Two Complete Mitochondrial Genomes of Leptobrachium (Anura: Megophryidae: Leptobrachiinae): Characteristics, Population Divergences, and Phylogenetic Implications
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. Sequence Assembly, Annotation, and Analysis
2.3. Phylogenetic Analysis
3. Results
3.1. Mitogenome Annotation and Nucleotide Composition
3.2. Characteristics of rRNAs, tRNAs, and the Control Region
3.3. Codon Usage and Genetic Distances
3.4. Phylogenetic Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahony, S.; Kamei, R.G.; Teeling, E.C.; Biju, S.D. Cryptic diversity within the Megophrys major species group (Amphibia: Megophryidae) of the Asian Horned Frogs: Phylogenetic perspectives and a taxonomic revision of South Asian taxa, with descriptions of four new species. Zootaxa 2018, 4523, 1–96. [Google Scholar] [CrossRef] [PubMed]
- Cannatella, D.C. A Phylogeny of Primitive Frogs (Archaeobatrachians) (Cladistics, Taxonomy, Morphology, Systematics, Evolution). Ph.D. Thesis, University of Kansas, Lawrence, Kansas, 1985. [Google Scholar]
- Ford, L.S.; Cannatella, D.C. The Major Clades of Frogs. Herpetol. Monogr. 1993, 7, 94–117. [Google Scholar] [CrossRef]
- Frost, D.R.; Grant, T.; Faivovich, J.; Bain, R.H.; Haas, A.; Haddad, C.F.B.; Desa, R.O.; Channing, A.; Wilkinson, M.; Donnellan, S.C.; et al. The amphibian tree of life. Bull. Am. Mus. Nat. Hist. 2006, 297, 1–370. [Google Scholar] [CrossRef]
- Pyron, R.A.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef]
- AmphibiaWeb. Available online: https://amphibiaweb.org/ (accessed on 8 February 2023).
- Fei, L.; Ye, C.Y.; Jiang, J.P.; Huang, Y.Z. Atlas of Amphibians in China (Field Edition), 1st ed.; Henan Science and Technology Press: Zhengzhou, China, 2020; pp. 254–262. [Google Scholar]
- Rao, D.Q.; Wilkinson, J.A. Phylogenetic relationships of the mustache toads inferred from mt DNA sequences. Mol. Phylogenet. Evol. 2008, 46, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Fei, L.; Ye, C.Y.; Jiang, J.P. Colored Atlas of Chinese Amphibians, 1st ed.; Sichuan Publishing House of Science and Technology: Chengdu, China, 2010; pp. 162–167. [Google Scholar]
- Tian, W.; Hu, S. Taxonomical studies on the primitive anurans of the Hengduan Mountains, with descriptions of a new subfamily and subdivision of Bombina. Acta Herpetol. Sinica 1985, 4, 219–224. [Google Scholar]
- Dubois, A.; Ohler, A. A new species of Leptobrachium (Vibrissaphora) from northern Vietnam, with a review of the taxonomy of the genus Leptobrachium (Pelobatidae, Megophryinae). Dumerilia 1998, 4, 1–32. [Google Scholar]
- Zhao, E.M.; Adler, K. Herpetology of China, 1st ed.; Society for the Study of Amphibians and Reptiles: Oxford, MI, USA, 1993; pp. 1–522. [Google Scholar]
- Zheng, Y.C.; Li, S.; Fu, J. A phylogenetic analysis of the frog genera Vibrissaphora and Leptobrachium, and the correlated evolution of nuptial spine and reversed sexual size dimorphism. Mol. Phylogenet. Evol. 2008, 46, 695–707. [Google Scholar] [CrossRef]
- Matsui, M.; Hamidy, A.; Murphy, R.W.; Khonsue, W.; Yambun, P.; Shimada, T.; Ahmad, N.; Belabut, D.M.; Jiang, J.P. Phylogenetic relationships of megophryid frogs of the genus Leptobrachium(Amphibia, Anura) as revealed by mtDNA gene sequences. Mol. Phylogenet. Evol. 2010, 56, 259–272. [Google Scholar] [CrossRef]
- Gao, J.M.; Zhang, Y.D. Retinal organization in Vibrissaphora liui. Acta Zool. Sin. 1996, 42, 405–412. [Google Scholar]
- Zheng, Y.C.; Zeng, X.M. A review of study on moustache toads, Vibrissaphora (Anura: Megophryidae). Sichuan J. Zool. 2003, 22, 268–271. [Google Scholar]
- Zhang, W.Y.; Lu, Y.Y.; Shi, J.S.; Zhu, L.; Li, P.P. Microstructure and Sub-Microstructure of the Keratinized Nuptial Spines of Male Leptobrachium boringii (Liu, 1945). Sichuan J. Zool. 2019, 38, 186–193. [Google Scholar]
- Hudson, C.M.; He, X.J.; Fu, J.Z. Keratinized Nuptial Spines Are Used for Male Combat in the Emei Moustache Toad (Leptobrachium boringii). Asian Herpetol. Res. 2011, 2, 142–148. [Google Scholar]
- Zheng, Y.C.; Deng, D.C.; Li, S.Q.; Fu, J.Z. Aspects of the breeding biology of the Omei mustache toad (Leptobrachium boringii): Polygamy and paternal care. Amphib. -Reptil. 2010, 31, 183–194. [Google Scholar] [CrossRef]
- Hudson, C.M.; Fu, J. Male-biased sexual size dimorphism, resource defense polygyny, and multiple paternity in the Emei moustache toad (Leptobrachium boringii). PLoS ONE 2013, 8, e67502. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.X.; Ren, Y.D.; Zhang, D.R.; Jiang, H.; Wang, Z.K.; Li, X.Y.; Rao, D.Q. Chromosome-level assembly of the mustache toad genome using third-generation DNA sequencing and Hi-C analysis. GigaScience 2019, 8, giz114. [Google Scholar] [CrossRef] [PubMed]
- Fei, L.; Hu, S.Q.; Ye, C.Y.; Huang, Y.Z. Amphibia. In Fauna Sinica, 1st ed.; Fauna Sinica Editorial Board; Science Press: Beijing, China, 2009; Volume 2, pp. 81–485. [Google Scholar]
- Liu, B.H. Vibrissaphora liui in Jiulongshan Mountain, Zhejiang province. Chin. J. Wildl. 1987, 6, 22–23. [Google Scholar]
- IUCN SSC Amphibian Specialist Group. Leptobrachium boringii. In The IUCN Red List of Threatened Species 2020. 2020. [Google Scholar] [CrossRef]
- Gao, Z.W.; Qian, T.Y.; Jiang, J.P.; Hou, D.J.; Deng, X.J.; Yang, D.D. Species diversity and distribution of amphibians and reptiles in Hunan Province, China. Biodivers. Sci. 2022, 30, 101–115. [Google Scholar] [CrossRef]
- IUCN SSC Amphibian Specialist Group. Leptobrachium liui. In The IUCN Red List of Threatened Species 2020. 2020. [Google Scholar] [CrossRef]
- Xu, Q.P.; Liu, S.L.; Wan, R.Z.; Yue, B.S.; Zhang, X.Y. The complete mitochondrial genome of the Vibrissaphora boringii (Anura: Megophryidae). Mitochondrial DNA Part A 2016, 27, 758–759. [Google Scholar] [CrossRef]
- Li, Y.W.; Li, C.; Chen, Q.L.; Liu, Z.H.; Shen, Y.J. The mitochondrial genome for tadpole of Emei moustache toad (Leptobrachium boringii: Anura:Megophryidae) from the Southwest China and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 2019, 4, 265–266. [Google Scholar] [CrossRef]
- Xiang, Z.Y.; Zhang, C.Y.; Zheng, W.C.; Yu, S.S.; Zhang, L.; Ding, G.H. Characterization of the complete mitochondrial genome of the Chong’an Moustache Toad, Leptobrachium liui (Pope, 1947) with a phylogenetic analysis of Megophryidae. Mitochondrial DNA Part B Resour. 2021, 6, 1061–1063. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, D.; Patrick, M.; Guillaume, S. NOVOPlasty: De Novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed]
- Zardoya, R.; Meyer, A. Phylogenetic performance of mitochondrial protein-coding genes in resolving relationships among vertebrates. Mol. Biol. Evol. 1996, 13, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.Y.; Chang, H.H.; Yuan, H.; Huang, Y.; Lu, H.M.; Li, X.; Gou, X.C. Comparative mitochondrial genomes of four species of Sinopodisma and phylogenetic implications (Orthoptera, Melanoplinae). ZooKeys 2020, 969, 23–42. [Google Scholar]
- Bao, X.L.; Cui, L.; Li, D.Y.; Fan, X.L.; Yang, M.Y.; Yang, D.Y.; Ni, Q.Y.; Yao, Y.F.; Xu, H.L.; Zeng, B.; et al. The near complete mitochondrial genome of Oreolalax schmidti (Anura: Megophryidae). Mitochondrial DNA Part B Resour. 2020, 5, 3536–3537. [Google Scholar] [CrossRef]
- Shu, G.C.; Yu, M.; He, Z.P.; Xie, F.; Liang, X.X. Complete mitochondrial genome of the Alpine Metacarpal-tubercled Toad Leptobrachella alpina (Amphibia, Anura, Megophryidae). Mitochondrial DNA B Resour. 2021, 6, 3242–3243. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Miao, G.P.; Hu, S.J.; Sun, Q.; Ding, H.W.; Ji, Z.C.; Guo, P.; Yan, S.B.; Wang, C.R.; Kan, X.Z.; et al. Quantification and evolution of mitochondrial genome rearrangement in Amphibians. BMC Ecol. Evol. 2021, 21, 19. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Van Dijk, E.L.; Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef]
- Ye, F.; Samuels, D.C.; Clark, T.; Guo, Y. High-throughput sequencing in mitochondrial DNA research. Mitochondrion 2014, 17, 157–163. [Google Scholar] [CrossRef]
- Jex, A.R.; Hall, R.S.; Littlewood, D.T.J.; Gasser, R.B. An integrated pipeline for next-generation sequencing and annotation of mitochondrial genomes. Nucleic Acids Res. 2010, 38, 522–533. [Google Scholar] [CrossRef]
- Schuster, S.C. Next-generation sequencing transforms today’s biology. Nat. Methods 2008, 5, 16–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Lan, X.Y.; Luo, Q.H.; Gu, Z.R.; Zhou, Q.; Zhang, M.Y.; Zhang, Y.X.; Jiang, W.S. Characterization, Comparison of Two New Mitogenomes of Crocodile Newts Tylototriton (Caudata: Salamandridae), and Phylogenetic Implications. Genes 2022, 13, 1878. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.X.; Shu, G.C.; Wang, B.; Jiang, J.P.; Cheng, L.; Xie, F. Complete mitochondrial genome of the Leishan moustache toad, Vibrissaphora leishanensis (Anura: Megophryidae). Mitochondrial DNA B Resour. 2016, 1, 275–276. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.D.; Labthavikul, S.T.; Otey, C.R.; Arnold, F.H. Protein stability promotes evolvability. Proc. Natl. Acad. Sci. USA 2006, 103, 5869–5874. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.H.; Bielawski, J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000, 15, 496–503. [Google Scholar] [CrossRef]
- Hurst, L.D. The Ka/Ks ratio: Diagnosing the form of sequence evolution. Trends Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef] [PubMed]
- Hanada, K.; Shiu, S.H.; Li, W.H. The nonsynonymous/synonymous substitution rate ratio versus the radical/conservative replacement rate ratio in the evolution of mammalian genes. Mol. Biol. Evol. 2007, 24, 2235–2241. [Google Scholar] [CrossRef]
- Yang, Y.W. The Genetic Diversity and Geographical Differentiation of V. boringii. Master’s Thesis, Guizhou Normal University, Guizhou, China, 2014. [Google Scholar]
- Hu, M.L. Research on Population Demographic History of Four Species in Vibrissaphora in Mountain Region of South China. Master’s Thesis, Central China Normal University, Wuhan, China, 2015. [Google Scholar]
- Chen, W.C.; Zhang, W.; Zhou, S.C.; Li, N.; Huang, Y.; Mo, Y.M. Insight into the validity of Leptobrachium guangxiense (Anura: Megophryidae): Evidence from mitochondrial DNA sequences and morphological characters. Zootaxa 2013, 3641, 31–40. [Google Scholar] [CrossRef]
- Hou, X.J.; Lin, H.D.; Tang, W.Q.; Liu, D.; Han, C.C.; Yang, J.Q. Complete mitochondrial genome of the freshwater fish Acrossocheilus longipinnis (Teleostei: Cyprinidae): Genome characterization and phylogenetic analysis. Biologia 2020, 75, 1871–1880. [Google Scholar] [CrossRef]
- Zhang, M.Y.; Zhou, Q.; Xiang, H.M.; Wang, J.X.; Lan, X.Y.; Luo, Q.H.; Jiang, W.S. Complete mitochondrial genome of Rectoris luxiensis (Teleostei, Cyprinidae): Characterisation and phylogenetic implications. Biodivers. Data J. 2023, 11, e96066. [Google Scholar] [CrossRef] [PubMed]

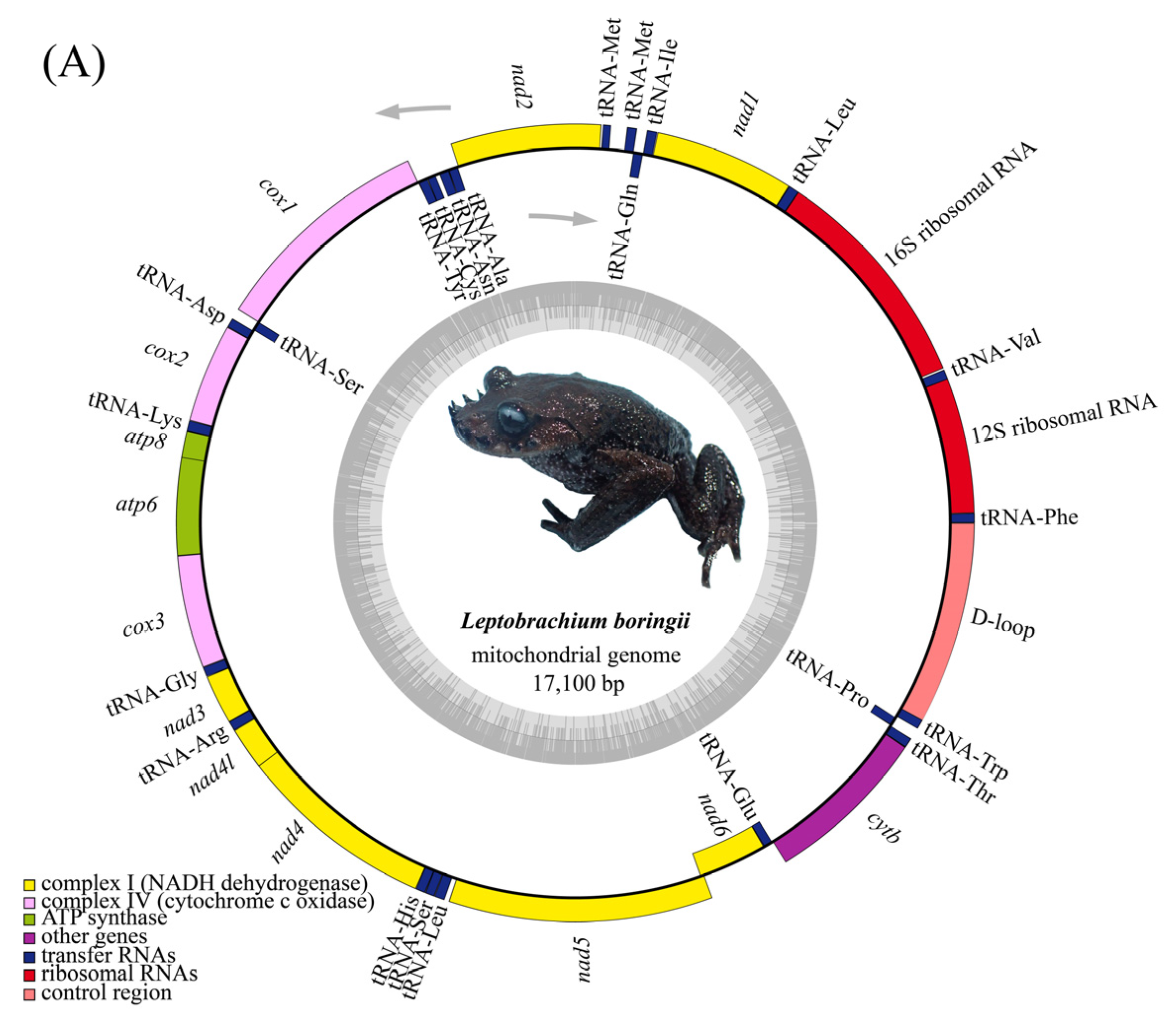
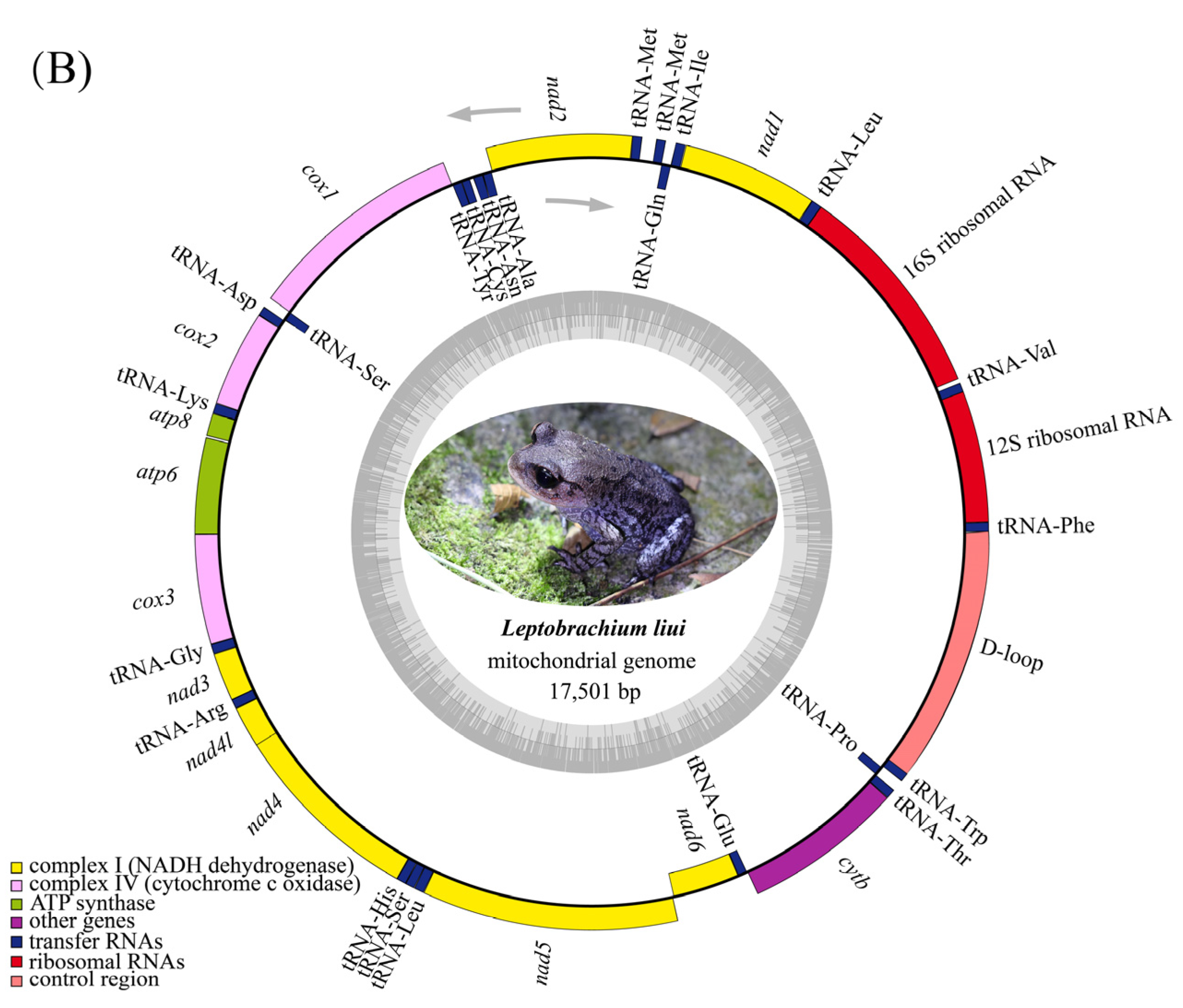




| Gene | Position | Length (bp) | Start Codon | Stop Codon | Anti Codon | Strand * | Intergenic Nucleotide # | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LB | LL | LB | LL | LB | LL | LB | LL | LB | LL | |||
| tRNAPhe | 1–67 | 1–67 | 67 | 67 | GAA | H | ||||||
| 12S rRNA | 68–1007 | 68–1004 | 940 | 937 | H | |||||||
| tRNAVal | 1004–1072 | 1001–1069 | 69 | 69 | TAC | H | −4 | −4 | ||||
| 16S rRNA | 1083–2662 | 1099–2661 | 1580 | 1563 | H | 10 | 29 | |||||
| tRNALeu | 2661–2735 | 2660–2734 | 75 | 75 | TAA | H | −2 | −2 | ||||
| nad1 | 2736–3709 | 2735–3708 | 974 | 974 | ATG | ATG | TA+ | TA+ | H | |||
| tRNAIle | 3712–3782 | 3711–3781 | 71 | 71 | CAT | H | 2 | 2 | ||||
| tRNAGln | 3782–3852 | 3781–3851 | 71 | 71 | TTG | L | −1 | −1 | ||||
| tRNAMet1 | 3852–3920 | 3851–3919 | 69 | 69 | CAT | H | −1 | −1 | ||||
| tRNAMet2 | 4029–4085 | 4019–4086 | 57 | 68 | CAT | H | 108 | 99 | ||||
| nad2 | 4096–5139 | 4087–5130 | 1044 | 1044 | ATG | ATG | TAA | TAA | H | 10 | ||
| tRNAAla | 5144–5213 | 5135–5204 | 70 | 70 | TGC | L | 4 | 4 | ||||
| tRNAAsn | 5214–5286 | 5205–5277 | 73 | 73 | GTT | L | ||||||
| NCR | 5289–5316 | 5280–5307 | 28 | 28 | H | 2 | 2 | |||||
| tRNACys | 5316–5379 | 5307–5370 | 64 | 64 | GCA | L | −1 | −1 | ||||
| tRNATyr | 5380–5449 | 5371–5440 | 70 | 70 | GTA | L | ||||||
| cox1 | 5451–7013 | 5442–7004 | 1563 | 1563 | GTG | GTG | TAA | TAA | H | 1 | 1 | |
| tRNASer | 7011–7081 | 7002–7072 | 71 | 71 | TGA | L | −3 | −3 | ||||
| tRNAAsp | 7086–7153 | 7077–7144 | 68 | 68 | GTC | H | 4 | 4 | ||||
| cox2 | 7154–7841 | 7145–7832 | 688 | 688 | ATG | ATG | T++ | T++ | H | |||
| tRNALys | 7842–7915 | 7833–7906 | 74 | 74 | TTT | H | ||||||
| atp8 | 7916–8101 | 7907–8071 | 186 | 165 | GTG | GTG | TAA | CAT | H | |||
| atp6 | 8092–8774 | 8083–8765 | 683 | 683 | ATG | ATG | TA+ | TA+ | H | −10 | 11 | |
| cox3 | 8774–9557 | 8765–9548 | 784 | 784 | ATG | ATG | T++ | T++ | H | −1 | −1 | |
| tRNAGly | 9558–9626 | 9549–9617 | 69 | 69 | TCC | H | ||||||
| nad3 | 9627–9971 | 9618–9962 | 345 | 345 | GTG | GTG | TAA | TAA | H | |||
| tRNAArg | 9970–10,038 | 9961–10,029 | 69 | 69 | TCG | H | −2 | −2 | ||||
| nad4l | 10,039–10,335 | 10,030–10,326 | 297 | 297 | ATG | ATG | TAA | TAA | H | |||
| nad4 | 10,329–11,706 | 10,320–11,697 | 1378 | 1378 | ATG | GTG | T++ | T++ | H | −7 | −7 | |
| tRNAHis | 11,707–11,775 | 11,698–11,766 | 69 | 69 | GTG | H | ||||||
| tRNASer | 11,776–11,842 | 11,767–11,833 | 67 | 67 | GCT | H | ||||||
| tRNALeu | 11,843–11,915 | 11,834–11,906 | 73 | 73 | TAG | H | ||||||
| nad5 | 11,947–13,779 | 11,908–13,737 | 1833 | 1830 | ATG | ATG | TAA | AGG | H | 31 | 1 | |
| nad6 | 13,752–14,261 | 13,743–14,252 | 510 | 510 | ATG | ATG | AGG | AGG | L | −28 | 5 | |
| tRNAGlu | 14,262–14,330 | 14,253–14,321 | 69 | 69 | TTC | L | ||||||
| cytb | 14,335–15,475 | 14,326–15,466 | 1141 | 1141 | ATG | ATG | T++ | T++ | H | 4 | 4 | |
| tRNAThr | 15,476–15,545 | 15,467–15,536 | 70 | 70 | TGT | H | ||||||
| tRNAPro | 15,548–15,616 | 15,539–15,607 | 69 | 69 | TGG | L | 2 | 2 | ||||
| tRNATrp | 15,626–15,694 | 15,618–15,686 | 69 | 69 | TCA | H | 9 | 10 | ||||
| D-loop | 15,695–17,100 | 15,687–17,501 | 1406 | 1815 | H | 0 | 0 | |||||
| Species | Total Length (bp) | T% | C% | G% | A% | A+T% | AT Skew | GC Skew | Accession Number |
|---|---|---|---|---|---|---|---|---|---|
| L. boringii (SZ) | 17,100 | 31.5 | 25.5 | 15.3 | 27.7 | 59.2 | −0.064 | −0.250 | OP373724 * |
| L. boringii (EM) | 17,085 | 31.5 | 25.5 | 15.4 | 27.6 | 59.1 | −0.066 | −0.247 | KJ630505 |
| L. boringii (PS) | 16,557 | 31.6 | 25.5 | 15.2 | 27.7 | 59.3 | −0.066 | −0.253 | MH643882 |
| L. liui (WLY) | 17,501 | 32.7 | 24.3 | 14.8 | 28.1 | 60.8 | −0.076 | −0.243 | OP503540 * |
| L. liui (JLS) | 17,190 | 32.6 | 24.4 | 14.9 | 28.1 | 60.7 | −0.074 | −0.242 | MW429348 |
| Oreolalax major | 17,786 | 32.6 | 24.3 | 14.3 | 28.7 | 61.3 | −0.064 | −0.259 | MN803320 |
| O. major | 15,469 | 32.1 | 24.7 | 14.4 | 28.7 | 60.8 | −0.056 | −0.263 | KU310894 |
| O. major | 17,431 | 32.4 | 24.5 | 14.4 | 28.8 | 61.2 | −0.059 | −0.260 | KU127230 |
| Oreolalax xiangchengensis | 17,110 | 33.0 | 23.6 | 14.2 | 29.2 | 62.2 | −0.061 | −0.249 | MH727696 |
| Oreolalax jingdongensis | 17,864 | 32.7 | 23.9 | 14.3 | 29.1 | 61.8 | −0.058 | −0.251 | MF953479 |
| Oreolalax omeimontis | 17,675 | 32.6 | 25.0 | 14.0 | 28.5 | 61.1 | −0.067 | −0.282 | MN803321 |
| Oreolalax multipunctatus | 17,358 | 32.0 | 24.2 | 14.3 | 28.5 | 60.5 | −0.058 | −0.257 | MF966382 |
| Oreolalax lichuanensis | 17,702 | 32.2 | 24.9 | 15.0 | 28.0 | 60.2 | −0.070 | −0.248 | KU096847 |
| Oreolalax schmidti | 18,481 | 32.8 | 24.5 | 14.4 | 28.3 | 61.1 | −0.074 | −0.260 | MT773151 |
| Oreolalax rhodostigmatus | 18,676 | 32.4 | 24.9 | 14.7 | 28.0 | 60.4 | −0.073 | −0.258 | MF770485 |
| Leptobrachium ailaonicum | 17,318 | 31.8 | 25.0 | 15.3 | 27.9 | 59.7 | −0.065 | −0.241 | MZ394043 |
| Leptobrachium leishanense | 17,485 | 32.6 | 24.4 | 14.9 | 28.1 | 60.7 | −0.074 | −0.242 | KU760082 |
| Scutiger ningshanensis | 17,265 | 32.8 | 24.2 | 14.0 | 29.1 | 61.9 | −0.060 | −0.267 | KX619450 |
| S. ningshanensis | 16,799 | 32.6 | 24.3 | 14.3 | 28.8 | 61.4 | −0.062 | −0.259 | KX352260 |
| Scutiger liupanensis | 16,890 | 32.2 | 24.9 | 14.5 | 28.4 | 60.6 | −0.063 | −0.264 | KX352261 |
| Leptobrachella oshanensis | 17,747 | 29.9 | 26.3 | 15.2 | 28.8 | 58.7 | −0.019 | −0.267 | KC460337 |
| Leptobrachella alpina | 17,763 | 30.8 | 25.6 | 15.1 | 28.5 | 59.3 | −0.039 | −0.258 | MW487804 |
| Leptobrachella pelodytoides | 14,682 | 29.1 | 27.8 | 15.5 | 27.7 | 56.8 | −0.025 | −0.284 | JX564874 |
| Atympanophrys shapingensis | 17,631 | 31.5 | 26.0 | 14.3 | 28.2 | 59.7 | −0.055 | −0.290 | JX458090 |
| Megophrys gigantica | 18,259 | 32.1 | 25.2 | 14.3 | 28.4 | 60.5 | −0.061 | −0.276 | MZ364157 |
| Brachytarsophrys carinense | 15,271 | 29.8 | 27.6 | 15.1 | 27.5 | 57.3 | −0.040 | −0.293 | JX564854 |
| Boulenophrys jingganggensis | 17,263 | 31.6 | 26.3 | 14.5 | 27.6 | 59.2 | −0.068 | −0.289 | MT683772 |
| Boulenophrys pingjianggensis | 17,866 | 32.0 | 25.9 | 14.3 | 27.8 | 59.8 | −0.070 | −0.289 | KT601071 |
| Boulenophrys omeimontis | 17,013 | 31.8 | 25.7 | 14.2 | 28.3 | 60.1 | −0.058 | −0.288 | KP728257 |
| Length (bp) | T% | C% | G% | A% | A+T% | AT Skew | GC Skew | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LB | LL | LB | LL | LB | LL | LB | LL | LB | LL | LB | LL | LB | LL | LB | LL | |
| nad1 | 974 | 974 | 34.7 | 35.3 | 24.8 | 23.8 | 14.8 | 14.5 | 25.7 | 26.4 | 60.4 | 61.7 | −0.149 | −0.145 | −0.253 | −0.244 |
| nad2 | 1044 | 1044 | 32.3 | 32.6 | 29.7 | 29.4 | 12.1 | 12.2 | 26.0 | 25.9 | 58.3 | 58.4 | −0.108 | −0.115 | −0.421 | −0.415 |
| cox1 | 1563 | 1563 | 31.6 | 33.1 | 24.7 | 22.5 | 18.0 | 17.8 | 25.7 | 26.6 | 57.3 | 59.7 | −0.103 | −0.110 | −0.157 | −0.117 |
| cox2 | 688 | 688 | 31.4 | 32.6 | 25.4 | 24.3 | 15.7 | 15.4 | 27.5 | 27.8 | 58.9 | 60.3 | −0.065 | −0.080 | −0.236 | −0.223 |
| atp8 | 186 | 165 | 38.7 | 43.6 | 24.2 | 20.6 | 10.2 | 9.7 | 26.9 | 26.1 | 65.6 | 69.7 | −0.180 | −0.252 | −0.407 | −0.360 |
| atp6 | 683 | 683 | 35.7 | 36.0 | 26.2 | 25.3 | 13.6 | 12.0 | 24.5 | 26.6 | 60.2 | 62.7 | −0.186 | −0.150 | −0.317 | −0.357 |
| cox3 | 784 | 784 | 33.9 | 33.8 | 25.1 | 25.8 | 16.7 | 16.8 | 24.2 | 23.6 | 58.1 | 57.4 | −0.167 | −0.178 | −0.201 | −0.210 |
| nad3 | 345 | 345 | 38.3 | 39.1 | 25.2 | 23.8 | 15.4 | 14.2 | 21.2 | 22.9 | 59.5 | 62.0 | −0.287 | −0.262 | −0.241 | −0.252 |
| nad4l | 297 | 297 | 35.4 | 37.4 | 27.9 | 25.9 | 12.8 | 12.8 | 23.9 | 23.9 | 59.3 | 61.3 | −0.194 | −0.220 | −0.371 | −0.339 |
| nad4 | 1378 | 1378 | 33.4 | 35.2 | 28.4 | 26.1 | 12.7 | 12.3 | 25.5 | 26.4 | 58.9 | 61.6 | −0.134 | −0.143 | −0.382 | −0.357 |
| nad5 | 1833 | 1830 | 33.8 | 34.8 | 26.0 | 25.0 | 13.1 | 12.9 | 27.0 | 27.4 | 60.8 | 62.1 | −0.112 | −0.119 | −0.330 | −0.319 |
| nad6 | 510 | 510 | 35.1 | 35.5 | 12.7 | 12.9 | 33.7 | 32.9 | 18.4 | 18.6 | 53.5 | 54.1 | −0.312 | −0.312 | 0.453 | 0.436 |
| cytb | 1141 | 1141 | 34.1 | 35.1 | 27.0 | 26.1 | 14.7 | 13.7 | 24.2 | 25.1 | 58.3 | 60.2 | −0.170 | −0.167 | −0.295 | −0.313 |
| PCGs | 11,426 | 11,402 | 33.7 | 34.7 | 25.8 | 24.6 | 15.3 | 14.9 | 25.2 | 25.8 | 58.9 | 60.5 | −0.144 | −0.148 | −0.255 | −0.246 |
| PCGs–1st | 3809 | 3801 | 33.9 | 35.0 | 27.0 | 26.0 | 14.9 | 14.3 | 24.2 | 24.6 | 58.1 | 59.6 | −0.167 | −0.174 | −0.289 | −0.290 |
| PCGs–2nd | 3809 | 3801 | 34.7 | 35.4 | 26.0 | 24.9 | 14.2 | 14.3 | 25.2 | 25.4 | 59.9 | 60.8 | −0.159 | −0.164 | −0.294 | −0.270 |
| PCGs–3rd | 3808 | 3800 | 32.6 | 33.7 | 24.3 | 22.9 | 16.8 | 16.1 | 26.2 | 27.3 | 58.8 | 61.0 | −0.109 | −0.105 | −0.182 | −0.174 |
| 12S rRNA | 940 | 937 | 25.0 | 25.5 | 23.7 | 23.5 | 19.9 | 19.9 | 31.4 | 31.2 | 56.4 | 56.7 | 0.113 | 0.100 | −0.087 | −0.084 |
| 16S rRNA | 1580 | 1563 | 27.0 | 27.2 | 21.8 | 21.8 | 17.5 | 17.1 | 33.7 | 34.0 | 60.7 | 61.2 | 0.110 | 0.111 | −0.109 | −0.120 |
| rRNAs | 2520 | 2500 | 26.3 | 26.6 | 22.5 | 22.4 | 18.2 | 18.1 | 33.0 | 32.9 | 59.3 | 59.5 | 0.113 | 0.107 | −0.106 | −0.106 |
| tRNAs | 1536 | 1536 | 28.6 | 28.6 | 20.4 | 20.8 | 22.1 | 21.7 | 28.8 | 29.0 | 57.4 | 57.6 | 0.003 | 0.007 | 0.040 | 0.021 |
| D-loop | 1406 | 1815 | 35.1 | 38.8 | 21.2 | 19.4 | 13.8 | 12.8 | 29.9 | 29.0 | 65.0 | 67.8 | −0.080 | −0.145 | −0.211 | −0.205 |
| Mitogenome | 17,100 | 17,501 | 31.5 | 32.7 | 25.5 | 24.3 | 15.3 | 14.8 | 27.7 | 28.1 | 59.2 | 60.8 | −0.064 | −0.076 | −0.250 | −0.242 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Q.; Xiang, H.-M.; Zhang, M.-Y.; Liu, Y.; Gu, Z.-R.; Lan, X.-Y.; Wang, J.-X.; Jiang, W.-S. Two Complete Mitochondrial Genomes of Leptobrachium (Anura: Megophryidae: Leptobrachiinae): Characteristics, Population Divergences, and Phylogenetic Implications. Genes 2023, 14, 768. https://doi.org/10.3390/genes14030768
Zhou Q, Xiang H-M, Zhang M-Y, Liu Y, Gu Z-R, Lan X-Y, Wang J-X, Jiang W-S. Two Complete Mitochondrial Genomes of Leptobrachium (Anura: Megophryidae: Leptobrachiinae): Characteristics, Population Divergences, and Phylogenetic Implications. Genes. 2023; 14(3):768. https://doi.org/10.3390/genes14030768
Chicago/Turabian StyleZhou, Qiang, Hong-Mei Xiang, Ming-Yao Zhang, Ying Liu, Zhi-Rong Gu, Xiang-Ying Lan, Jin-Xiu Wang, and Wan-Sheng Jiang. 2023. "Two Complete Mitochondrial Genomes of Leptobrachium (Anura: Megophryidae: Leptobrachiinae): Characteristics, Population Divergences, and Phylogenetic Implications" Genes 14, no. 3: 768. https://doi.org/10.3390/genes14030768
APA StyleZhou, Q., Xiang, H.-M., Zhang, M.-Y., Liu, Y., Gu, Z.-R., Lan, X.-Y., Wang, J.-X., & Jiang, W.-S. (2023). Two Complete Mitochondrial Genomes of Leptobrachium (Anura: Megophryidae: Leptobrachiinae): Characteristics, Population Divergences, and Phylogenetic Implications. Genes, 14(3), 768. https://doi.org/10.3390/genes14030768






