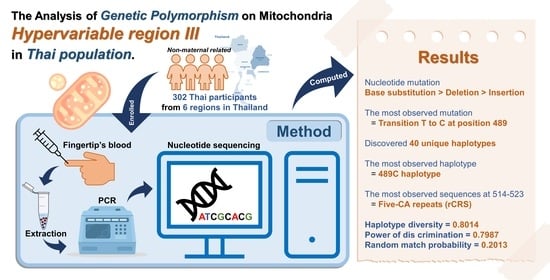
The Analysis of Genetic Polymorphism on Mitochondrial Hypervariable Region III in Thai Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. Hypervariable Region III (HVRIII) Amplification
2.4. Nucleotide Sequencing
2.5. Data Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Court, D.S. Mitochondrial DNA in forensic use. Emerg. Top. Life Sci. 2021, 5, 415–426. [Google Scholar] [CrossRef]
- Torchilin, V.P.; Khaw, B.A.; Weissig, V. Intracellular targets for DNA delivery: Nuclei and mitochondria. Somat. Cell Mol. Genet. 2002, 27, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Stoneking, M. Hypervariable Sites in the mtDNA Control Region Are Mutational Hotspots. Am. J. Hum. Genet. 2000, 67, 1029–1032. [Google Scholar] [CrossRef] [PubMed]
- Lutz, S.; Weisser, H.J.; Heizmann, J.; Pollak, S. A third hypervariable region in the human mitochondrial D-loop. Hum. Genet. 1997, 101, 384. [Google Scholar]
- Hutchison, C.A.; Newbold, J.E.; Potter, S.S.; Edgell, M.H. Maternal inheritance of mammalian mitochondrial DNA. Nature 1974, 251, 536–538. [Google Scholar] [CrossRef]
- Shokolenko, I.; Alexeyev, M. Mitochondrial DNA: Consensuses and Controversies. DNA 2022, 2, 131–148. [Google Scholar] [CrossRef]
- Pakendorf, B.; Stoneking, M. Mitochondrial DNA and human evolution. Annu. Rev. Genom. Hum. Genet. 2005, 6, 165–183. [Google Scholar] [CrossRef]
- Brown, W.M.; George, M., Jr.; Wilson, A.C. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1967–1971. [Google Scholar] [CrossRef]
- Lutz, S.; Weisser, H.J.; Heizmann, J.; Pollak, S. Location and frequency of polymorphic positions in the mtDNA control region of individuals from Germany. Int. J. Leg. Med. 1998, 111, 67–77. [Google Scholar] [CrossRef]
- Wilson, M.R.; DiZinno, J.A.; Polanskey, D.; Replogle, J.; Budowle, B. Validation of mitochondrial DNA sequencing for forensic casework analysis. Int. J. Leg. Med. 1995, 108, 68–74. [Google Scholar] [CrossRef]
- Lutz, S.; Weisser, H.J.; Heizmann, J.; Pollak, S. mtDNA as a tool for identification of human remains. Identification using mtDNA. Int. J. Leg. Med. 1996, 109, 205–209. [Google Scholar] [CrossRef]
- Budowle, B.; Allard, M.W.; Wilson, M.R.; Chakraborty, R. Forensics and mitochondrial DNA: Applications, debates, and foundations. Annu. Rev. Genom. Hum. Genet. 2003, 4, 119–141. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C.; Brown, M.D.; Lott, M.T. Mitochondrial DNA variation in human evolution and disease. Gene 1999, 238, 211–230. [Google Scholar] [CrossRef]
- Bär, W.; Brinkmann, B.; Budowle, B.; Carracedo, A.; Gill, P.; Holland, M.; Lincoln, P.J.; Mayr, W.; Morling, N.; Olaisen, B.; et al. DNA Commission of the International Society for Forensic Genetics: Guidelines for mitochondrial DNA typing. Int. J. Leg. Med. 2000, 113, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Lutz, S.; Wittig, H.; Weisser, H.J.; Heizmann, J.; Junge, A.; Dimo-Simonin, N.; Parson, W.; Edelmann, J.; Anslinger, K.; Jung, S.; et al. Is it possible to differentiate mtDNA by means of HVIII in samples that cannot be distinguished by sequencing the HVI and HVII regions? Forensic Sci. Int. 2000, 113, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Fridman, C.; Gonzalez, R.S. HVIII discrimination power to distinguish HVI and HVII common sequences. Forensic Sci. Int. Genet. Suppl. Ser. 2009, 2, 320–321. [Google Scholar] [CrossRef]
- Pischedda, S.; Barral-Arca, R.; Gómez-Carballa, A.; Pardo-Seco, J.; Catelli, M.; Alvarez-Iglesias, V.; Cárdenas, J.; Nguyen Duc, N.; Ha, H.; Le, A.; et al. Phylogeographic and genome-wide investigations of Vietnam ethnic groups reveal signatures of complex historical demographic movements. Sci. Rep. 2017, 7, 12630. [Google Scholar] [CrossRef]
- Nagai, A.; Nakamura, I.; Bunai, Y. Sequence analysis of mitochondrial DNA HVIII region in a Japanese population. Int. Congr. Ser. 2004, 1261, 410–412. [Google Scholar] [CrossRef]
- Nur Haslindawaty, A.R.; Panneerchelvam, S.; Edinur, H.A.; Norazmi, M.N.; Zafarina, Z. Sequence polymorphisms of mtDNA HV1, HV2, and HV3 regions in the Malay population of Peninsular Malaysia. Int. J. Leg. Med. 2010, 124, 415–426. [Google Scholar] [CrossRef]
- Thongngam, P.; Leewattanapasuk, W.; Bhoopat, T.; Sangthong, P. Nucleotide sequence analysis of the hypervariable region III of mitochondrial DNA in Thais. J. Forensic Leg. Med. 2016, 41, 10–14. [Google Scholar] [CrossRef]
- Comas, D.; Calafell, F.; Mateu, E.; Pérez-Lezaun, A.; Bertranpetit, J. Geographic variation in human mitochondrial DNA control region sequence: The population history of Turkey and its relationship to the European populations. Mol. Biol. Evol. 1996, 13, 1067–1077. [Google Scholar] [CrossRef]
- He, Y.; Ren, L.Y.; Shan, K.R.; Zhang, T.; Wang, C.J.; Guan, Z.Z. Characterization of polymorphisms in the mitochondrial DNA of twelve ethnic groups in the Guizhou province of China. Mitochondrial DNA Part A 2016, 27, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Areesirisuk, P.; Srikulnath, K.; Onsod, P.; Jaroensuk, J.; Rerkamnuaychoke, B. Haplogroup Distribution of 309 Thais from Admixed Populations across the Country by HVI and HVII Sanger-Type Sequencing. Diversity 2021, 13, 496. [Google Scholar] [CrossRef]
- Walsh, P.S.; Metzger, D.A.; Higuchi, R. Chelex 100 as a Medium for Simple Extraction of DNA for PCR-Based Typing from Forensic Material. Biotechniques 2013, 54, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Corporation, L.T. SeqScape® Software 3 User Guide; Life Technologies Corporation: Carlsbad, CA, USA, 2012; Available online: https://www.thermofisher.com/order/catalog/product/4474978 (accessed on 2 March 2023).
- Andrews, R.M.; Kubacka, I.; Chinnery, P.F.; Lightowlers, R.N.; Turnbull, D.M.; Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999, 23, 147. [Google Scholar] [CrossRef] [PubMed]
- Desmyter, S.; Hoste, B. Influence of the electrophoresis-resequencing method on the forensic mtDNA profiling quality. Forensic Sci. Int. Genet. 2007, 1, 199–200. [Google Scholar] [CrossRef]
- Stoneking, M.; Hedgecock, D.; Higuchi, R.G.; Vigilant, L.; Erlich, H.A. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am. J. Hum. Genet. 1991, 48, 370–382. [Google Scholar]
- Szibor, R.; Michael, M.; Spitsyn, V.A.; Plate, I.; Ginter, E.K.; Krause, D. Mitochondrial D-loop 3′ (CA)n repeat polymorphism: Optimization of analysis and population data. Electrophoresis 1997, 18, 2857–2860. [Google Scholar] [CrossRef]
- Bermisheva, M.A.; Viktorova, T.V.; Khusnutdinova, E.K. Polymorphism of Human Mitochondrial DNA. Russ. J. Genet. 2003, 39, 849–859. [Google Scholar] [CrossRef]
- Sharma, S.; Saha, A.; Rai, E.; Bhat, A.; Bamezai, R. Human mtDNA hypervariable regions, HVR I and II, hint at deep common maternal founder and subsequent maternal gene flow in Indian population groups. J. Hum. Genet. 2005, 50, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA; Chichester, UK; West Sussex, UK, 1987. [Google Scholar] [CrossRef]
- Elkins, K.M. Chapter 13—Computing Random Match Probability from DNA Profile Data Using Population Databases. In Forensic DNA Biology; Elkins, K.M., Ed.; Academic Press: San Diego, CA, USA, 2013; pp. 115–120. [Google Scholar] [CrossRef]
- Zupanič Pajnič, I.; Balažic, J.; Komel, R. Sequence polymorphism of the mitochondrial DNA control region in the Slovenian population. Int. J. Leg. Med. 2004, 118, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Hoong, L.; Lek, K. Genetic polymorphisms in mitochondrial DNA hypervariable regions I, II and III of the Malaysian population. Asia Pac. J. Mol. Biol. Biotechnol. 2005, 13, 79–85. [Google Scholar]
- Sylvester, C.; Krishna, M.; Rao, J.; Chandrasekar, A. Allele frequencies of mitochondrial DNA HVR III 514–524 (CA)n dinucleotide repeats in the Urali Kuruman tribal population of South India. Egypt. J. Forensic Sci. 2018, 8, 52. [Google Scholar] [CrossRef]
- Fan, H.; Chu, J.-Y. A Brief Review of Short Tandem Repeat Mutation. Genom. Proteom. Bioinform. 2007, 5, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Chung, U.; Lee, H.Y.; Yoo, J.-E.; Park, M.J.; Shin, K.-J. Mitochondrial DNA CA dinucleotide repeats in Koreans: The presence of length heteroplasmy. Int. J. Leg. Med. 2005, 119, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Welikala, A.H.J.; Ranasinghe, R.; Tennekoon, K.H.; Kotelawala, J.T.; Weerasooriya, P.R. Mitochondrial DNA (CA)n dinucleotide repeat variations in Sinhalese and Vedda populations in Sri Lanka. Genetica 2022, 150, 145–150. [Google Scholar] [CrossRef]
- Agrawal, G.; Singh, D.; Goyal, V.K. Extent of heterogeneity in the poly-nucleotide stretches of mtDNA hypervariable regions in the Indian population. J. Forensic Med. Toxicol. 2008, 25, 1–8. [Google Scholar]
- Bandelt, H.J.; Parson, W. Consistent treatment of length variants in the human mtDNA control region: A reappraisal. Int. J. Leg. Med. 2008, 122, 11–21. [Google Scholar] [CrossRef]
| Characteristics | Number of Participants | ||||||
|---|---|---|---|---|---|---|---|
| Northern | Northeastern | Western | Central | Eastern | Southern | ||
| Sex | Male | 5 | 18 | 17 | 23 | 6 | 16 |
| Female | 39 | 43 | 36 | 39 | 25 | 35 | |
| Total | 44 | 61 | 53 | 62 | 31 | 51 | |
| Type of Nucleotide Change | Observed Number | Different Nucleotide Positions in HVRIII (Observed Number) |
|---|---|---|
| Total number of variant sites | 28 | |
| Total number of base substitutions | 197 | |
| Transition | 194 | |
| A→G | 8 | 444 (1), 488 (3), 503 (1), 508 (1), 533 (1), 547 (1) |
| G→A | 6 | 499 (2), 513 (4) |
| T→C | 164 | 454 (1), 482 (1), 485 (4), 489 (157), 539 (1) |
| C→T | 16 | 456 (9), 461 (1), 498 (2), 501 (1), 518 (3) |
| Transversion | 3 | |
| A→C | 1 | 574 (1) |
| C→G | 2 | 447 (1), 530 (1) |
| Total number of base insertions | 13 | |
| +AC | 1 | 514 (1) |
| +C | 2 | 573 (2) |
| +CC | 1 | 573 (1) |
| +CA | 7 | 523 (7) |
| +CACA | 2 | 523 (2) |
| Total number of base deletions | 271 | |
| −A | 133 | 521 (3), 523 (130) |
| −G | 2 | 513 (2) |
| −C | 136 | 459 (1), 514 (2), 520 (3), 522 (130) |
| Type of Nucleotide Change | Different Nucleotide Position in HVRIII (Observed Number) | |||||
|---|---|---|---|---|---|---|
| Northern (n = 44) | Northeast (n = 61) | Western (n = 53) | Central (n = 62) | Eastern (n = 31) | Southern (n = 51) | |
| Total number of base substitutions | ||||||
| Transition | ||||||
| A→G | 488 (2) | 488 (1), 547 (1) | 508 (1) | 444 (1), 503 (1), 533 (1) | ||
| G→A | 499 (1) | 513 (2) | 513 (1) | 499 (1), 513 (1) | ||
| T→C | 485 (1), 489 (23) | 489 (25) | 489 (34), 454 (1) | 489 (35) | 485 (2), 489 (14), 539 (1) | 482 (1), 485 (1), 489 (26) |
| C→T | 456 (2), 498 (2), 518 (1) | 501 (1) | 456 (2), 518 (1) | 456 (4), 518 (1) | 461 (1) | 456 (1) |
| Transversion | ||||||
| A→C | 574 (1) | |||||
| C→G | 530 (1) | 447 (1) | ||||
| Total number of base insertions | ||||||
| +AC | 514 (1) | |||||
| +C | 573 (1) | 573 (1) | ||||
| +CC | 573 (1) | |||||
| +CA | 523 (1) | 523 (1) | 523 (3) | 523 (1) | 523 (1) | |
| +CACA | 523 (1) | 523 (1) | ||||
| Total number of base deletions | ||||||
| −A | 523 (17) | 523 (30) | 523 (18) | 521 (1), 523 (27) | 521 (2), 523 (15) | 523 (23) |
| −G | 513 (2) | |||||
| −C | 459 (1), 514(2), 522 (17) | 522 (30) | 522 (18) | 520 (1), 522 (27) | 520 (2), 522 (15) | 522 (23) |
| 444 | 447 | 454 | 456 | 459 | 461 | 482 | 485 | 488 | 489 | 498 | 499 | 501 | 503 | 508 | 513 | 514 | 514.1 | 514.2 | 518 | 520 | 521 | 522 | 523 | 523.1 | 523.2 | 523.3 | 523.4 | 530 | 533 | 539 | 547 | 573 | 573.1 | 573.2 | 574 | n |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | T | C | C | C | T | T | A | T | C | G | C | A | A | G | C | - | - | C | C | A | C | A | - | - | - | - | C | A | T | A | C | - | - | A | |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 92 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 87 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 34 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 32 |
| * | * | * | T | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 7 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | C | A | * | * | * | * | * | * | * | * | * | * | 4 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | C | A | * | * | * | * | * | * | * | * | * | * | 4 |
| * | * | * | * | * | * | C | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 3 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | del | del | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 3 |
| * | * | * | * | * | * | * | * | G | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 2 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | T | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 2 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | C | * | * | 2 |
| * | * | * | * | * | * | * | * | * | * | * | A | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 2 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | A | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 2 |
| G | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | G | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | C | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | T | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | T | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | del | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | T | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | G | C | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | T | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | G | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | A | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | C | A | C | A | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | G | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | C | C | * | 1 |
| * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | C | 1 |
| * | * | * | * | * | * | * | * | * | * | T | A | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | G | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | A | * | A | C | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | T | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | G | * | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | del | del | * | * | * | * | * | * | * | * | * | * | * | * | G | * | * | * | * | * | * | 1 |
| * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | * | C | * | * | * | * | * | 1 |
| Nucleotide Polymorphism in HVRIII at Positions 514–523 | Total Number of Nucleotide Changes | % Insertion and Deletion from Samples |
|---|---|---|
| CACACA (−CACA) | 3 | 0.99 |
| CACACACA (−CA) | 127 | 42.05 |
| CACACACACA | 163 | 53.97 |
| CACACACACACA (+CA) | 7 | 2.32 |
| CACACACACACACA (+CACA) | 2 | 0.66 |
| Nucleotide Polymorphism in HVRIII at Positions 514–523 | Northern | Northeastern | Western | Central | Eastern | Southern |
|---|---|---|---|---|---|---|
| CACACA (−CACA) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1.61%) | 2 (6.45%) | 0 (0%) |
| CACACACA (−CA) | 17 (38.63%) | 30 (49.18%) | 18 (33.96%) | 26 (41.93%) | 13 (41.94%) | 23 (45.10%) |
| CACACACACA | 26 (59.09%) | 29 (47.54%) | 32 (60.38%) | 35 (56.45%) | 15 (48.39%) | 26 (50.98%) |
| CACACACACACA (+CA) | 1 (2.27%) | 1 (1.64%) | 3 (5.66%) | 0 (0%) | 1 (3.23%) | 1 (1.96%) |
| CACACACACACACA (+CACA) | 0 (0%) | 1 (1.64%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (1.96%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanichanukulyakit, J.; Khacha-ananda, S.; Monum, T.; Mahawong, P.; Moophayak, K.; Penkhrue, W.; Khumpook, T.; Thongsahuan, S. The Analysis of Genetic Polymorphism on Mitochondrial Hypervariable Region III in Thai Population. Genes 2023, 14, 682. https://doi.org/10.3390/genes14030682
Vanichanukulyakit J, Khacha-ananda S, Monum T, Mahawong P, Moophayak K, Penkhrue W, Khumpook T, Thongsahuan S. The Analysis of Genetic Polymorphism on Mitochondrial Hypervariable Region III in Thai Population. Genes. 2023; 14(3):682. https://doi.org/10.3390/genes14030682
Chicago/Turabian StyleVanichanukulyakit, Jirat, Supakit Khacha-ananda, Tawachai Monum, Phatcharin Mahawong, Kittikhun Moophayak, Watsana Penkhrue, Taddaow Khumpook, and Sorawat Thongsahuan. 2023. "The Analysis of Genetic Polymorphism on Mitochondrial Hypervariable Region III in Thai Population" Genes 14, no. 3: 682. https://doi.org/10.3390/genes14030682
APA StyleVanichanukulyakit, J., Khacha-ananda, S., Monum, T., Mahawong, P., Moophayak, K., Penkhrue, W., Khumpook, T., & Thongsahuan, S. (2023). The Analysis of Genetic Polymorphism on Mitochondrial Hypervariable Region III in Thai Population. Genes, 14(3), 682. https://doi.org/10.3390/genes14030682