 43.04 | |||
| 140561713 | rs183105785 | A | 69 | 98.57 | 25 | 89.29 | 1 | 0.037 | ||
| C | 1 | 1.43 | 3 | 10.71 | 6.359 | 0.89 | 45.41 | |||
| AA | 34 | 97.14 | 11 | 78.57 | 1 | 0.034 | ||||
| AC+CC | 1 | 2.86 | 3 | 21.43 | 7.000 | 0.92 | 53.08 | |||
| 140563477 | unknown | T | 69 | 98.57 | 23 | 82.14 | 1 | 0.002 | ||
| C | 1 | 1.43 | 5 | 17.86 | 10.84 | 1.68 | 70.01 | |||
| TT | 34 | 97.14 | 9 | 64.29 | 1 | 0.002 | ||||
| CT+CC | 1 | 2.86 | 5 | 35.71 | 13.32 | 1.91 | 92.94 | |||
| 140564690 | unknown | T | 55 | 78.57 | 15 | 53.57 | 1 | 0.014 | ||
| C | 15 | 21.43 | 13 | 46.43 | 3.119 | 1.24 | 7.84 | |||
| TT | 20 | 57.14 | 1 | 7.14 | 1 | 0.002 | ||||
| CT+CC | 15 | 42.86 | 13 | 92.86 | 11.90 | 1.95 | 72.83 | |||
| 140567294 | unknown | A | 63 | 90.00 | 21 | 75.00 | 1 | 0.056 | ||
| C | 7 | 10.00 | 7 | 25.00 | 2.953 | 0.96 | 9.09 | |||
| AA | 28 | 80.00 | 7 | 50.00 | 1 | 0.038 | ||||
| AC+CC | 7 | 20.00 | 7 | 50.00 | 3.800 | 1.04 | 13.84 | |||
| 140568108 | rs3138733 | CGTGTGTGT | 44 | 62.86 | 23 | 82.14 | 1 | 0.065 | ||
| GTGTGTGT | C | 26 | 37.14 | 5 | 17.86 | 0.393 | 0.14 | 1.12 | ||
| insertion | (CGTGTGTGT)2 | 14 | 40.00 | 10 | 71.43 | 1 | 0.049 | |||
| CCGTGTGTGT+CC | 21 | 60.00 | 4 | 28.57 | 0.289 | 0.08 | 1.05 | |||
| 140568337 | unknown | A | 52 | 74.29 | 15 | 53.57 | 1 | 0.047 | ||
| T | 18 | 25.71 | 13 | 46.43 | 2.472 | 1.00 | 6.09 | |||
| AA | 19 | 54.29 | 1 | 7.143 | 1 | 0.003 | ||||
| AT+TT | 16 | 45.71 | 13 | 92.86 | 10.64 | 1.74 | 64.98 | |||
| 140569265 | rs3811787 | T | 57 | 91.94 | 42 | 77.78 | 1 | 0.032 | ||
| G | 5 | 8.07 | 12 | 22.22 | 3.075 | 1.05 | 9.04 | |||
| TT | 26 | 83.87 | 17 | 62.96 | 1 | 0.072 | ||||
| GT+GG | 5 | 16.13 | 10 | 37.04 | 2.891 | 0.87 | 9.55 | |||
| 140569304 | rs36207410 | G | 55 | 88.71 | 53 | 98.15 | 1 | 0.046 | ||
| A | 7 | 11.29 | 1 | 1.85 | 0.207 | 0.03 | 1.25 | |||
| GG | 25 | 80.65 | 26 | 96.3 | 1 | 0.070 | ||||
| AG+AA | 6 | 19.35 | 1 | 3.70 | 0.222 | 0.03 | 1.42 | |||
| 140573371 | rs6536992 | G | 55 | 88.71 | 39 | 72.22 | 1 | 0.024 | ||
| A | 7 | 11.29 | 15 | 27.78 | 2.904 | 1.11 | 7.60 | |||
| GG | 25 | 80.65 | 16 | 59.26 | 1 | 0.077 | ||||
| AG+AA | 6 | 19.35 | 11 | 40.74 | 2.734 | 0.87 | 8.58 | |||
| 140573392 | rs6536993 | C | 55 | 88.71 | 39 | 72.22 | 1 | 0.024 | ||
| T | 7 | 11.29 | 15 | 27.78 | 2.904 | 1.11 | 7.60 | |||
| CC | 25 | 80.65 | 16 | 59.26 | 1 | 0.077 | ||||
| CT+TT | 6 | 19.35 | 11 | 40.74 | 2.734 | 0.87 | 8.58 | |||
| 140573493 | rs6536994 | A | 55 | 88.71 | 39 | 72.22 | 1 | 0.024 | ||
| T | 7 | 11.29 | 15 | 27.78 | 2.904 | 1.11 | 7.60 | |||
| AA | 25 | 80.65 | 16 | 59.26 | 1 | 0.077 | ||||
| AT+TT | 6 | 19.35 | 11 | 40.74 | 2.734 | 0.87 | 8.58 | |||
| rsID | Position (GRCh38.p13) | Location | Allele Frequency | Studied Previously? | Conservation (Score) | MetS ** | MetS ** +T2DM |
|---|---|---|---|---|---|---|---|
| rs77149926 | chr4:140558375 | 3′UTR | C = 93.93% | No | −0.627 | + | + |
| unknown | chr4:140560701 | Intron 5 | - | No | - | + | + |
| unknown | chr4:140561203 | Intron 5 | - | No | - | - | + |
| unknown | chr4:140561340 | Intron 5 | - | No | - | - | + |
| rs183105785 | chr4:140561713 | Intron 5 | A ~ 100% | No | 0.328 | + | + |
| unknown | chr4:140563477 | Exon 3 | - | No | - | + | + |
| unknown | chr4:140564690 | Intron 2 | - | No | - | - | + |
| unknown | chr4:140567294 | Intron 2 | - | No | - | - | + |
| rs3138733 (Indel) | chr4:140568109-140568156 | Intron 1 | (GT)24 = 12.14% | No | 2.089 | - | + |
| unknown | chr4:140568337 | Intron 1 | - | No | - | - | + |
| rs3811787 | chr4:140569265 | 5′UTR | T = 74.97% | Yes * | −2.682 | - | + |
| rs36207410 | chr4:140569304 | 5′UTR | G = 93.38% | No | −2.179 | + | + |
| rs36207408 | chr4:140570776 | 5′UTR | G = 93.72% | No | −0.223 | + | - |
| rs76129861 | chr4:140571476 | 5′UTR | G = 93.70% | No | 0.005 | + | - |
| rs56724370 | chr4:140572036 | 5′UTR | A = 93.70% | No | −0.244 | + | - |
| rs77178927 | chr4:140572113 | 5′UTR | T = 98.18% | No | 0.042 | + | - |
| rs6536992 | chr4:140573371 | 5′UTR | G = 68.23% | No | −0.639 | - | + |
| rs6536993 | chr4:140573392 | 5′UTR | C = 67.31% | No | −0.372 | - | + |
| rs6536994 | chr4:140573493 | 5′UTR | A = 78.14% | No | −1.211 | - | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrzejczak, A.; Witkowicz, A.; Kujawa, D.; Skrypnik, D.; Szulińska, M.; Bogdański, P.; Łaczmański, Ł.; Karabon, L. NGS Sequencing Reveals New UCP1 Gene Variants Potentially Associated with MetS and/or T2DM Risk in the Polish Population—A Preliminary Study. Genes 2023, 14, 789. https://doi.org/10.3390/genes14040789
Andrzejczak A, Witkowicz A, Kujawa D, Skrypnik D, Szulińska M, Bogdański P, Łaczmański Ł, Karabon L. NGS Sequencing Reveals New UCP1 Gene Variants Potentially Associated with MetS and/or T2DM Risk in the Polish Population—A Preliminary Study. Genes. 2023; 14(4):789. https://doi.org/10.3390/genes14040789
Chicago/Turabian StyleAndrzejczak, Anna, Agata Witkowicz, Dorota Kujawa, Damian Skrypnik, Monika Szulińska, Paweł Bogdański, Łukasz Łaczmański, and Lidia Karabon. 2023. "NGS Sequencing Reveals New UCP1 Gene Variants Potentially Associated with MetS and/or T2DM Risk in the Polish Population—A Preliminary Study" Genes 14, no. 4: 789. https://doi.org/10.3390/genes14040789
APA StyleAndrzejczak, A., Witkowicz, A., Kujawa, D., Skrypnik, D., Szulińska, M., Bogdański, P., Łaczmański, Ł., & Karabon, L. (2023). NGS Sequencing Reveals New UCP1 Gene Variants Potentially Associated with MetS and/or T2DM Risk in the Polish Population—A Preliminary Study. Genes, 14(4), 789. https://doi.org/10.3390/genes14040789










