Comparative Analyses of Chloroplast Genomes for Parasitic Species of Santalales in the Light of Two Newly Sequenced Species, Taxillus nigrans and Scurrula parasitica
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Sequencing and Chloroplast Genome Assembly
2.2. Gene Annotation and Sequence Analyses
2.3. Genome Comparison
2.4. Phylogenetic Analysis
3. Results
3.1. Characteristics of T. nigrans and S. parasitica Chloroplast Genomes
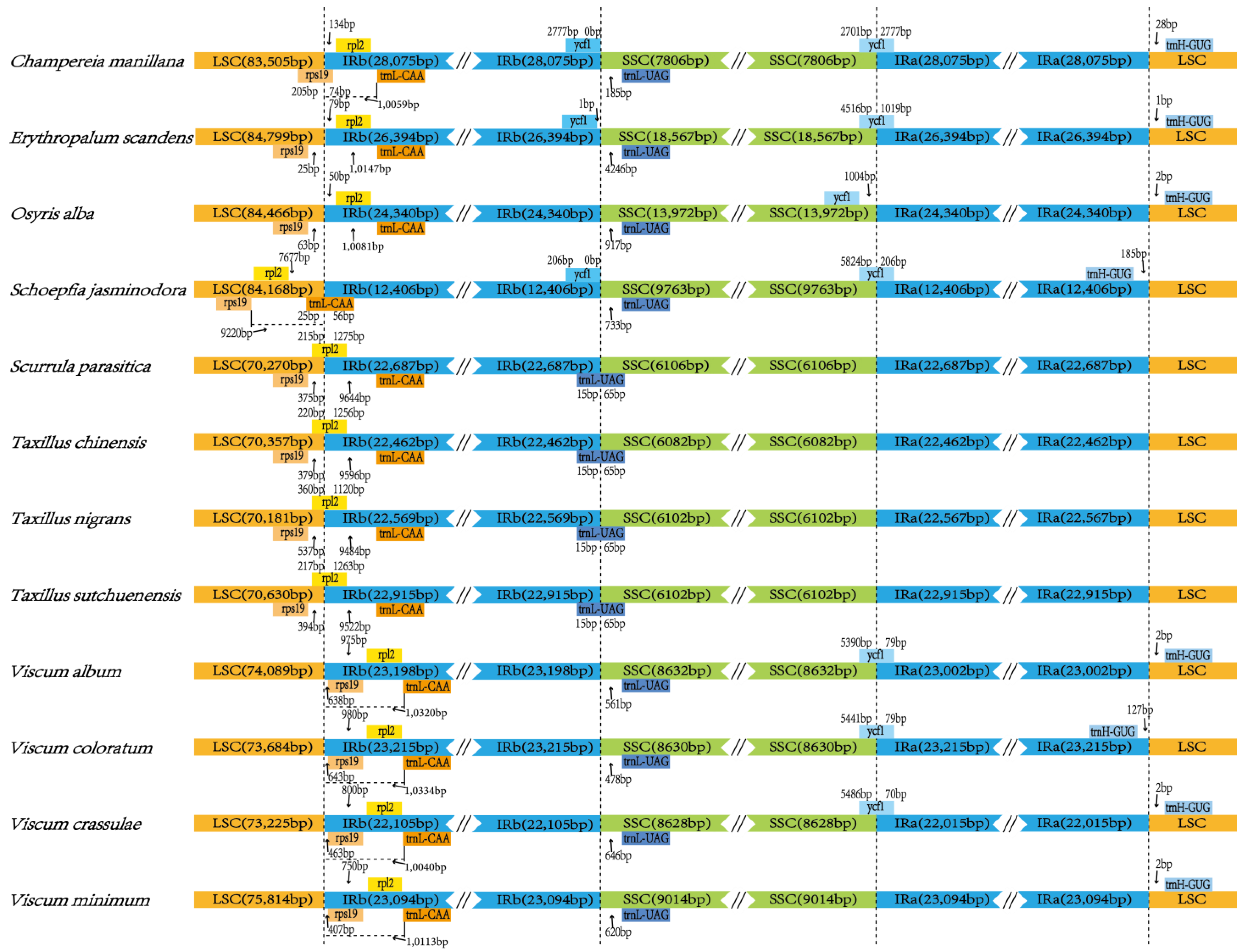
3.2. Comparative Chloroplast Genomic Analysis
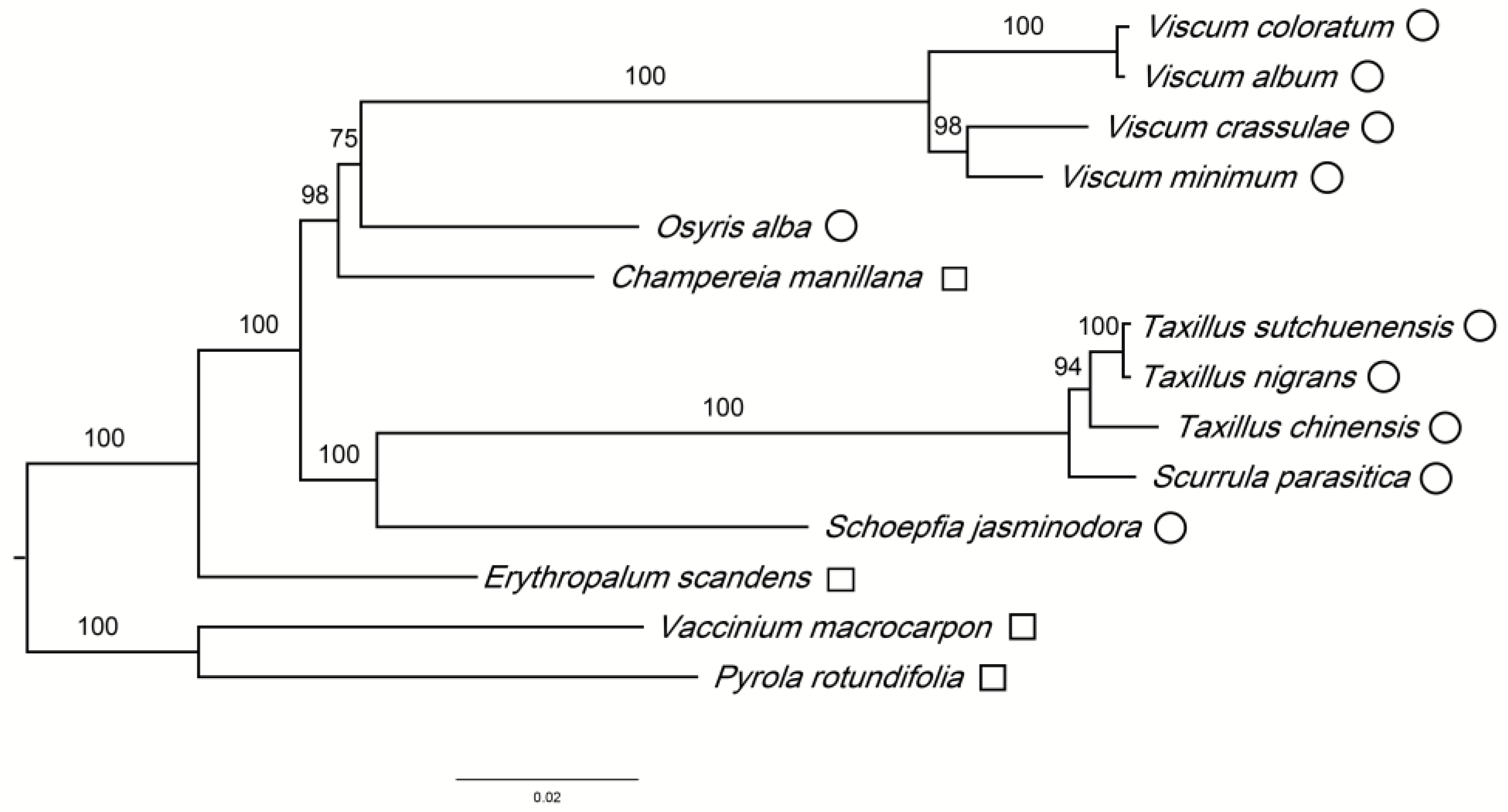
3.3. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heide-Jørgensen, H.S. Parasitic Flowering Plants, 1st ed.; Brill: Leiden, The Netherlands, 2008; Volume 10, pp. 6–7. [Google Scholar]
- Xu, Y.; Zhang, J.; Ma, C.; Lei, Y.; Shen, G.; Jin, J.; Eaton, D.A.R.; Wu, J. Comparative genomics of orobanchaceous species with different parasitic lifestyles reveals the origin and stepwise evolution of plant parasitism. Mol. Plant 2022, 15, 1384–1399. [Google Scholar] [CrossRef] [PubMed]
- Young, N.D.; Steiner, K.E.; dePamphilis, C.W. The Evolution of Parasitism in Scrophulariaceae/Orobanchaceae: Plastid Gene Sequences Refute an Evolutionary Transition Series. Ann. Mo. Bot. Gard. 1999, 86, 876–893. [Google Scholar] [CrossRef]
- Twyford, A.D. Parasitic plants. Curr. Biol. 2018, 28, R857–R859. [Google Scholar] [CrossRef] [PubMed]
- Westwood, J.H.; Yoder, J.I.; Timko, M.P.; dePamphilis, C.W. The evolution of parasitism in plants. Trends Plant Sci. 2010, 15, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.F.; Wicke, S.; Sass, C. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 2018, 218, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Barrett, C.F.; Freudenstein, J.V.; Li, J.; Mayfield-Jones, D.R.; Perez, L.; Pires, J.C.; Santos, C. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol. Biol. Evol. 2014, 31, 3095–3112. [Google Scholar] [CrossRef] [PubMed]
- Petersen, G.; Anderson, B.; Braun, H.-P.; Meyer, E.H.; Møller, I.M. Mitochondria in parasitic plants. Mitochondrion 2020, 52, 173–182. [Google Scholar] [CrossRef]
- Su, H.J.; Liang, S.L.; Nickrent, D.L. Plastome variation and phylogeny of Taxillus (Loranthaceae). PLoS ONE 2021, 16, e0256345. [Google Scholar] [CrossRef]
- Yoshida, S.; Kee, Y.J. Large-scale sequencing paves the way for genomic and genetic analyses in parasitic plants. Curr. Opin. Biotechnol. 2021, 70, 248–254. [Google Scholar] [CrossRef]
- Li, X.; Zhang, T.C.; Qiao, Q.; Ren, Z.M.; Zhao, J.Y.; Yonezawa, T.; Hasegawa, M.; Crabbe, M.J.C.; Li, J.Q.; Zhong, Y. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 2013, 8, e58747. [Google Scholar] [CrossRef]
- Palmer, J.D. Comparative organization of chloroplast genomes. Ann. Rev. Genet. 1985, 19, 325–354. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, J.g.; Chen, X.l.; Cui, Y.x.; Xu, Z.c.; Li, Y.h.; Song, J.y.; Duan, B.z.; Yao, H. Gene losses and partial deletion of small single-copy regions of the chloroplast genomes of two hemiparasitic Taxillus species. Sci. Rep. 2017, 7, 12834. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, H.L. The hows and whys of cytoplasmic inheritance in seed plants. Am. J. Bot. 1996, 83, 383–404. [Google Scholar] [CrossRef]
- Nagata, N. Mechanisms for independent cytoplasmic inheritance of mitochondria and plastids in angiosperms. J. Plant Res. 2010, 123, 193–199. [Google Scholar] [CrossRef]
- Barnard-Kubow, K.B.; McCoy, M.A.; Galloway, L.F. Biparental chloroplast inheritance leads to rescue from cytonuclear incompatibility. New Phytol. 2017, 213, 1466–1476. [Google Scholar] [CrossRef]
- Zheng, H.; Fan, L.; Wang, T.; Zhang, L.; Ma, T.; Mao, K. The complete chloroplast genome of Populus rotundifolia (Salicaceae). Conserv. Genet. Resour. 2016, 8, 399–401. [Google Scholar] [CrossRef]
- Wang, L.; Dong, W.P.; Zhou, S.L. Structural mutations and reorganizations in chloroplast genomes of flowering plants. Acta Bot. Boreali-Occident. Sin. 2012, 32, 1282–1288. (In Chinese) [Google Scholar]
- Bungard, R.A. Photosynthetic evolution in parasitic plants: Insight from the chloroplast genome. Bioessays 2004, 26, 235–247. [Google Scholar] [CrossRef]
- Gruzdev, E.V.; Mardanov, A.V.; Beletsky, A.V.; Kochieva, E.Z.; Ravin, N.V.; Skryabin, K.G. The complete chloroplast genome of parasitic flowering plant Monotropa hypopitys: Extensive gene losses and size reduction. Mitochondrial DNA B 2016, 1, 212–213. [Google Scholar] [CrossRef]
- Guo, X.R.; Zhang, G.F.; Fan, L.Y.; Liu, C.K.; Ji, Y.H. Highly degenerate plastomes in two hemiparasitic dwarf mistletoes: Arceuthobium chinense and A. pini (Viscaceae). Planta 2021, 253, 125. [Google Scholar] [CrossRef]
- Petersen, G.; Cuenca, A.; Seberg, O. Plastome evolution in hemiparasitic mistletoes. Genome Biol. Evol. 2015, 7, 2520–2532. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Morden, C.W.; Ems, S.C.; Palmer, J.D. Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant—Loss or accelerated sequence evolution of transfer-rna and ribosomal-protein genes. J. Mol. Evol. 1992, 35, 304–317. [Google Scholar] [CrossRef] [PubMed]
- dePamphilis, C.W.; Palmer, J.D. Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 1990, 348, 337. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Morden, C.W.; Palmer, J.D. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 1992, 89, 10648–10652. [Google Scholar] [CrossRef] [PubMed]
- Miao, N.; Zhang, L.; Li, M.; Fan, L.; Mao, K. Development of EST-SSR markers for Taxillus nigrans (Loranthaceae) in southwestern China using next-generation sequencing. Appl. Plant Sci. 2017, 5, 1700010. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.L.; Doyle, J.A.; Doyle, F.J. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.I.; Cronk, Q.C. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015, 3, 1500026. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef]
- Braukmann, T.W.A.; Broe, M.B.; Stefanović, S.; Freudenstein, J.V. On the brink: The highly reduced plastomes of nonphotosynthetic Ericaceae. New Phytol. 2017, 216, 254–266. [Google Scholar] [CrossRef]
- Han, R.L.; Hao, G.; Zhang, D.X. Interfamilial relationships of Santalales as revealed by chloroplast trnL intron sequences. J. Trop. Subtrop. Bot. 2004, 12, 393–398. (In Chinese) [Google Scholar]
- Freyer, R.; Neckermann, K.; Maier, R.M.; Kossel, H. Structural and Functional-Analysis of Plastid Genomes from Parasitic Plants—Loss of an Intron within the Genus Cuscuta. Curr. Genet. 1995, 27, 580–586. [Google Scholar] [CrossRef]
- Wu, C.S.; Wang, T.J.; Wu, C.W.; Wang, Y.N.; Chaw, S.M. Plastome Evolution in the Sole Hemiparasitic Genus Laurel Dodder (Cassytha) and Insights into the Plastid Phylogenomics of Lauraceae. Genome Biol. Evol. 2017, 9, 2604–2614. [Google Scholar] [CrossRef]
- Wicke, S.; Muller, K.F.; dePamphilis, C.W.; Quandt, D.; Bellot, S.; Schneeweiss, G.M. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 9045–9050. [Google Scholar] [CrossRef] [PubMed]
- Ruhlman, T.A.; Chang, W.J.; Chen, J.J.W.; Huang, Y.T.; Chan, M.T.; Zhang, J.; Liao, D.C.; Blazier, J.C.; Jin, X.H.; Shih, M.C.; et al. NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol. 2015, 15, 100. [Google Scholar] [CrossRef]
- Xue, Z.Q.; Xue, J.H.; Victorovna, K.M.; Ma, K.P. The complete chloroplast DNA sequence of Trapa maximowiczii Korsh. (Trapaceae), and comparative analysis with other Myrtales species. Aquat. Bot. 2017, 143, 54–62. [Google Scholar] [CrossRef]
- Li, X.; Yang, J.B.; Wang, H.; Song, Y.; Corlett, R.T.; Yao, X.; Li, D.Z.; Yu, W.B. Plastid NDH Pseudogenization and Gene Loss in a Recently Derived Lineage from the Largest Hemiparasitic Plant Genus Pedicularis (Orobanchaceae). Plant Cell Physiol. 2021, 62, 971–984. [Google Scholar] [CrossRef] [PubMed]
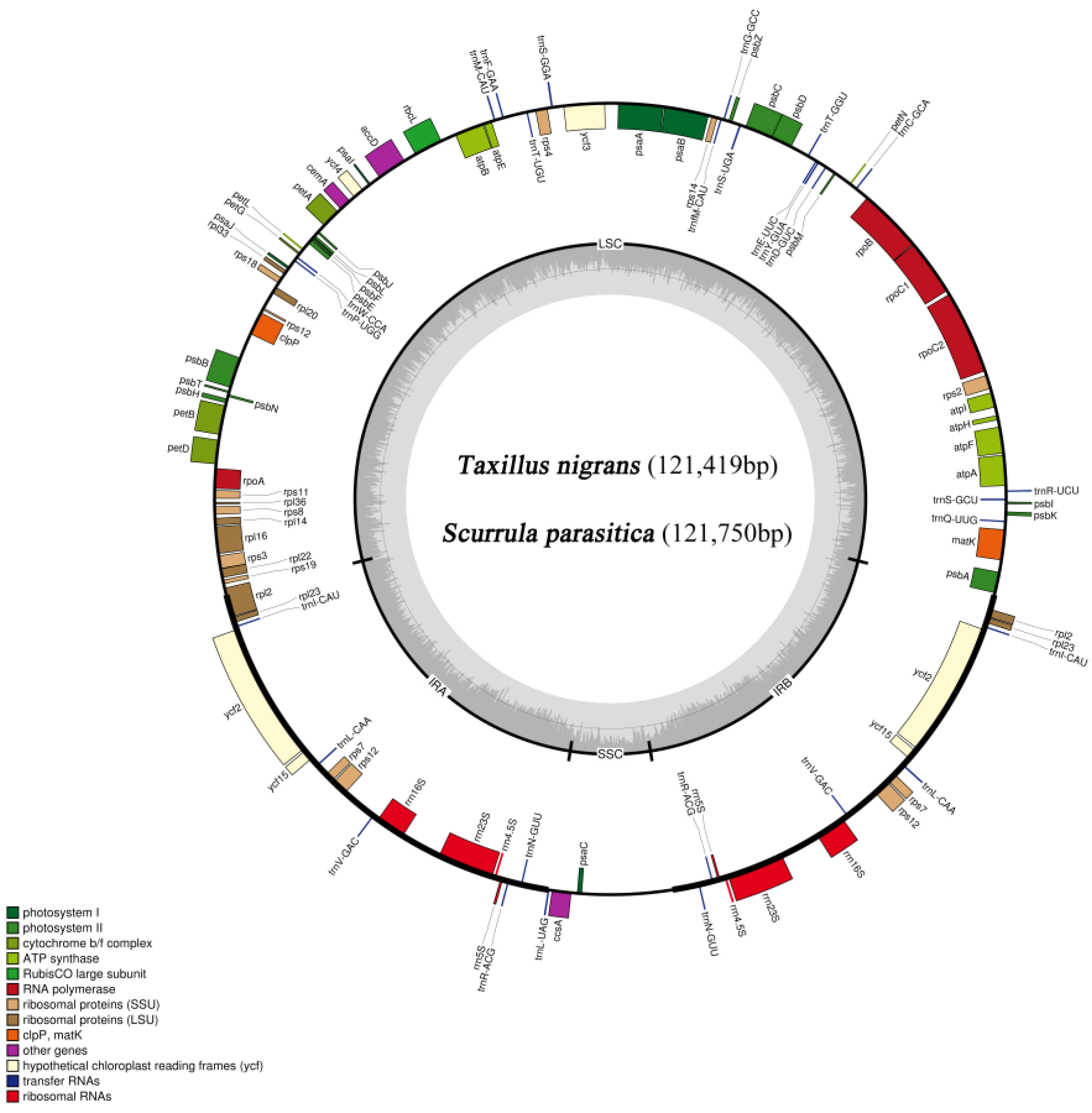


| Species | T(U) (%) | C (%) | A (%) | G (%) | Length (bp) | |
|---|---|---|---|---|---|---|
| T. nigrans | LSC | 33.4 | 18.0 | 31.8 | 16.8 | 70,181 |
| SSC | 41.2 | 13.3 | 32.5 | 13.0 | 6100 | |
| IRa | 28.8 | 20.6 | 28.3 | 22.3 | 22,569 | |
| IRb | 28.3 | 22.3 | 28.7 | 20.7 | 22,569 | |
| Total | 32.0 | 19.0 | 30.6 | 18.4 | 121,419 | |
| S. parasitica | LSC | 33.5 | 17.7 | 31.9 | 16.7 | 70,270 |
| SSC | 41.5 | 12.8 | 32.6 | 13.1 | 6106 | |
| IRa | 28.8 | 20.5 | 28.3 | 22.4 | 22,687 | |
| IRb | 28.3 | 22.4 | 28.8 | 20.5 | 22,687 | |
| Total | 32.1 | 18.9 | 30.7 | 18.3 | 121,750 |
| Genes | ||
|---|---|---|
| 1 | Photosystem I | psaA, B, C, I, J, ycf3, 4 |
| 2 | Photosystem II | psbA, B, C, D, E, F, H, I, J, K, L, M, N, T, Z |
| 3 | Cytochrome b6/f | petA, B, D, G, L, N |
| 4 | ATP synthase | atpA, B, E, F, H, I |
| 5 | Rubisco | rbcL |
| 6 | RNA polymerase | rpoA, B, C1, C2 |
| 7 | Small subunit ribosomal proteins | rps2, 3, 4, 7, 8, 11, 12, 14, 18, 19 |
| 8 | Large subunit ribosomal proteins | rpl2, 14, 20, 22, 23, 33, 36 |
| 9 | Other proteins | accD, ccsA, cemA, clpP, matK |
| 10 | Proteins of unknown function | ycf2 |
| 11 | Ribosomal RNAs | rrn4.5S, rrn5S, rrn16S, rrn23S |
| 12 | Transfer RNAs | trnC(GCA), trnD(GUC), trnE(UUC), trnF(GAA), trnfM(CAU), trnG(GCC), trnI(CAU), trnL(CAA), trnL(UAG), trnM(CAU), trnN(GUU), trnP(UGG), trnQ(UUG), trnR(ACG), trnR(UCU), trnS(GGA), trnS(GCU), trnS(UGA), trnT(GGU), trnT(UGU), trnV(GAC), trnW(CCA), trnY(GUA) |
| Species | E. scandens | S. jasminodora | S. parasitica | T. chinensis | T. nigrans | T. sutchuenensis | C. manillana | O. alba | V. album | V. coloratum | V. crassulae | V. minimum |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family | Erythropalaceae | Schoepfiaceae | Loranthaceae | Loranthaceae | Loranthaceae | Loranthaceae | Opiliaceae | Santalaceae | Santalaceae | Santalaceae | Santalaceae | Santalaceae |
| Accession No. | NC_036759.1 | NC_034228.1 | MH101514 | NC_036306.1 | MH095982 | NC_036307.1 | NC_034931.1 | NC_027960.1 | NC_028012.1 | NC_035414.1 | NC_027959.1 | NC_027829.1 |
| Genome size (bp) | 156,154 | 118,743 | 121,750 | 121,363 | 121,419 | 122,562 | 147,461 | 147,253 | 128,921 | 128,744 | 126,064 | 131,016 |
| LSC length (bp) | 84,799 | 84,168 | 70,270 | 70,357 | 70,181 | 70,630 | 83,505 | 84,466 | 73,893 | 73,684 | 73,225 | 75,814 |
| LSC length (%) | 54.3 | 70.9 | 57.7 | 58.0 | 57.8 | 57.6 | 56.6 | 57.4 | 57.3 | 57.2 | 58.1 | 57.9 |
| SSC length (bp) | 18,567 | 9763 | 6106 | 6082 | 6100 | 6102 | 7806 | 13,972 | 8632 | 8630 | 8628 | 9014 |
| SSC length (%) | 11.9 | 8.2 | 5.0 | 5.0 | 5.0 | 5.0 | 5.3 | 9.5 | 6.7 | 6.7 | 6.8 | 6.9 |
| IR length (bp) | 26,394 | 12,406 | 22,687 | 22,462 | 22,569 | 22,915 | 28,075 | 24,340 | 23,198 | 23,215 | 22,105 | 23,094 |
| IR length (%) | 33.8 | 20.9 | 37.3 | 37.0 | 37.2 | 37.4 | 38.1 | 33.1 | 36.0 | 36.1 | 35.1 | 35.2 |
| GC content (%) | 38.0 | 38.1 | 37.2 | 37.3 | 37.4 | 37.3 | 37.4 | 37.7 | 36.4 | 36.3 | 36.4 | 36.2 |
| Number of genes | 130 | 121 | 106 | 106 | 106 | 106 | 120 | 117 | 115 | 119 | 116 | 104 |
| Number of protein coding genes | 86 | 72 | 66 | 66 | 66 | 66 | 73 | 74 | 71 | 70 | 73 | 66 |
| Number of tRNAs | 36 | 34 | 28 | 28 | 28 | 28 | 37 | 35 | 36 | 36 | 35 | 29 |
| Number of rRNAs | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 | 8 |
| Species | Amino Acid | Codon | No. | RSCU | tRNA | Amino Acid | Codon | No. | RSCU | tRNA | Amino Acid | Codon | No. | RSCU | tRNA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T. nigrans | Phe | UUU | 1821 | 1.22 | Tyr | UAU | 1227 | 1.38 | Stop | UAG | 584 | 0.75 | |||
| Phe | UUC | 1153 | 0.78 | trnF-GAA | Tyr | UAC | 545 | 0.62 | trnY-GUA | Leu | UUG | 863 | 1.3 | trnL-CAA | |
| Leu | UUA | 877 | 1.32 | Stop | UAA | 961 | 1.24 | Leu | CUC | 497 | 0.75 | ||||
| Leu | CUU | 807 | 1.21 | His | CAU | 773 | 1.41 | His | CAC | 321 | 0.59 | ||||
| Leu | CUA | 579 | 0.87 | trnL-UAG | Gln | CAA | 785 | 1.33 | trnQ-UUG | Asn | AAC | 600 | 0.59 | trnN-GUU | |
| Leu | CUG | 370 | 0.56 | Gln | CAG | 397 | 0.67 | Asn | AAU | 1419 | 1.41 | ||||
| Ile | AUU | 1387 | 1.21 | Ile | AUC | 905 | 0.79 | Ile | AUA | 1148 | 1 | ||||
| Asp | GAU | 789 | 1.44 | Lys | AAA | 1617 | 1.37 | Lys | AAG | 748 | 0.63 | ||||
| Met | AUG | 726 | 1 | trn(f)M-CAU trnI-CAU | Val | GUC | 369 | 0.74 | trnV-GAC | Asp | GAC | 304 | 0.56 | trnD-GUC | |
| Val | GUU | 659 | 1.32 | Val | GUA | 599 | 1.2 | Val | GUG | 370 | 0.74 | ||||
| Ser | UCC | 593 | 1.03 | trnS-GGA | Glu | GAA | 997 | 1.37 | trnE-UUC | Cys | UGC | 316 | 0.74 | trnC-GCA | |
| Ser | UCU | 816 | 1.42 | Cys | UGU | 535 | 1.26 | Glu | GAG | 455 | 0.63 | ||||
| Ser | UCG | 490 | 0.85 | Stop | UGA | 777 | 1 | Pro | CCU | 536 | 1.08 | ||||
| Pro | CCC | 499 | 1.01 | Arg | CGC | 228 | 0.56 | Arg | CGA | 461 | 1.14 | ||||
| Trp | UGG | 581 | 1 | trnW-CCA | Arg | CGU | 267 | 0.66 | trnR-ACG | Ser | UCA | 684 | 1.19 | trnS-UGA | |
| Pro | CCA | 589 | 1.19 | trnP-UGG | Thr | ACC | 478 | 1.01 | trnT-GGU | Ser | AGC | 367 | 0.64 | trnS-GCU | |
| Pro | CCG | 358 | 0.72 | Arg | CGG | 301 | 0.75 | Ser | AGU | 491 | 0.86 | ||||
| Thr | ACU | 532 | 1.13 | Arg | AGG | 466 | 1.15 | Thr | ACG | 331 | 0.7 | ||||
| Thr | ACA | 545 | 1.16 | trnT-UGU | Arg | AGA | 700 | 1.73 | trnR-UCU | Gly | GGC | 267 | 0.63 | trnG-GCC | |
| Ala | GCU | 376 | 1.28 | Ala | GCC | 277 | 0.94 | Ala | GCA | 314 | 1.07 | ||||
| Ala | GCG | 206 | 0.7 | Gly | GGA | 527 | 1.23 | Gly | GGU | 444 | 1.04 | ||||
| Gly | GGG | 469 | 1.1 | ||||||||||||
| S. parasitica | Phe | UUU | 1842 | 1.23 | Tyr | UAU | 1157 | 1.33 | Stop | UAA | 985 | 1.32 | |||
| Phe | UUC | 1151 | 0.77 | trnF-GAA | Tyr | UAC | 588 | 0.67 | trnY-GUA | Arg | AGA | 730 | 1.76 | trnR-UCU | |
| Leu | UUA | 883 | 1.32 | Stop | UAG | 542 | 0.72 | Leu | CUU | 777 | 1.16 | ||||
| Leu | CUC | 477 | 0.71 | His | CAC | 325 | 0.66 | His | CAU | 664 | 1.34 | ||||
| Leu | CUA | 578 | 0.86 | trnL-UAG | Gln | CAA | 738 | 1.35 | trnQ-UUG | Leu | UUG | 909 | 1.35 | trnL-CAA | |
| Leu | CUG | 403 | 0.6 | Gln | CAG | 354 | 0.65 | Ile | AUU | 1468 | 1.27 | ||||
| Ile | AUC | 781 | 0.67 | Asn | AAU | 1438 | 1.39 | Lys | AAG | 732 | 0.6 | ||||
| Met | AUG | 701 | 1 | trn(f)M-CAU trnI-CAU | Asn | AAC | 624 | 0.61 | trnN-GUU | Glu | GAA | 941 | 1.34 | trnE-UUC | |
| Ile | AUA | 1223 | 1.06 | Lys | AAA | 1725 | 1.4 | Val | GUA | 571 | 1.19 | ||||
| Val | GUU | 673 | 1.4 | Asp | GAU | 832 | 1.4 | Val | GUG | 350 | 0.73 | ||||
| Val | GUC | 328 | 0.68 | trnV-GAC | Asp | GAC | 360 | 0.6 | trnD-GUC | Ser | UCA | 709 | 1.15 | trnS-UGA | |
| Ser | UCU | 861 | 1.4 | Cys | UGU | 603 | 1.28 | Glu | GAG | 468 | 0.66 | ||||
| Ser | UCC | 685 | 1.11 | trnS-GGA | Cys | UGC | 336 | 0.72 | trnC-GCA | Trp | UGG | 524 | 1 | trnW-CCA | |
| Ser | UCG | 508 | 0.83 | Stop | UGA | 716 | 0.96 | Arg | CGA | 457 | 1.1 | ||||
| Pro | CCU | 480 | 1.01 | Arg | CGU | 305 | 0.74 | trnR-ACG | Pro | CCA | 569 | 1.2 | trnP-UGG | ||
| Pro | CCC | 519 | 1.1 | Pro | CCG | 324 | 0.68 | Arg | CGC | 217 | 0.52 | ||||
| Thr | ACU | 510 | 1.11 | Ser | AGU | 540 | 0.88 | Arg | CGG | 317 | 0.77 | ||||
| Thr | ACC | 470 | 1.03 | trnT-GGU | Ser | AGC | 388 | 0.63 | trnS-GCU | Gly | GGC | 278 | 0.65 | trnG-GCC | |
| Thr | ACA | 530 | 1.16 | trnT-UGU | Ala | GCC | 279 | 0.94 | Arg | AGG | 460 | 1.11 | |||
| Thr | ACG | 321 | 0.7 | Ala | GCG | 186 | 0.62 | Ala | GCA | 352 | 1.18 | ||||
| Ala | GCU | 374 | 1.26 | Gly | GGA | 563 | 1.31 | Gly | GGU | 421 | 0.98 | ||||
| Gly | GGG | 462 | 1.07 |
| Species | Gene | Location | Exon I(bp) | Intron I(bp) | Exon II(bp) | Intron II(bp) | Exon III(bp) |
|---|---|---|---|---|---|---|---|
| T. nigrans | atpF | LSC | 150 | 786 | 390 | ||
| rpoC1 | LSC | 450 | 756 | 1602 | |||
| ycf3 | LSC | 127 | 755 | 230 | 785 | 153 | |
| rps12 * | LSC, IR | 114 | - | 232 | 543 | 26 | |
| petB | LSC | 6 | 798 | 642 | |||
| petD | LSC | 9 | 696 | 483 | |||
| rpl2 | LSC, IR | 394 | 652 | 434 | |||
| S. parasitica | atpF | LSC | 150 | 759 | 390 | ||
| rpoC1 | LSC | 456 | 778 | 1626 | |||
| ycf3 | LSC | 153 | 712 | 230 | 759 | 127 | |
| petB | LSC | 6 | 700 | 642 | |||
| petD | LSC | 9 | 652 | 483 | |||
| rpl2 | LSC, IR | 394 | 665 | 431 | |||
| rps12 * | LSC, IR | 114 | - | 232 | 534 | 26 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yue, X.; Miao, N.; Fan, Z.; Mao, K. Comparative Analyses of Chloroplast Genomes for Parasitic Species of Santalales in the Light of Two Newly Sequenced Species, Taxillus nigrans and Scurrula parasitica. Genes 2023, 14, 560. https://doi.org/10.3390/genes14030560
Yue X, Miao N, Fan Z, Mao K. Comparative Analyses of Chloroplast Genomes for Parasitic Species of Santalales in the Light of Two Newly Sequenced Species, Taxillus nigrans and Scurrula parasitica. Genes. 2023; 14(3):560. https://doi.org/10.3390/genes14030560
Chicago/Turabian StyleYue, Ximing, Ning Miao, Zilu Fan, and Kangshan Mao. 2023. "Comparative Analyses of Chloroplast Genomes for Parasitic Species of Santalales in the Light of Two Newly Sequenced Species, Taxillus nigrans and Scurrula parasitica" Genes 14, no. 3: 560. https://doi.org/10.3390/genes14030560
APA StyleYue, X., Miao, N., Fan, Z., & Mao, K. (2023). Comparative Analyses of Chloroplast Genomes for Parasitic Species of Santalales in the Light of Two Newly Sequenced Species, Taxillus nigrans and Scurrula parasitica. Genes, 14(3), 560. https://doi.org/10.3390/genes14030560






