Characterization of phi112, a Molecular Marker Tightly Linked to the o2 Gene of Maize, and Its Utilization in Multiplex PCR for Differentiating Normal Maize from QPM
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval
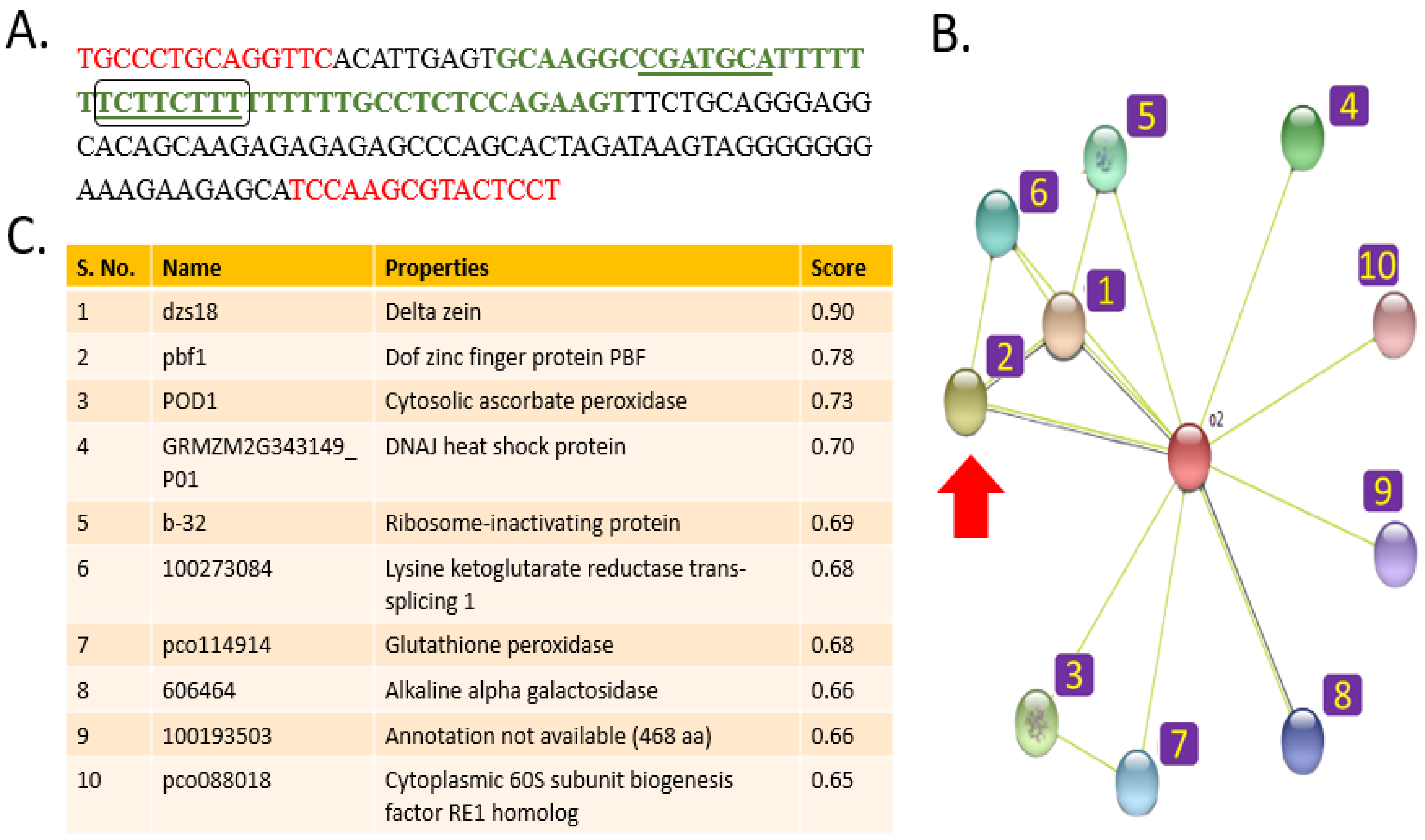
2.2. DNA Motif Discovery
2.3. Protein Interaction Analysis
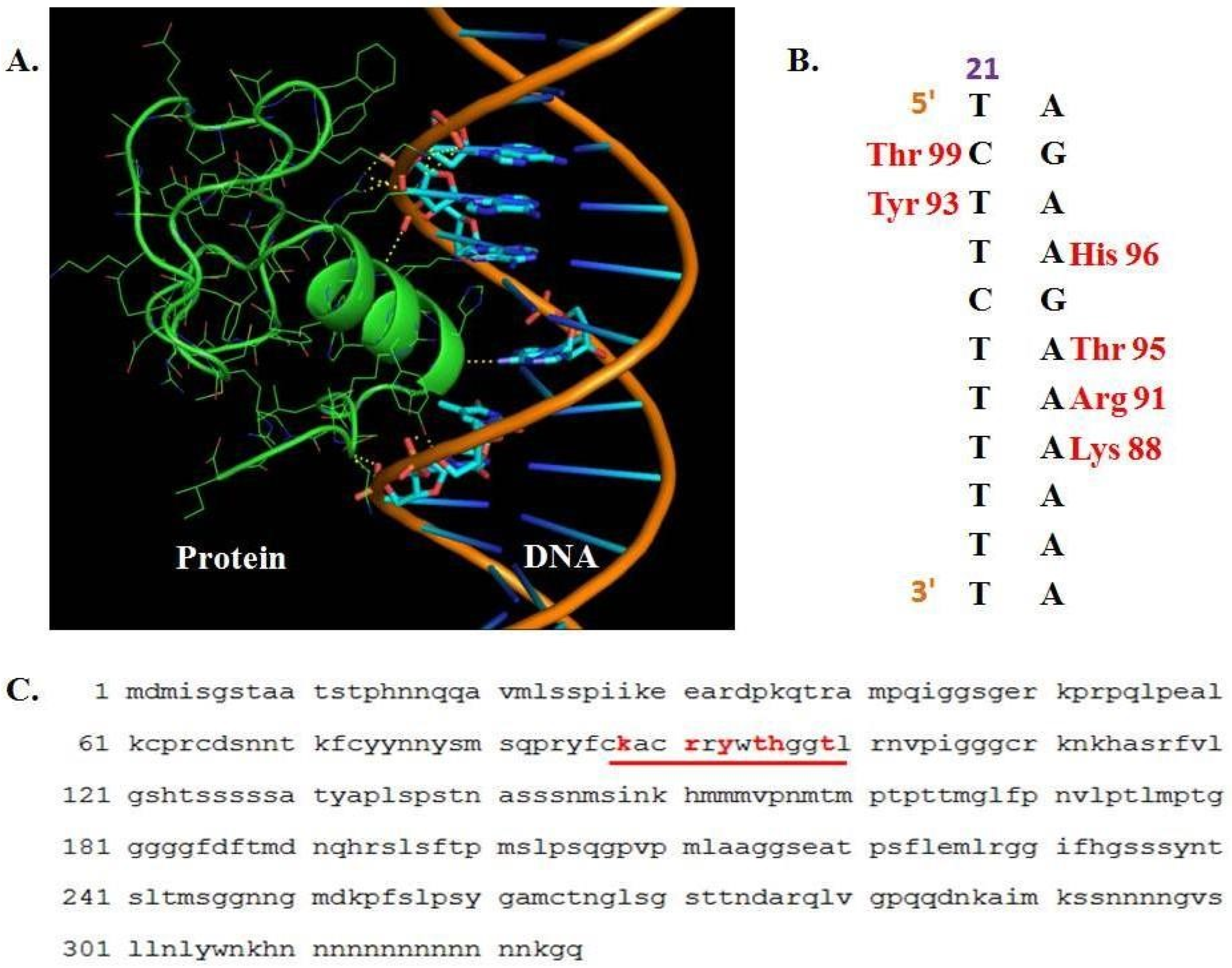
2.4. Generation of a Homology Model of Zea Mays PBF1
2.5. Elucidation of Protein–DNA Interaction
2.6. PCR Amplification
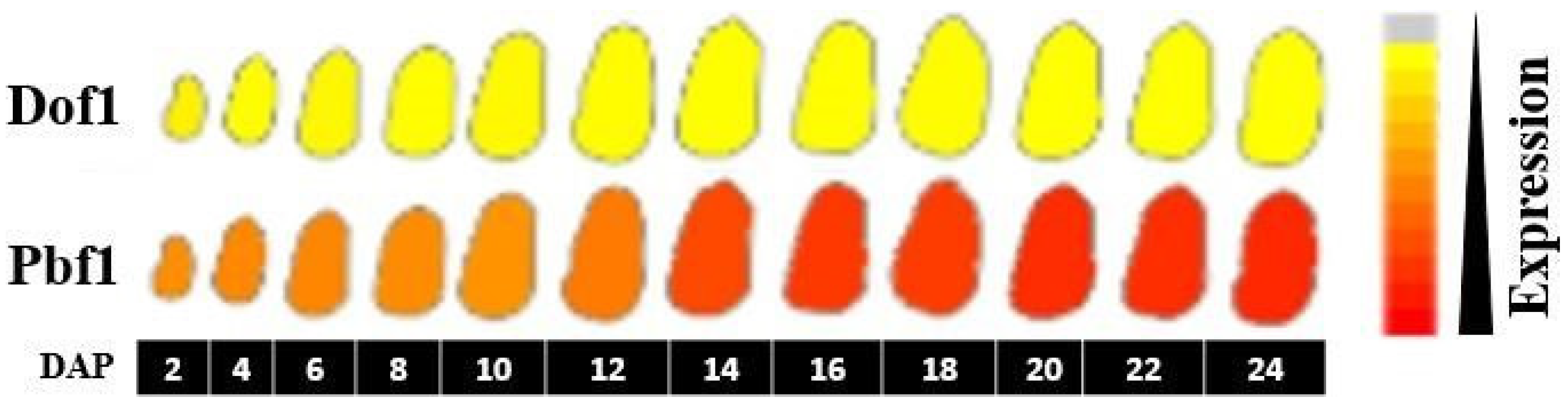
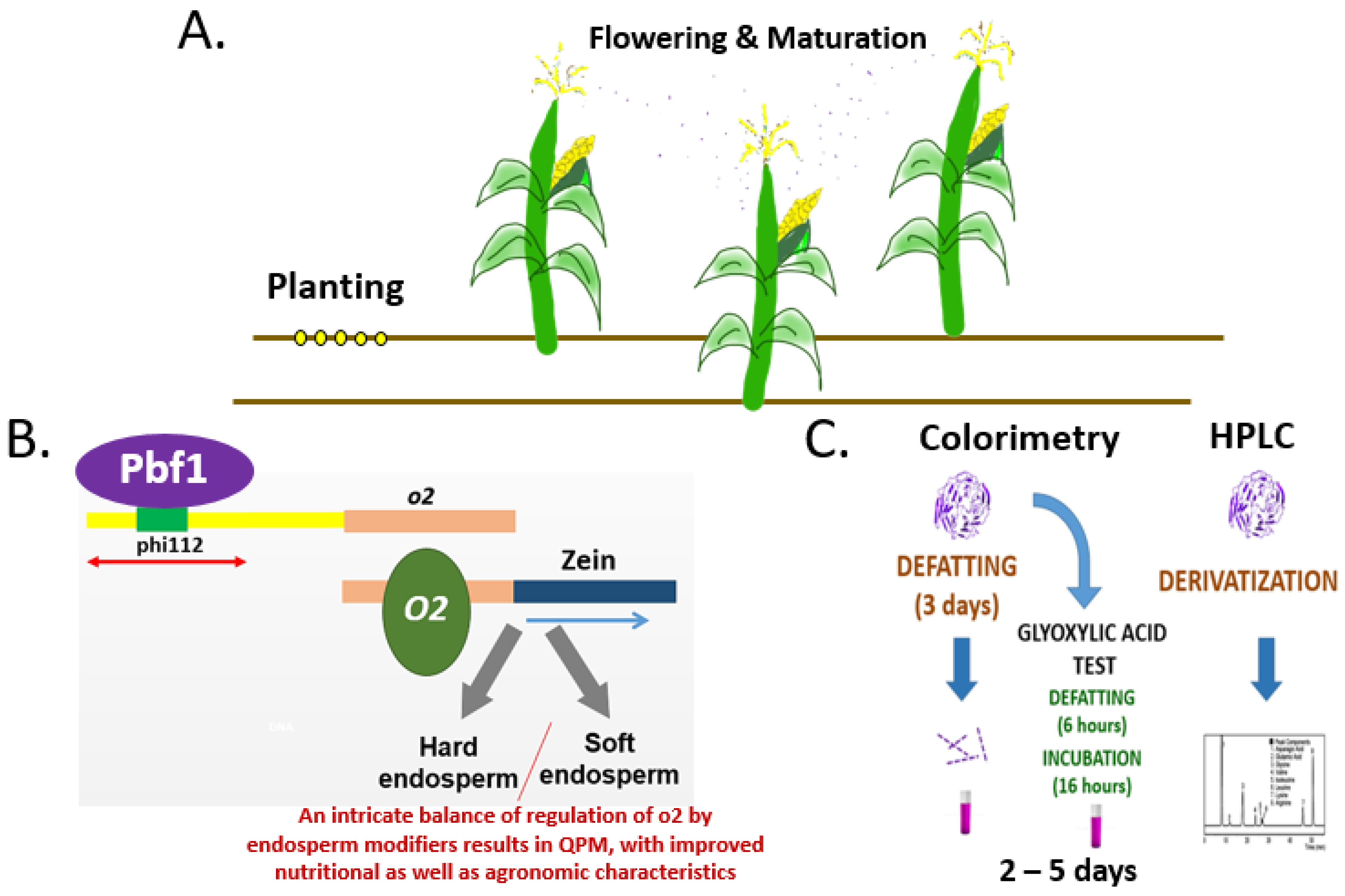
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rouf Shah, T.; Prasad, K.; Kumar, P. Maize—A potential source of human nutrition and health: A review. Cogent Food Agric. 2016, 2, 1166995. [Google Scholar] [CrossRef]
- Kaul, J.; Jain, K.; Olakh, D. An Overview on Role of Yellow Maize in Food, Feed and Nutrition Security. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 3037–3048. [Google Scholar] [CrossRef]
- Choudhary, M.; Singh, A.; Gupta, M.; Rakshit, S. Enabling technologies for utilization of maize as a bioenergy feedstock. Biofuels Bioprod. Biorefining 2020, 14, 402–416. [Google Scholar] [CrossRef]
- Nuss, E.T.; Tanumihardjo, S.A. Maize: A Paramount Staple Crop in the Context of Global Nutrition. Compr. Rev. Food Sci. Food Saf. 2010, 9, 417–436. [Google Scholar] [CrossRef] [PubMed]
- Vietmeyer, N.D. A drama in three long acts: The story behind the story of the development of quality-protein maize. Diversity 2000, 16, 29–32. [Google Scholar]
- Mertz, E.T.; Bates, L.S.; Nelson, O.E. Mutant Gene That Changes Protein Composition and Increases Lysine Content of Maize Endosperm. Science 1964, 145, 279–280. [Google Scholar] [CrossRef]
- Azevedo, R.A.; Arruda, P. High-lysine maize: The key discoveries that have made it possible. Amino Acids 2010, 39, 979–989. [Google Scholar] [CrossRef] [PubMed]
- Vasal, S.K. The Quality Protein Maize Story. Food Nutr. Bull. 2000, 21, 445–450. [Google Scholar] [CrossRef]
- De Groote, H.; Gunaratna, N.S.; Okuro, J.O.; Wondimu, A.; Chege, C.K.; Tomlins, K. Consumer acceptance of quality protein maize (QPM) in East Africa. J. Sci. Food Agric. 2014, 94, 3201–3212. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Qiao, Z.; Qi, W.; Wang, Q.; Yuan, Y.; Yang, X.; Tang, Y.; Mei, B.; Lv, Y.; Zhao, H.; et al. Genome-Wide Characterization of cis-Acting DNA Targets Reveals the Transcriptional Regulatory Framework of Opaque2 in Maize. Plant Cell 2015, 27, 532–545. [Google Scholar] [CrossRef]
- Lopes, M.A.; Takasaki, K.; Bostwick, D.E.; Helentjaris, T.; Larkins, B.A. Identification of two opaque2 modifier loci in Quality Protein Maize. Mol. Genet. Genom. 1995, 247, 603–613. [Google Scholar] [CrossRef]
- Geetha, K.B.; Lending, C.R.; Lopes, M.A.; Wallace, J.C.; Larkins, B.A. Opaque-2 modifiers increase α-zein synthesis and alter its spatial distribution in maize endosperm. Plant Cell. 1991, 3, 1207–1219. [Google Scholar]
- Holding, D.R. Recent advances in the study of prolamin storage protein organization and function. Front. Plant Sci. 2014, 5, 276. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xiang, X.; Huang, Y.; Zhou, Y.; An, D.; Dong, J.; Zhao, C.; Liu, H.; Li, Y.; Wang, Q.; et al. Long-read sequencing reveals genomic structural variations that underlie creation of quality protein maize. Nat. Commun. 2020, 11, 17. [Google Scholar] [CrossRef]
- Hunter, B.G.; Beatty, M.K.; Singletary, G.W.; Hamaker, B.R.; Dilkes, B.P.; Larkins, B.A.; Jung, R. Maize Opaque Endosperm Mutations Create Extensive Changes in Patterns of Gene Expression. Plant Cell 2002, 14, 2591–2612. [Google Scholar] [CrossRef] [PubMed]
- Gibbon, B.C.; Larkins, B.A. Molecular genetic approaches to developing quality protein maize. Trends Genet. 2005, 21, 227–233. [Google Scholar] [CrossRef]
- Zhan, J.; Li, G.; Ryu, C.H.; Ma, C.; Zhang, S.; Lloyd, A.; Hunter, B.G.; Larkins, B.A.; Drews, G.N.; Wang, X.; et al. Opaque-2 regulates a complex gene network associated with cell differentiation and storage functions of maize endosperm. Plant Cell 2018, 30, 2425–2446. [Google Scholar] [CrossRef]
- Huang, S.; Frizzi, A.; Florida, C.A.; Kruger, D.E.; Luethy, M.H. High Lysine and High Tryptophan Transgenic Maize Resulting from the Reduction of Both 19- and 22-kD α-zeins. Plant Mol. Biol. 2006, 61, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Bicar, E.H.; Woodman-Clikeman, W.; Sangtong, V.; Peterson, J.M.; Yang, S.S.; Lee, M.; Scott, M.P. Transgenic maize endosperm containing a milk protein has improved amino acid balance. Transgenic Res. 2008, 17, 59–71. [Google Scholar] [CrossRef]
- Yu, J.; Peng, P.; Zhang, X.; Zhao, Q.; Zhy, D.; Sun, X.; Liu, J.; Ao, G. Seed-specific expression of a lysine rich protein sb401 gene significantly increases both lysine and total protein content in maize seeds. Mol. Breed. 2004, 14, 1–7. [Google Scholar] [CrossRef]
- Yue, J.; Li, C.; Zhao, Q.; Zhu, D.; Yu, J. Seed-specific expression of a lysine-rich protein gene, GhLRP, from cotton significantly increases the lysine content in maize seeds. Int. J. Mol. Sci. 2014, 15, 5350–5365. [Google Scholar] [CrossRef]
- Chang, Y.; Shen, E.; Wen, L.; Yu, J.; Zhu, D.; Zhao, Q. Seed-Specific Expression of the Arabidopsis AtMAP18 Gene Increases both Lysine and Total Protein Content in Maize. PLoS ONE 2015, 10, e0142952. [Google Scholar] [CrossRef]
- Liu, C.; Li, S.; Yue, J.; Xiao, W.; Zhao, Q.; Zhu, D.; Yu, J. Microtubule-Associated Protein SBgLR Facilitates Storage Protein Deposition and Its Expression Leads to Lysine Content Increase in Transgenic Maize Endosperm. Int. J. Mol. Sci. 2015, 16, 29772–29786. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, C.; Li, S.; Zhu, D.; Zhao, Q.; Yu, J. Improved nutritive quality and salt resistance intransgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1. Int. J. Mol. Sci. 2013, 14, 9459–9474. [Google Scholar] [CrossRef] [PubMed]
- Nerling, D.; Coelho, C.M.M.; Brümmer, A. Biochemical profiling and its role in physiological quality of maize seeds. J. Seed Sci. 2018, 40, 7–15. [Google Scholar] [CrossRef]
- Yadava, P.; Singh, A.; Kumar, K.; Sapna; Singh, I. Plant Senescence and Agriculture. In Senescence Signalling and Control in Plants, 1st ed.; Elsevier: London, UK, 2019; pp. 283–302. [Google Scholar] [CrossRef]
- Dhir, A.; Kaur, C.; Devi, V.; Singh, A.; Das, A.K.; Rakshit, S.; Chaudhary, D.P. A Rapid Single Kernel Screening Method for Preliminary Estimation of Amylose in Maize. Food Anal. Methods 2022, 15, 2163–2171. [Google Scholar] [CrossRef]
- Meacham-Hensold, K.; Fu, P.; Wu, J.; Serbin, S.; Montes, C.M.; Ainsworth, E.; Guan, K.; Dracup, E.; Pederson, T.; Driever, S.; et al. Plot-level rapid screening for photosynthetic parameters using proximal hyperspectral imaging. J. Exp. Bot. 2020, 71, 2312–2328. [Google Scholar] [CrossRef]
- Tembo, M.; Adediji, A.O.; Bouvaine, S.; Chikoti, P.C.; Seal, S.E.; Silva, G. A quick and sensitive diagnostic tool for detection of Maize streak virus. Sci. Rep. 2020, 10, 19633. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhu, Y.; Tao, X.; Chen, X.; Li, X. Rapid prediction of winter wheat yield and nitrogen use efficiency using consumer-grade unmanned aerial vehicles multispectral imagery. Front. Plant Sci. 2022, 13, 1032170. [Google Scholar] [CrossRef]
- Sethi, M.; Kumar, S.; Singh, A.; Chaudhary, D.P. Temporal profiling of essential amino acids in developing maize kernel of normal, opaque-2 and QPM germplasm. Physiol. Mol. Biol. Plants 2020, 26, 341–351. [Google Scholar] [CrossRef]
- Sethi, M.; Singh, A.; Kaur, H.; Phagna, R.K.; Rakshit, S.; Chaudhary, D.P. Expression profile of protein fractions in the developing kernel of normal, Opaque-2 and quality protein maize. Sci. Rep. 2021, 11, 2469. [Google Scholar] [CrossRef]
- Kaur, C.; Singh, A.; Sethi, M.; Devi, V.; Chaudhary, D.P.; Phagna, R.K.; Langyan, S.; Bhushan, B.; Rakshit, S. Optimization of Protein Quality Assay in Normal, opaque-2, and Quality Protein Maize. Front. Sustain. Food Syst. 2022, 6, 743019. [Google Scholar] [CrossRef]
- Harper, L.; Gardiner, J.; Andorf, C.; Lawrence, C.J. MaizeGDB: The Maize Genetics and Genomics Database. Plant Bioinform. Methods Protoc. 2016, 1374, 187–202. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37 (Suppl. S2), w202–w208. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Snel, B.; Lehmann, G.; Bork, P.; Huynen, M.A. STRING: A web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000, 28, 3442–3444. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wen, Z.; Wang, X.; Huang, S.Y. Addressing recent docking challenges: A hybrid strategy to integrate template-based and free protein-protein docking. Proteins Struct. Funct. Bioinform. 2017, 85, 497–512. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein–protein and protein–DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Doyle, J.J. A rapid total DNA preparation procedure for fresh plant tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Wang, Z.; Ueda, T.; Messing, J. Characterization of the maize prolamin box-binding factor-1 (PBF-1) and its role in the developmental regulation of the zein multigene family. Gene 1998, 223, 321–332. [Google Scholar] [CrossRef]
- Yanagisawa, S. A novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 1995, 23, 3403–3410. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof Domain Proteins: Plant-Specific Transcription Factors Associated with Diverse Phenomena Unique to Plants. Plant Cell Physiol. 2004, 45, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, H.; Bates, L.S. A Modified Method for Rapid Tryptophan Analysis of Maize; CIMMYT: Mexico City, Mexico, 1969. [Google Scholar]
- Nurit, E.; Tiessen, A.; Pixley, K.V.; Palacios-Rojas, N. Reliable and Inexpensive Colorimetric Method for Determining Protein-Bound Tryptophan in Maize Kernels. J. Agric. Food Chem. 2009, 57, 7233–7238. [Google Scholar] [CrossRef] [PubMed]
- Çevikkalp, S.A.; Löker, G.B.; Yaman, M.; Amoutzopoulos, B. A simplified HPLC method for determination of tryptophan in some cereals and legumes. Food Chem. 2016, 193, 26–29. [Google Scholar] [CrossRef]
- Kaur, N.; Kumar, R.; Singh, A.; Shobha, D.; Das, A.K.; Chaudhary, D.; Kaur, Y.; Kumar, P.; Sharma, P.; Singh, B. Improvement in nutritional quality of traditional unleavened flat bread using Quality Protein Maize. Front. Nutr. 2022, 9, 963368. [Google Scholar] [CrossRef]
- Magulama, E.E.; Sales, E.K. Marker-assisted introgression of opaque 2 gene into elite maize inbred lines. USM R D 2009, 17, 131–135. [Google Scholar]
- Babu, R.; Nair, S.; Kumar, A.; Venkatesh, S.; Sekhar, J.C.; Singh, N.N.; Srinivasan, G.; Gupta, H.S. Two-generation marker-aided backcrossing for rapid conversion of normal maize lines to quality protein maize (QPM). Theor. Appl. Genet. 2005, 111, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Dreher, K.; Khairallah, M.; Ribaut, J.-M.; Morris, M. Money matters (I): Costs of field and laboratory procedures associated with conventional and marker-assisted maize breeding at CIMMYT. Mol. Breed. 2003, 11, 221–234. [Google Scholar] [CrossRef]
- Jilo, T. Nutritional benefit and development of quality protein maize (QPM) in Ethiopia. Cereal Res. Commun. 2021, 50, 559–572. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, A.; Karjagi, C.; Kaur, S.; Jeet, G.; Bhamare, D.; Gupta, S.; Kumar, S.; Das, A.; Gupta, M.; Chaudhary, D.P.; et al. Characterization of phi112, a Molecular Marker Tightly Linked to the o2 Gene of Maize, and Its Utilization in Multiplex PCR for Differentiating Normal Maize from QPM. Genes 2023, 14, 531. https://doi.org/10.3390/genes14020531
Singh A, Karjagi C, Kaur S, Jeet G, Bhamare D, Gupta S, Kumar S, Das A, Gupta M, Chaudhary DP, et al. Characterization of phi112, a Molecular Marker Tightly Linked to the o2 Gene of Maize, and Its Utilization in Multiplex PCR for Differentiating Normal Maize from QPM. Genes. 2023; 14(2):531. https://doi.org/10.3390/genes14020531
Chicago/Turabian StyleSingh, Alla, Chikkappa Karjagi, Sehgeet Kaur, Gagan Jeet, Deepak Bhamare, Sonu Gupta, Sunil Kumar, Abhijit Das, Mamta Gupta, D. P. Chaudhary, and et al. 2023. "Characterization of phi112, a Molecular Marker Tightly Linked to the o2 Gene of Maize, and Its Utilization in Multiplex PCR for Differentiating Normal Maize from QPM" Genes 14, no. 2: 531. https://doi.org/10.3390/genes14020531
APA StyleSingh, A., Karjagi, C., Kaur, S., Jeet, G., Bhamare, D., Gupta, S., Kumar, S., Das, A., Gupta, M., Chaudhary, D. P., Bhushan, B., Jat, B. S., Kumar, R., Dagla, M. C., & Kumar, M. (2023). Characterization of phi112, a Molecular Marker Tightly Linked to the o2 Gene of Maize, and Its Utilization in Multiplex PCR for Differentiating Normal Maize from QPM. Genes, 14(2), 531. https://doi.org/10.3390/genes14020531






