Abstract
The Lygaeoidea comprise about 4660 species in 790 genera and 16 families. Using standard chromosome staining and FISH with 18S rDNA and telomeric (TTAGG)n probes, we studied male karyotypes and meiosis in 10 species of Lygaeoidea belonging to eight genera of the families Blissidae, Cymidae, Heterogastridae, Lygaeidae, and Rhyparochromidae. Chromosome numbers were shown to range from 12 to 28, with 2n = 14 being predominant. All species have an XY system and all but one have a pair of m-chromosomes. The exception is Spilostethus saxatilis (Lygaeidae: Lygaeinae); in another species of Lygaeinae, Thunbergia floridulus, m-chromosomes were present, which represents the first finding for this subfamily. All species have an inverted sequence of sex chromosome divisions (“post-reduction”). The 18S rDNA loci were observed on one or both sex chromosomes in Kleidocerys resedae and Th. floridulus, respectively (Lygaeidae), while on an autosomal bivalent in all other species. The rDNA loci tended to be close to the end of the chromosome. Using (TTAGG)n—FISH, we were able to show for the first time that the Lygaeoidea lack the canonical “insect” telomere motif (TTAGG)n. We speculate that this ancestral motif is absent from the entire infraorder Pentatomomorpha being replaced by some other telomere repeat motif sequences.
1. Introduction
The hemipteran superfamily Lygaeoidea (Heteroptera: Pentatomomorpha) is one of the largest and most diverse true bug groups comprising more than 4660 described species worldwide classified into about 790 genera and 16 families [1,2]. The superfamily is one of the cytogenetically better-studied groups of Pentatomomorpha, with basic cytogenetic data available for about 440 species [3,4,5,6,7,8,9,10,11,12]. The studied species represent about 9% of the total number of described lygaeoid species. Most of the species belong to the families Rhyparochromidae (175 species), Lygaeidae (128), and Blissidae (41), while for other families, data are sparse (Artheneidae, Berytidae, Colobathristidae, Cymidae, Heterogastridae, Geocoridae, Malcidae, Oxycarenidae, Pachygronthidae, and Piesmatidae) or absent altogether (Cryptorhamphidae, Meschiidae, and Ninidae).
Based on the current state of knowledge, lygaeoids exhibit a wide cytogenetic diversity in chromosome numbers and sex chromosome systems, sometimes even between closely related species. In general, chromosome numbers range from 10 to 46 (with some gaps) in diploid karyotypes; however, these extreme counts are not characteristic of the Lygaeoidea being found only sporadically, the lowest in Rhyparochromidae and Arhteneidae, while the highest in Berytidae [3,6,13,14]. In different lygaeoid families, both simple sex chromosome systems, XX/X(0) and XX/XY, and multiple systems, XnXn/XnY and XX/XYn, occur. In addition, a pair of m-chromosomes is encountered in all explored lygaeoid families, but are not found in the Berytidae and Piesmatidae families [8,14,15]. Despite the aforementioned diversity, the great majority of lygaeoids have 2n = 16, XY or 14, XY, and in many cases, the 16-chromosome karyotypes appear to be derivative of the original ones with 2n = 14, XY [4]. The course of male meiosis in lygaeoids conforms to the general heteropteran type as described in [3], being characterized, among other things, by the so-called sex chromosome “post-reduction”, with the sex chromosomes segregating equationally during the first round of meiosis, while separating reductionally during the second round of meiosis.
With rare exceptions, karyotype analysis was done using the routine chromosomal staining technique. Just a few lygaeoid species have been studied after DAPI/CMA3 fluorochrome staining and/or chromosome banding, including C-banding or NOR-banding [8,9,11,16,17,18]. Fluorescence in situ hybridization (FISH) has proven to be an effective and precise tool for physical mapping specific DNA sequences throughout the chromosomes [19]. There are also isolated examples of using FISH to detect and localize the major rDNA (28/26S, 18S, and 5.8S, spliced from a single precursor) on chromosomes of some lygaeoid species [11,17]. In all cases reported, rDNA loci were detected on one of the autosomal bivalents, but presumably not the same in different species. Finally, Grozeva et al. [17] undertook a dot-blotting experiment on Oxycarenus lavaterae (Fabricius, 1787) (Oxycarenidae) with different telomeric probes, the “insect” (TTAGG)n probe, and six alternative telomerase-based repeats; however, none of the tested probes hybridized with telomeres of the studied species.
In the present research, we obtained new data for ten species belonging to five families of Lygaeoidea. We studied for the first time the standard karyotypes of Elasmolomus squalidus (Gmelin, 1790) from the family Rhyparochromidae; Nerthus sp. 1 and Nerthus sp. 2 from the Heterogastridae; Thunbergia floridulus (Distant, 1918) and Nysius cymoides (Spinola, 1837) from the Lygaeidae. To identify new cytogenetic markers, we used FISH with the 18S rDNA probe to characterize the distribution profile of 18S rDNA in all but one (Nerthus sp. 1) above species and, additionally, in Cymus claviculus (Fallen, 1807) (Cymidae), Dimorphopterus spinolae (Signoret, 1857) (Blissidae), Kleidocerys resedae (Panzer, 1793), Spilostethus saxatilis (Scopoli, 1763), and Nysius helveticus (Herrich-Schäffer, 1850) (Lygaeidae). It is currently believed that Pentatomomoprha as a whole lack the canonical “insect” telomeric repeat TTAGG and, most likely, have a different mechanism of telomere elongation [20,21]; however, data for the superfamily Lygaeoidea, with the exception of the aforementioned dot-blotting experiment, are still missing. To fill this gap, we performed FISH mapping of the insect-type telomeric sequence (TTAGG)n on the chromosomes of all species explored.
2. Materials and Methods
The material used for karyotype analysis is listed in Table 1. Specimens (only males) were fixed immediately after collection in ethanol/acetic acid fixative (3:1) and stored in the fixative at 4 °C until slides were made. Species identification was made by Viktor B. Golub and Dmitry A. Gapon. Chromosome preparations were obtained from the testes of adult males using a squash technique as described elsewhere (e.g., [22]). The number of specimens karyotyped varied from one to three in the studied species, and several slides were prepared from the testes of each male. The standard karyotypes were studied after staining by the Schiff–Giemsa method [22,23]. To study the chromosomal distribution of the major rDNA, and to determine if the species display or not the “insect” telomere (TTAGG)n motif, double-target FISH with a firebug (Pyrrhocoris apterus (Linneus, 1758)) 18S rDNA probe and the (TTAGG)n probe was carried out according to the protocol described by Grozeva et al. [24]. As a positive control for the TTAGG telomeric probe, a barklouse (Psocomorpha) species, Psococerastis gibbosa (Sulzer, 1766), which is proven to be TTAGG-positive [25], was used. The whole procedure can be found in Golub et al. [23,25]. Standard preparations were photographed under oil immersion (X100 objective) using an Olympus BX 51 light microscope with an Olympus C-35 AD-4 camera (Olympus Optical Co., Tokyo, Japan). FISH preparations were photographed under oil immersion (X100 objective) using a Leica DM 6000 B microscope, Leica DFC 345 FX camera, and Leica Application Suite 3.7 software with an Image Overlay module (Leica Microsystems, Wetzlar, Germany). The filter sets applied were A, L5, and N21 (Leica Microsystems). The specimens, from which chromosome preparations were made, and the preparations themselves are stored at the Zoological Institute RAS (St. Petersburg, Russia).

Table 1.
Localities of the material used for chromosome analysis.
3. Results
3.1. Family Rhyparochromidae
Elasmolomus squalidus, 2n = 12(8A + 2m + XY) (Figure 1a,b)
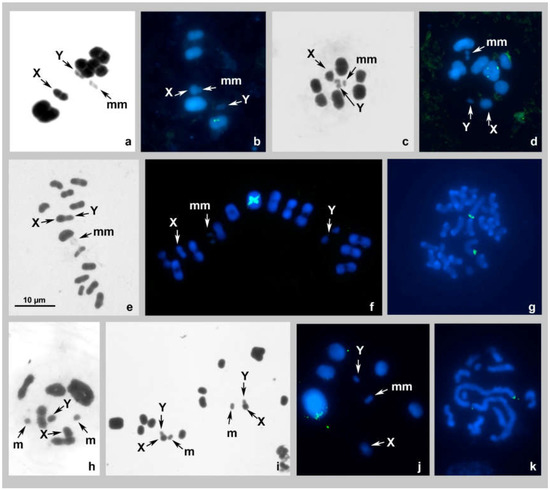
Figure 1.
(a–k). Karyotypes of species of the families Rhyparochromidae (a,b), Heterogastridae (c,d), Cymidae (e–g), and Blissidae (h–k) after Schiff–Giemsa and double-target FISH with 18S rDNA and telomeric (TTAGG)n probes. (a,b) Elasmolomus squalidus MI standard staining (a), MI FISH (b); (c) Nerthus sp. 1 MI standard staining; (d) Nerthus sp. 2 MI FISH; (e–g) Cymus claviculus MI standard staining (e), MI FISH (f) spermatogonial metaphase FISH (g); (h–k) Dimorphopterus spinolae late diakinesis standard staining (h), MII standard staining (i), MI FISH (j), and spermatogonial metaphase FISH (k). 18S rDNA signals are green; TTAGG signals are absent. Bar = 10 µm in all figures.
The karyotype was studied for the first time. At spermatocyte metaphase I (MI), there are five bivalents of autosomes, including a very small and negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y (meioformula: n = 4AA + mm + X + Y). One of the bivalents is noticeably larger than the others, which, with the exception of the m-chromosomes, make up a decreasing size range. The sex chromosomes are of different sizes—the larger one is taken as the X and the smaller one as the Y; they are located separately from each other, and each is split into chromatids (Figure 1a,b). The FISH using the 18S rDNA probe provided distinct signals on the second-largest bivalent (AA2); their intrachromosomal location remained unclear; no signals of the TTAGG telomeric probe were detected (Figure 1b).
3.2. Family Heterogastridae
3.2.1. Nerthus sp. 1, 2n = 16(12A + 2m + XY) (Figure 1c)
The karyotype was studied for the first time; only Schiff-Giemsa staining was applied. At spermatocyte MI, there are 7 bivalents of autosomes, including a negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y (meioformula: n = 6AA + mm + X + Y). The autosomal bivalents, with the exception of the m-chromosomes, make up a decreasing size range. The sex chromosomes are of different sizes—they are located separately from each other, and each is split into chromatids (Figure 1c).
3.2.2. Nerthus sp. 2, 2n = 18(14A + 2m + XY) (Figure 1d)
The karyotype was studied for the first time and only the FISH technique was applied. At spermatocyte MI, there are eight bivalents of autosomes, including a negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y (meioformula: n = 7AA + mm + X + Y). The autosomal bivalents, with the exception of the m-chromosomes, make up a decreasing size range. The sex chromosomes are of different size; they are located separately from each other and each is split into chromatids. FISH using the 18S probe provided distinct signals on the largest bivalent (AA1); their intrachromosomal location remained unclear; no signals of the TTAGG telomeric probe were detected (Figure 1d).
3.3. Family Cymidae
Cymus claviculus, 2n = 28(24A + 2m + XY) (Figure 1e–g)
The karyotype was previously studied by Pfaler-Collander from Finland [26], and our observations corroborate with those data. At spermatocyte MI, there are 13 bivalents of autosomes, including a negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y (meioformula: n = 12AA + mm + X + Y). The autosomal bivalents, with the exception of the m-chromosomes, make up a decreasing size range. The sex chromosomes are of different size sizes—they are split into chromatids and could be seen either next to each other (Figure 1e) or separately from one another (Figure 1f). FISH using the 18S probe provided distinct terminal signals on the AA1 bivalent at MI (Figure 1f) and on the two largest chromosomes in a spermatogonial cell (Figure 1g); no signals of the TTAGG telomeric probe were detected (Figure 1f,g).
3.4. Family Blissidae
Dimorphopterus spinolae 2n = 14(10A + 2m + XY) (Figure 1h–k)
The karyotype of D. spinolae was previously studied by Muramoto [27] who described the karyotype of males from Japan as “2n = 16 (and XY type)”, which most likely means 2n = 16(14 + XY). According to our observations, males collected in Voronezh region (Russia) have a different karyotype. At spermatocyte diakinesis/MI of these males, we observed six bivalents of autosomes, including a minute-sized m-chromosome pair, and two sex chromosomes, X and Y (meioformula: n = 5AA + mm + X + Y). One of the bivalents is noticeably larger than the others, which in turn decrease in size linearly. The sex chromosomes are of different sizes—they are split into chromatids and located separately from one another (Figure 1h,j). The sister metaphases II (MII) show six autosomes each, including the m-chromosome, and an XY-pseudobivalent; this latter and the m-chromosome are located in the center of the radial metaphase plate (Figure 1i). FISH using the 18S probe provided distinct terminal signals on the AA1 bivalent at MI (Figure 1j) and on the two largest chromosomes in spermatogonial cells (Figure 1k); no signals of the TTAGG telomeric probe were detected (Figure 1j,k).
3.5. Family Lygaeidae
3.5.1. Subfamily Ischnorhynchinae
Kleidocerys resedae, 2n = 14(10A + 2m + XY) (Figure 2a–d)
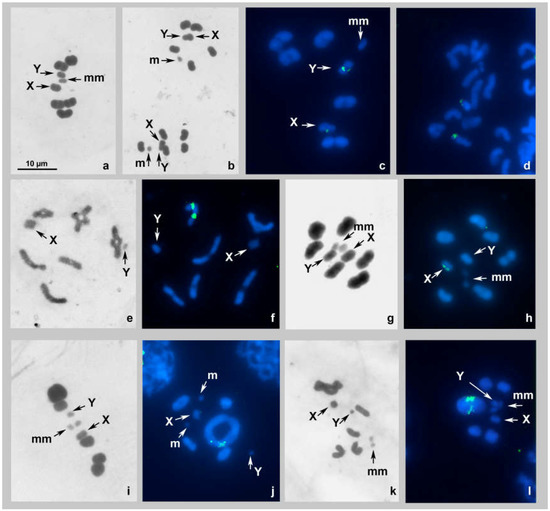
Figure 2.
(a–l). Karyotypes of species of the family Lygaeidae after Schiff–Giemsa and double-target FISH with 18S rDNA and telomeric (TTAGG)n probes. (a–d) Kleidocerys resedae MI standard staining (a), MII standard staining (b), MI FISH (c), spermatogonial metaphase FISH (d); (e,f) Spilostethus saxatilis diakinesis/MI transition standard staining (e), diakinesis/MI transition FISH (f); (g,h) Thunbergia floridulus MI standard staining (g) MI FISH (h); (i,j) Nysius cymoides MI standard staining (i), diakinesis FISH (j); (k,l) Nysius helveticus diakinesis/MI transition standard staining (k), MI FISH (l). 18S rDNA signals are green; TTAGG signals are absent. Bar = 10 µm in all figures.
The karyotype was previously studied by Pfaler-Collander from Finland [26], and our observations corroborate with those data. At spermatocyte MI, there are 6 bivalents of autosomes, including a negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y (meioformula: n = 5AA + mm + X + Y). The autosomal bivalents, with the exception of very small m-chromosomes, are poorly differentiated in size. The sex chromosomes are of different sizes—they are located separately from each other, and each is split into chromatids (Figure 2a,c). The sister metaphases II (MII) show each 6 autosomes, including the m-chromosome, and an XY-pseudobivalent; this latter and the m-chromosome are located in the center of the radial metaphase plate (Figure 2b). FISH using the 18S probe provided distinct terminal signals on each of the sex chromosomes at MI (Figure 2c) and in the spermatogonial nucleus (Figure 2d); no signals of the TTAGG telomeric probe were detected (Figure 2c,d).
3.5.2. Subfamily Lygaeinae
Spilostethus saxatilis 2n = 14(12A + XY) (Figure 2e,f)
The karyotype was previously studied by Geitler [28], and our observations corroborate with those data. During the spermatocyte diakinesis/MI transition (Figure 2e,f), six bivalents of autosomes and two sex chromosomes, X and Y, were observed. No m-chromosomes were detected (meioformula: n = 6AA + X + Y). The bivalents make up a decreasing size range. The sex chromosomes are of different sizes—they are located separately from each other, and each is split into chromatids. FISH using the 18S probe provided distinct interstitial signals on each of the homologues of a medium-sized bivalent (AA); no signals of the TTAGG telomeric probe were detected (Figure 2f).
Thunbergia floridulus 2n = 16(12A + 2m + XY) (Figure 2g,h)
The karyotype was studied for the first time. At spermatocyte MI, there are seven bivalents of autosomes, including a negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y, each being split into the chromatids (meioformula: n = 6AA + mm + X + Y). The bivalents make up a decreasing size range; the sex chromosomes are of similar size and they are located separately from one another (Figure 2g,h). FISH, when using the 18S probe provided distinct signals on the putative X-chromosome; their intrachromosomal location remained unclear; no signals of the TTAGG telomeric probe were detected (Figure 2h).
3.5.3. Subfamily Orsillinae
Nysius cymoides, 2n = 14(10A + 2m + XY) (Figure 2i,j)
The karyotype was studied for the first time. At spermatocyte MI, there are six bivalents of autosomes, including a negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y, each being split into the chromatids (meioformula: n = 5AA + mm + X + Y) (Figure 2i). One of the bivalents is very large, much larger than the second-largest bivalent. The sex chromosomes differ in size, and the putative Y chromosome is quite the same size as m-chromosomes. The latter were found to place separately in prophases (Figure 2j) while forming a bivalent at MI (Figure 2i). Sex chromosomes were located separately from each other at both MI and diakinesis (Figure 2i,j). Fluorescent signals of the 18S probe could be seen at terminal position on each homologue of the AA1 bivalent; no signals of the TTAGG telomeric probe were detected (Figure 2j).
Nysius helveticus, 2n = 14(10A + 2m + XY) (Figure 2k,l)
The karyotype was previously studied by Pfaler-Collander from Finland [26], and our observations corroborate with those data. Like in N. cymoides, at spermatocyte MI of this species, there are six bivalents of autosomes, including a negatively heteropycnotic pair of m-chromosomes, and two sex chromosomes, X and Y, each of which is split into the chromatids (meioformula: n = 5AA + mm + X + Y) (Figure 2k,l). One of the bivalents is very large, much larger than the second-largest bivalent. The sex chromosomes noticeably differ in size, and the smaller one, the putative Y chromosome, is quite the same size as the m-chromosomes. Sex chromosomes were located separately from each other at both diakinesis/MI transition and MI (Figure 2k,l). Fluorescent signals of the 18S probe could be seen at terminal position on each homologue of the AA1 bivalent; no signals of the TTAGG telomeric probe were detected (Figure 2l).
4. Discussion
4.1. Standard Karyotypes
In the present study, we have contributed to cytogenetics of the superfamily Lygaeoidea, the second-largest superfamily in the infraorder Pentatomomorpha, by examining ten different lygaeoid species from five families (Table 2). Our study confirms the previously published information that Cymus claviculus (Cymidae) has 28(24A + 2m + XY), Kleidocerys resedae (Lygaeidae: Ischnorhynchinae) − 14(10A + 2m + XY), Spilostethus saxatilis (Lygaeidae: Lygaeinae) − 14(12A + XY), and Nysius helveticus (Lygaeidae: Orsillinae) − 14(10A + 2m + XY). The high-numbered karyotype in C. claviculus was expected, since the family Cymidae is characterized by one of the highest chromosome numbers in the Lygaeoidea. In this family, chromosome numbers vary between 22 and 30, with the exception of the monotypic genus Ninus Stal, 1860 showing 2n = 16(12A + 2m + XY), which, along with 2n = 14(10A + 2m + XY), is the most commonly found in the family as a whole [3,4,6]. K. resedae and S. saxatilis have the same karyotypes as all other so far studied species within their genera [3,4]. Nysius helveticus has a karyotype found in the vast majority of species of the genus [3,5]. On the other hand, Muramoto [27] recorded Dimorphopterus spinolae in Japan as having “2n = 16 (and XY type)”, which we interpret as 16(14A + XY); however, this does not match our observations of this species karyotype. In all studied males of D. spinolae collected from Voronezh region of Russia, we found 2n = 14(10A + 2m + XY). Since Muramoto did not provide a photograph or a drawing of the karyotype, it is difficult to explain the discrepancy between the data.

Table 2.
Karyotypes of the studied species, with data on chromosome number and diploid karyotype formulae, rDNA loci location, TTAGG repeats, and references. A—autosome, AA—autosomal bivalent.
The karyotypes of five species were studied herein for the first time. Elasmolomus squalidus (Rhyparochromidae) was found to share 2n = 12(8A + 2m + XY) with E. sordidus (Fabricius, 1787) [29] and Elasmolomus sp. [30], while two other species studied in this genus, E. mendicus Stål, 1872 and E. transversus (Signoret, 1860), have 2n = 14(10A + 2m + XY) [4]. In the subfamily Orsillinae (Lygaeidae), Nysius cymoides have 2n = 14(10A + 2m + XY), just like the aforementioned N. helveticus. The same karyotype was previously recorded for all studied species of this genus, with only two exceptions: N. senecionis (Schilling, 1829) and N. tennelus Barber, 1947 were reported to have a karyotype 2n = 22(18A + 2m + XY) diverging much from the modal one in terms of the number of autosomes [3,5].
Our data on Thunbergia floridulus (Lygaeidae: Lygaeinae) and Nerthus spp. (Heterogastridae) are the first for their genera. Th. floridulus was found to have 2n = 16(12A + 2m + XY), and it is the first species with m-chromosomes discovered in the subfamily Lygaeinae. Our data discredit the early idea that this subfamily does not have, unlike all other subfamilies of the Lygaeidae, m-chromosomes [3,5,13]. Moreover, given the presence of m-chromosomes in all families of the Lygaeoidea, we can now suggest that m-chromosomes were already present in the ancestral lygaeoid karyotype and disappeared independently in individual species and higher taxa of this superfamily.
The data obtained herein on the small genus Nerthus Distant, 1909 turned out to be intriguing. The two males collected from an undetermined bush in the Popa mount, Myanmar, and fixed for chromosome analysis were identified as N. dudgeioni Distant, 1909. However, these males showed different karyotypes, 16(14A + 2m + XY) and 18(14A + 2m + XY), thus casting doubt on whether they belong to the same species. At this stage, we therefore designated them as Nerthus sp. 1 and Nerthus sp. 2. The solution to this problem remains a challenge for future research.
Our data cover five different families of the Lygaeoidea and all three recent subfamilies of the largest family Lygaeidae. Despite the small number of studied species, we discovered five different chromosome counts: 12 (1 species), 14 (5), 16 (2), 18 (1), and 28 (1), and six different karyotypes: 12(8A + 2m + XY), 14(12A + XY), 14(10A + 2m + XY), 16(12A + 2m + XY), 18(14A + 2m + XY), and 28(24A + 2m + XY). This indicates that the Lygaeoidea have a wide diversity in chromosome numbers, which is consistent with previous analyses that, in total, used far more species than the current research (reviewed in [6]). As discerned in previous reviews, the karyotypes 2n = 14(10A + 2m + XY) and 2 = 16(12A + 2m + XY), which predominate in our study, clearly predominate in the Lygaeoidea as a whole [3,5,6,13,16]. In many cases, the 16-chromosome karyotypes appear to be derivatives of the putative ancestral karyotype with 2n = 14 [5,13].
4.1.1. Spermatocyte Meiosis
As stated in the Introduction, male meiosis in the Heteroptera is unique, showing an inverted sequence of sex chromosome divisions (“post-reduction”), with sex chromosomes undergoing equational separation of sister chromatids during the first division and reductional segregation of sex chromosomes during the second division; the autosomes remain their conventional sequence of divisions [3]. Although exceptions are known and some true bug taxa have sex chromosome “pre-reduction” (see for references [3]; see also, e.g., [12,22,23,31,32]), all the studied species of the Lygaeoidea demonstrate the “post-reduction”. This is also the case with the species studied in the present work, which is most convincingly shown in Dimorphopterus spinolae and Kleidocerys resedae, in which it was possible to trace the behavior of sex chromosomes up to MII. In these species, X- and Y-chromosomes were located separately from each other and split into chromatids at MI (as with all other species). At MII, the X and Y were observed to come together and form a pseudobivalent in the spindle. Unfortunately, in our material, there was not a second anaphase (AII) in which the sex chromosomes segregated, thus demonstrating the whole process called the “touch-and-go pairing” [3].
4.1.2. 45S rDNA-FISH
The 45S major rDNA cistron is known to be highly dynamic in size, number, and distribution of chromosomal loci in the genome. This can be accomplished through different mechanisms, including an increase in the number of copies, chromosomal rearrangements, such as translocations, fusions, and inversions, ectopic recombination between non-homologous chromosomes, unequal exchanges, and transposition of rDNA repeats to new locations followed by amplification or deletion of original loci [33,34]. The number and chromosomal distribution of rDNA loci in the karyotype detected by FISH provide additional chromosomal landmarks useful for understanding the genome structure and evolution in different insect taxa. True bugs have holokinetic chromosomes in which centromeres are absent, so the search for chromosomal markers is of particular importance in this group. In several species, FISH-based karyotyping, specifically rDNA mapping, has pointed to chromosomal differences between species with similar karyotypes [22,35]. In recent years, studies using FISH, mainly for analyzing the 18S region of the major 45S rDNA, have become very popular in true bugs (reviewed in [24,25,36,37,38,39] and references therein). However, such studies have rarely been attempted in the Lygaeoidea. So far, physical mapping of 18S has only been done in Oxycarenus lavaterae (Fabricius, 1787) from the family Oxycarenidae [17] and in three species from the family Lygaeidae, Ochrimnus sagax Brailovsky, 1982, Oncopeltus femoralis Stal, 1874, and Lygaeus peruvianus Brailovsky, 1978, all from the subfamily Lygaeinae [11]. In all these species, 18S loci were detected on autosomal bivalents, and their chromosomal location was not uniform. For example, in O. sagax, it was the largest bivalent, while in all other species, the 18S-bearing bivalent has been identified as “an autosomal pair”, meaning it was most likely not the largest one. It is also stated that O. femoralis and L. peruvianus have an interstitial chromosomal position of 18S repeats.
In the present study, we analyzed 18S chromosomal positions in nine more species (Nerthus sp. 1 has remained unstudied), eight more genera, four more families of the Lygaeoidea, and, in addition, we obtained new data for each of the three subfamilies of the family Lygaeidae (Table 2). Our data demonstrated that the spatial positioning of ribosomal genes in the lygaeoid true bugs is sufficiently diverse. Autosomal location was found to predominate being found in seven of the nine studied species and in each of the families, but the autosomes bearing rRNA genes turned out to be different in different species. Specifically, the rDNA loci are located on a medium-sized bivalent in Spilostethus saxatilis, on the second-largest bivalent in Elasmolomus squalidus and on the largest bivalent in all other species (regardless of the number of chromosomes they possess). Moreover, we have identified two deviant patterns of the rDNA markings, both in the family Lygaeidae, namely, on both X- and Y- chromosomes in K. resedae (Ischnorhynchinae) and on one of the sex chromosomes, supposedly the X, in Th. floridulus (Lygaeinae).
In addition, the intrachromosomal location of rRNA genes does not seem to be the same in different species, although the location close to the chromosome ends predominates. In those species in which we had the opportunity to observe mitotic chromosomes, such as Cymus claviculus, Dimorphopterus spinolae, and Kleidocerys resedae, the ribosomal gene clusters were visible at the terminal position. In the other six species, the analysis was done mainly at MI when the chromosomes are highly condensed, preventing the accurate determination of the position of hybridization signals. However, at least in Spilostethus saxatilis, they seem to be located in an interstitial region in the chromosomes.
The autosomal distribution of rRNA genes is found in different non-related families, such as Heterogastridae, Cymidae, Blissidae, and Lygaeidae, as well as previously studied Oxycarenidae. Despite the limited sample of studied species, we can hypothesize that the autosomal location is an ancestral trait in the superfamily Lygaeoidea. Moreover, the autosomal rDNA location pattern predominates over all other patterns in other families of Pentatomomorpha (see, e.g., [37,40]); therefore, we can assume its ancestry in the infraorder as a whole. The divergent patterns found in our study in the subfamilies Ischnorhynchinae (K. resedae) and Lygaeinae (Th. floridulus) can be regarded as having arisen de novo and independently in the evolution of the family Lygaeidae. Future studies involving more lygaeoid taxa will allow for testing this hypothesis.
In the current literature, it is widely debated whether ribosomal genes are distributed randomly within chromosomes or, on the contrary, their distribution is regulated by some special mechanisms, and the rDNA loci tend, therefore, to occur preferentially in one or another region on the chromosome [41,42]. Recent analyses show that, although rDNA may actually occur in different chromosomal positions, there are some trends in particular groups of animals. For example, within three very large and most extensively studied “monocentric” insect orders, Coleoptera, Hymenoptera, and Orthoptera, the terminal (in the first) and pericentromeric or interstitial (in the last two) locations of the rDNA clusters seem to be more frequent [40,43,44,45]. In insects with holokinetic chromosomes, only two alternative intrachromosomal positions of rDNA, terminal and interstitial, can be expected and both have been detected [46,47,48,49], with terminal position (always on X chromosomes) being a general feature of aphids ([50] and references therein). In the superfamily Lygaeoidea, both patterns were observed or, at least, suspected, although in all more conclusively confirmed cases, it is a terminal location. Some reviews and some original studies (e.g., [24,36,39]) suggest the terminal pattern to be more abundant in true bugs; however, there has never been any targeted study of this problem and, therefore, the question of whether this pattern is a real trend in these insects remains open. As already noted above, all rDNA-FISH analyses in true bugs have almost exclusively been carried out in meiosis, mainly at MI, and the compressed structure and small size of meiotic bivalents could prevent the accurate determination of loci positions in the majority of cases [22].
4.1.3. (TTAGG)n-FISH
In none of the species studied herein, the (TTAGG)n probe labeled any chromosome areas, indicating the absence of the 5 bp repeat (TTAGG)n, the commonest and supposedly an ancestral DNA motif of insect telomeres [20,21]. Although data are only available for nine species, they cover five differently related families of the superfamily Lygaeoidea. No data are yet available for 11 minor families. Earlier, based on data available at that time for only a few species of the family Pentatomidae [17,51,52,53], it was speculated that the insect motif (TTAGG)n is absent (lost) in the entire clade Pentatomomorpha [17,20,53]. Our data reinforce thus this hypothesis. Until recently, the question remained unanswered as to whether this motif had been replaced with any other motif or whether Pentatomomorpha had acquired some an alternative telomerase-independent mechanism of telomere maintenance. Recently, the first chromosome-level telomere-to-telomere genome assemblies have been achieved for Acanthosoma haemorrhoidale (Linnaeus, 1758) from the family Acanthosomatidae and Aelia acuminata (Linnaeus, 1758) from the family Pentatomidae, both from the infraorder Pentatomomorpha, and the distribution of telomere repeat sequences in the assembled genomes of these species was analyzed [54]. The results showed that A. haemorrhoidale and A. acuminata contained at the telomeres the 10 bp repeats, TTAGGGATGG and TTAGGGTGGT, respectively. The DNA-based approach proved to be an efficient tool for identifying yet unknown telomere motifs. Specifically with regard to the Pentatomomorpha true bugs, it is now possible to assume that they have, instead of the consensus “insect” telomere motif (TTAGG)n, some other repeat motifs at their chromosome termini.
5. Conclusions
A study of 10 species of Lygaeoidea (Pentatomomorpha) showed that they have a considerable variety of chromosome numbers ranging from 12 to 28 (specifically, 12–18, and 28) in male diploid karyotypes, with 2n = 14 being predominant. A pair of m-chromosomes is present in all but one species, and m-chromosomes are, for the first time, found in the subfamily Lygaeinae (Lygaeidae). All species have an XY sex chromosome system and an inverted sequence of sex chromosome divisions (“post-reduction”) in male meiosis. The major rDNA can be located on both autosomes and sex chromosomes while the former pattern is predominant. The species studied do not have the “insect” telomere motif (TTAGG)n, which reinforces the hypothesis that this motif is not present in the entire infraorder Pentatomomorpha, being replaced in this group with some other telomere repeats.
Author Contributions
Conceptualization, V.G.K. and N.V.G.; methodology, B.A.A., A.M.-N., N.V.G. and V.G.K.; formal analysis, V.G.K., N.V.G. and A.M.-N.; investigation, V.G.K., N.V.G. and A.M.-N.; writing—original draft preparation, V.G.K. and N.V.G.; writing—review and editing, V.G.K., N.V.G. and A.M.-N.; visualization, B.A.A., N.V.G., A.M.-N. and V.G.K.; supervision, V.G.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon request from the corresponding author.
Acknowledgments
The present study was supported by the state research projects nos. 122031100272-3 and 122031100275-4. We thank Viktor B. Golub (Voronezh State University, Russia) and Dmitry A. Gapon (Zoological Institute RAS, Russia) for collecting and identifying lygaeoid bugs.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Henry, T.J.; Dellapé, P.M.; de Paula, A.S. The big-eyed bugs, chinch bugs, and seed bugs (Lygaeoidea). In True Bugs (Heteroptera) of the Neotropics, Entomology in Focus 2; Panizzi, A.R., Grazia, J., Eds.; Springer: Dordrecht, The Netherlands; Springer: Heidelberg, Germany; Springer: New York, NY, USA; Springer: London, UK, 2015; pp. 459–514. [Google Scholar] [CrossRef]
- Dellapé, P.M.; Henry, T.J. Lygaeoidea Species File. Version 5.0/5.0. Available online: http://Lygaeoidea.SpeciesFile.org/ (accessed on 10 November 2022).
- Ueshima, N. Animal Cytogenetics. Insecta 6. Hemiptera II: Heteroptera; Gebrüder Borntraeger: Berlin/Stuttgart, Germany, 1979; Volume 117, 528p. [Google Scholar]
- Ueshima, N.; Ashlock, P.D. Cytotaxonomy of the Lygaeidae (Hemiptera-Heteroptera). Univ. Kans. Sci. Bull. 1980, 51, 717–801. [Google Scholar] [CrossRef]
- Grozeva, S.; Kuznetsova, V.G. Notes on the karyotypes of some lygaeid bugs (Heteroptera, Pentatomomorpha, Lygaeidae). Folia Biol. 1993, 41, 65–75. [Google Scholar]
- Papeschi, A.G.; Bressa, M.J. Evolutionary cytogenetics in Heteroptera. J. Biol. Res. 2006, 5, 3–21. [Google Scholar]
- Kaur, H.; Suman, V. Chromosomes and their meiotic behavior in two species of Dieuches Dohrn, 1860 (Heteroptera: Lygaeidae: Rhyparochrominae). Comp. Cytogenet. 2009, 3, 43–50. [Google Scholar] [CrossRef]
- Suman, V.; Kaur, H. Meiotic studies in seven Heteropteran species. Cytologia 2012, 77, 311–322. [Google Scholar] [CrossRef]
- Souza, H.V.; Bicudo, H.E.M.C.; Itoyama, M.M. Study of chromosomal and nucleolar aspects in testes of Nysius californicus (Heteroptera-Lygaediae). Genet. Mol. Res. 2007, 6, 33–40. [Google Scholar]
- Souza, H.V.; Castanhole, M.M.U.; Gomes, M.O.; Murakami, A.S.; Souza-Firmino, T.S.; Saran, P.S.; Banho, C.A.; Monteiro, L.D.; Silva, J.C.; Itoyama, M.M. Meiotic behavior of 18 species from eight families of terrestrial Heteroptera. Insect Sci. 2014, 14, 149. [Google Scholar] [CrossRef]
- Bardella, V.B.; Sampaio, T.R.; Venturelli, N.B.; Dias, A.; Giuliano-Caetano, L.; Fernandes, J.A.M.; da Rosa, R. Physical mapping of 18S rDNA and heterochromatin in species of family Lygaeidae (Hemiptera: Heteroptera). Genet. Mol. Res. 2014, 13, 2186–2199. [Google Scholar] [CrossRef]
- Toscani, M.A.; Pigozzi, M.I.; Papeschi, A.G.; Bressa, M.J. Histone H3 methylation and autosomal vs. sex chromosome segregation during male meiosis in Heteroptera. Front. Ecol. Evol. 2022, 10, 836786. [Google Scholar] [CrossRef]
- Grozeva, S.; Kuznetsova, V.G. Karyotypes and some structural properties of the reproductive system of bugs of the subfamily Artheneinae (Heteroptera, Pentatomomorpha, Lygaeidae). Entomol. Rev. 1990, 69, 14–26. [Google Scholar]
- Grozeva, S. Karyotypes, male reproductive system, and abdominal trichobothria of the Berytidae (Heteroptera) with phylogenetic considerations. Syst. Entomol. 1995, 20, 207–216. [Google Scholar] [CrossRef]
- Grozeva, S. Karyotype and structure of the reproductive system in Piesma (Heteroptera, Piesmatidae). Entomol. Rev. 1991, 70, 157–166. [Google Scholar]
- Kaur, H.; Suman, V.; Kaur, R. First report on C-banding and fluorescent banding in species of Dieuches (Rhyparochrominae: Lygaeidae: Heteroptera). Entomol. Res. 2010, 40, 1–7. [Google Scholar] [CrossRef]
- Grozeva, S.; Kuznetsova, V.G.; Anokhin, B.A. Karyotypes, male meiosis and comparative FISH mapping of 18S ribosomal DNA and telomeric (TTAGG)n repeat in eight species of true bugs (Hemiptera, Heteroptera). Comp. Cytogenet. 2011, 5, 355–374. [Google Scholar] [CrossRef]
- Suman, V.; Kaur, H. First report on C-banding, fluorochrome staining and NOR location in holocentric chromosomes of Elasmolomus (Aphanus) sordidus Fabricius, 1787 (Heteroptera, Rhyparochromidae). Adv. Hemipterology 2013, 319, 283–291. [Google Scholar] [CrossRef]
- Levsky, J.F.; Singer, R.H. Fluorescence in situ hybridization: Past, present and future. J. Cell Sci. 2003, 116, 2833–2838. [Google Scholar] [CrossRef]
- Frydrychová, R.; Grossmann, P.; Trubac, P.; Vitková, M.; Marec, F.E. Phylogenetic distribution of TTAGG telomeric repeats in insects. Genome 2004, 47, 16–178. [Google Scholar] [CrossRef]
- Kuznetsova, V.; Grozeva, S.; Gokhman, V. Telomere structure in insects: A review. J. Zool. Syst. Evol. Res. 2020, 58, 127–158. [Google Scholar] [CrossRef]
- Golub, N.V.; Golub, V.B.; Anokhin, B.A.; Kuznetsova, V.G. Comparative Cytogenetics of Lace Bugs (Tingidae, Heteroptera): New Data and a Brief Overview. Insects 2022, 13, 608. [Google Scholar] [CrossRef]
- Grozeva, S.; Nokkala, S. Chromosomes and their meiotic behavior in two families of the primitive infraorder Dipsocoromorpha (Heteroptera). Hereditas 1996, 125, 189–192. [Google Scholar] [CrossRef]
- Grozeva, S.; Anokhin, B.; Kuznetsova, V.G. Bed bugs (Hemiptera). In Protocols for Cytogenetic Mapping of Arthropod Genomes; Sharachov, I., Ed.; CRC Press, Taylor and Francis: Boca Raton, FL, USA, 2015; pp. 285–326. [Google Scholar] [CrossRef]
- Golub, N.; Anokhin, B.; Kuznetsova, V. Comparative FISH mapping of ribosomal DNA clusters and TTAGG telomeric sequences to holokinetic chromosomes of eight species of the insect order Psocoptera. Comp. Cytogenet. 2019, 13, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Pfaler-Collander, E. Vergleichend-Karyologische Untersuchungen an Lygaeiden. Acta Zool. Fenn. 1941, 30, 1–119. [Google Scholar]
- Muramoto, N. A chromosome study of five hemipterans (Hemiptera) and two hymenopterans (Hymenoptera). La Kromosomo 1993, 69, 2336–2341. [Google Scholar]
- Geitler, L. Heterochromatin der Geschlechtschromosomen bei Heteropteren. Chromosoma 1939, 1, 197–229. [Google Scholar] [CrossRef]
- Parshad, R. Cytological studies in Heteroptera IV. Chromosome complement and meiosis in twenty-six species of the Pentatomidae, Lygaeidae and Coreidae with the consideration of the cytological bearing on the status of these three superfamilies. Res. Bull. Punjab Univ. 1957, 133, 521–559. [Google Scholar]
- Satapathy, S.N.; Patnaik, S.C. Chromosome numbers in forty-one species of Indian Heteroptera. Chromosome Inform. Serv. 1989, 47, 3–5. [Google Scholar]
- Grozeva, S.; Nokkala, S.; Simov, N. First evidence of sex chromosomes pre-reduction in male meiosis in the Miridae bugs (Heteroptera). Folia Biol. 2006, 54, 9–12. [Google Scholar] [CrossRef]
- Grozeva, S.; Kuznetsova, V.G.; Simov, N.; Langourov, M.; Dalakchieva, S. Sex chromosome pre-reduction in male meiosis of Lethocerus patruelis (Stal, 1854) (Heteroptera, Belostomatidae) with some notes on the distribution of the species. ZooKeys 2013, 319, 119–135. [Google Scholar] [CrossRef]
- Ferretti, A.; Ruiz-Ruano, F.J.; Milani, D.; Loreto, V.; Marti, D.A.; Ramos, E.; Martins, C.; Cabral-de-Mello, D.C. How dynamic could be the 45S rDNA cistron? An intriguing variability in a grasshopper species revealed by integration of chromosomal and genomic data. Chromosoma 2019, 128, 165–175. [Google Scholar] [CrossRef]
- Cabral-de-Mello, D.C.; Oliveira, S.G.; de Moura, R.C.; Martins, C. Chromosomal organization of the 18S and 5S rRNAs and histone H3 genes in Scarabaeinae coleopterans: Insights into the evolutionary dynamics of multigene families and heterochromatin. BMC Genet. 2011, 12, 88. [Google Scholar] [CrossRef]
- Panzera, Y.; Pita, S.; Ferreiro, M.J.; Ferrandis, I.; Lages, C.; Pérez, R.; Silva, A.E.; Guerra, M.; Panzera, F. High dynamics of rDNA cluster location in kissing bug holocentric chromosomes (Triatominae, Heteroptera). Cytogenet. Genome Res. 2012, 38, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Bardella, V.B.; Fernandes, J.A.M.; Cabral-de-Mello, D.C. Chromosomal evolutionary dynamics of four multigene families in Coreidae and Pentatomidae (Heteroptera) true bugs. Mol. Genet. Genomics 2016, 291, 1919–1925. [Google Scholar] [CrossRef]
- Souza-Firmino, T.S.; Alevi, K.C.C.; Itoyama, M.M. Chromosomal divergence and volutionary inferences in Pentatomomorpha infraorder (Hemiptera, Heteroptera) based on the chromosomal location of ribosomal genes. PLoS ONE 2020, 15, e0228631. [Google Scholar] [CrossRef]
- Panzera, F.; Pita, S.; Lorite, P. Chromosome structure and evolution of Triatominae: A review. In Triatominae—The Biology of Chagas Disease Vectors. Entomology in Focus; Guarneri, A., Lorenzo, M., Eds.; Springer: New York, NY, USA, 2021; Volume 5, pp. 65–99. [Google Scholar] [CrossRef]
- Gapon, D.A.; Kuznetsova, V.G.; Maryańska-Nadachowska, A. A new species of the genus Rhaphidosoma Amyot et Serville, 1843 (Heteroptera, Reduviidae), with data on its chromosome complement. Comp. Cytogenet. 2021, 15, 467–505. [Google Scholar] [CrossRef] [PubMed]
- Bardella, V.B.; Fernandes, T.; Vanzela, A.L.L. The conservation of number and location of 18S sites indicates the relative stability of rDNA in species of Pentatomomorpha (Heteroptera). Genome 2013, 56, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef]
- Teixeira, G.A.; de Aguiar, H.J.A.C.; Petitclerc, F.; Orivel, J.; Lopes, D.M.; Barros, L.A.C. Evolutionary insights into the genomic organization of major ribosomal DNA in ant chromosomes. Insect Mol. Biol. 2021, 30, 340–354. [Google Scholar] [CrossRef]
- Teixeira, G.A.; Barros, L.A.C.; de Aguiar, H.J.A.C.; Lopes, D.M. Distribution of GC-rich heterochromatin and ribosomal genes in three fungus-farming ants (Myrmicinae, Attini, Attina): Insights on chromosomal evolution. Comp. Cytogenet. 2021, 15, 413–428. [Google Scholar] [CrossRef]
- Sochorová, J.; Gálvez, F.; Matyášek, R.; Garcia, S.; Kovařík, A. Analyses of the updated “Animal rRDNA loci database” with an emphasis on its new features. Int. J. Mol. Sci. 2021, 22, 11403. [Google Scholar] [CrossRef]
- Buleu, O.G.; Jetybayev, I.Y.; Chobanov, D.P.; Bugrov, A.G. Comparative analysis of C-heterochromatin, ribosomal and telomeric DNA markers in chromosomes of Pamphagidae grasshoppers from Morocco. Comp. Cytogenet. 2019, 13, 61–74. [Google Scholar] [CrossRef]
- Nguyen, P.; Sahara, K.; Yoshido, A.; Marec, F. Evolutionary dynamics of rDNA clusters on chromosomes of moths and butterflies (Lepidoptera). Genetica 2010, 138, 343–354. [Google Scholar] [CrossRef]
- Anjos, A.; Paladini, A.; Evangelista, O.; Cabral-de-Mello, D.C. Insights into chromosomal evolution of Cicadomorpha using fluorochrome staining and mapping 18S rRNA and H3 histone genes. J. Zool. Syst. Evol. Res. 2018, 57, 314–322. [Google Scholar] [CrossRef]
- Kuznetsova, V.; Maryańska-Nadachowska, A.; Anokhin, B.; Shapoval, N.; Shapoval, A. Chromosomal analysis of eight species of dragonflies (Anisoptera) and damselflies (Zygoptera) using conventional cytogenetics and fluorescence in situ hybridization: Insights into the karyotype evolution of the ancient insect order Odonata. J. Zool. Syst. Evol. Res. 2021, 59, 387–399. [Google Scholar] [CrossRef]
- Pita, S.; Lorite, P.; Cuadrado, A.; Panzera, Y.; De Oliveira, J.; Alevi, K.C.C.; Rosa, J.A.; Freitas, S.P.C.; Gómez-Palacio, A.; Solari, A.; et al. High chromosomal mobility of rDNA clusters in holocentric chromosomes of Triatominae, vectors of Chagas disease (Hemiptera-Reduviidae). Med. Vet Entomol. 2022, 36, 66–80. [Google Scholar] [CrossRef]
- Vela, J.; Montiel, E.E.; Mora, P.; Lorite, P.; Palomeque, T. Aphids and ants, mutualistic species, share a mariner element with an unusual location on aphid chromosomes. Genes 2021, 12, 1966. [Google Scholar] [CrossRef]
- Okazaki, S.; Tsuchida, K.; Maekawa, H.; Ishikawa, H.; Fujiwara, H. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silk warm Bombyx mori and in other insects. J. Mol. Cell Biol. 1993, 15, 4545–4552. [Google Scholar] [CrossRef]
- Sahara, K.; Marec, F.; Traut, W. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 1999, 7, 449–460. [Google Scholar] [CrossRef]
- Mason, J.M.; Randall, T.A.; Frydrychova, R.C. Telomerase lost? Chromosoma 2016, 125, 65–73. [Google Scholar] [CrossRef]
- Lukhtanov, V.A. Diversity and evolution of telomere and subtelomere DNA sequences in insects. bioRxiv 2022. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).