Abstract
Some families of mobile elements in bacterial genomes encode not only a transposase but also an accessory TnpB gene. This gene has been shown to encode an RNA-guided DNA endonuclease, co-evolving with Y1 transposase and serine recombinase in mobile elements IS605 and IS607. In this paper, we reveal the evolutionary relationships among TnpB-containing mobile elements (TCMEs) in well-assembled genomes of six bacterial species: Bacillus cereus, Clostridioides difficile, Deinococcus radiodurans, Escherichia coli, Helicobacter pylori and Salmonella enterica. In total, 9996 TCMEs were identified in 4594 genomes. They belonged to 39 different insertion sequences (ISs). Based on their genetic structures and sequence identities, the 39 TCMEs were classified into three main groups and six subgroups. According to our phylogenetic analysis, TnpBs include two main branches (TnpB-A and TnpB-B) and two minor branches (TnpB-C and TnpB-D). The key TnpB motifs and the associated Y1 and serine recombinases were highly conserved across species, even though their overall sequence identities were low. Substantial variation was observed for the rate of invasion across bacterial species and strains. Over 80% of the genomes of B. cereus, C. difficile, D. radiodurans and E. coli contained TCMEs; however, only 64% of the genomes of H. pylori and 44% of S. enterica genomes contained TCMEs. IS605 showed the largest rate of invasion in these species, while IS607 and IS1341 had a relatively narrow distribution. Co-invasions of IS605, IS607 and IS1341 elements were observed in various genomes. The largest average copy number was observed for IS605b elements in C. difficile. The average copy numbers of most other TCMEs were smaller than four. Our findings have important implications for understanding the co-evolution of TnpB-containing mobile elements and their biological roles in host genome evolution.
1. Introduction
Insertion sequences (ISs) are the simplest mobile genetic elements and only contain the gene required for transposition and regulation. Notably, IS elements are widely spread in prokaryotes, and some families invade eukaryotes [1,2]. Furthermore, ISs play an essential role in shaping the genome’s evolution and its hosts’ adaptability [3]. The IS605 and IS607 are recognized as two distinct IS elements with a similar structural organization. They have two open reading frames (ORFA and ORFB) and encode two nuclease genes flanked with subterminal left end (LE) and right end (RE) palindromic elements. The ORFBs from IS605 and IS607 encode the TnpB nuclease, harboring a conserved RuvC motif. In contrast, the ORFAs of IS605 and IS607 encode substantially different nuclease genes belonging to the distinct superfamily of enzymes and do not harbor the classical “DDE” domain of IS transposases [4]. The ORFAs of IS605 encode single-domain proteins containing a single catalytic tyrosine recombinase (called Y1) which belongs to the HuH enzyme superfamily, containing a conserved amino-acid triad composed of Histidine (H)-bulky hydrophobic residue (u)-Histidine (H) [4]. Additionally, the ORFAs of IS607 encode a serine recombinase (SR) which carries a predicted helix-turn-helix DNA binding domain at the N-terminus and a catalytic domain at the C-terminus [5,6].
The transposition activity of IS605 and IS608 from H. pylori [7,8,9,10] and ISDra2 from D. radiodurans have been characterized [11,12], and their transposition mechanism has been analyzed extensively [4]. The transposition of IS605 members can be described as a “Peel and Paste” single-strand transposition mechanism [4]; this distinguishes them from classical ISs, which move via double-strand DNA intermediates. Although the mechanism for the SR of IS607 is still obscure, the transposition of IS607 has been detected in E. coli using a mating-out assay [13]. It is commonly accepted that the Y1 transposase of IS605 plays a crucial role in its transposition and is sufficient to promote its mobility in vivo and in vitro. While TnpB, designated as the accessory gene of IS605, is not required for the transposition of either IS608 or ISDra2 in E. coli and D. radiodurans [8,13,14,15], it has been suggested that TnpBs might play a regulatory role in transposition in bacteria [11]. The main function of TnpBs remains largely unknown. In very recent studies, both evolutionary analysis and biochemical experiments have suggested that TnpBs are RNA-guided endonucleases [16,17].
At least three groups (IS605, IS607, and IS1341) of TnpB-containing mobile elements (TCMEs) have been recognized. IS605 and IS607 carry two nuclease-encoding genes flanked with LE and RE elements. Although IS1341 displays fundamental differences in its organization, it only harbors one nuclease encoding gene (TnpB) flanked by the end elements of ISs [4]. However, it is still unclear whether members of IS1341 are autonomous or decay copies from IS605 or IS607 elements. These groups are widely distributed in prokaryotes and have been reported in eukaryotes [4,18]. IS605, IS607, and IS1341 have been detected in 71 species, 34 species, and 76 species, respectively, based on ISfinder database (http://www-is.biotoul.fr). The structure and activity of IS605, IS606, ISHp608 and ISHp609 from Helicobacter pylori, belonging to IS605 groups, have been well defined [15,18,19,20]. However, the evolution landscape of TCMEs is still poorly understood.
Here, the evolution profiles, including diversity, structure organization, and copy number in the genome of TCMEs across six species including Bacillus cereus (B. cereus), Clostridioides difficile (C. difficile), Deinococcus radiodurans (D. radiodurans), Escherichia coli (E. coli), Helicobacter pylori (H. pylori), and Salmonella enterica (S. enterica), were systematically investigated using the available genomes of high assembly levels (chromosome and complete). These data provide a basic framework for future evolutionary study, and the characteristics describing each subgroup may help in further experimental studies.
2. Materials and Methods
2.1. TnpB-Containing Mobile Elements Mining
All known TnpB proteins were collected from the ISfinder (https://www-is.biotoul.fr/index.php) accessed on October 2022 (including TCMEs: IS605, IS607, and IS1341) and Pfma (http://pfam.xfam.org/ accessed on 6 October 2022) databases with access numbers of PF12323, PF07282, and PF01385. Sequence lengths of less than 200 aa or longer than 600 aa were discarded. In total, 21,733 sequences were obtained and submitted for clustering using the cd-hit program with an 80% identity; 4399 clusters were obtained. The conserved RuvC domains (about 120 aa) of these TnpB representative proteins from each cluster were aligned to the RuvC hidden Markov model (HMM) profile of TnpB (ORFB_IS605.hmm, PF01385), which was downloaded from the Pfma database (https://pfam.xfam.org 6 October 2022), via the hmmalign program of HMMER3 [21]. The sequences with truncated RuvC domains (greater than 20% gaps) were discarded. Finally, 3661 cluster-representative sequences with well-conserved RuvC domains of TnpB were obtained and submitted for rebuilding a new HMM profile of the TnpB-conserved domain with the hmmbuild program of HMMER3 [21]. The hmmsearch program of HMM3 was then used to search against the non-redundant protein sequences (NRs), which were downloaded from the NCBI database (https://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz, accessed on 6 October 2021), with the newly constructed HMM profile. Target hits (altogether 150, 497 proteins) with default parameters are reported. Four species (Escherichia coli, Clostridioides difficile, Bacillus cereus, Salmonella enterica) with the most abundant hits (Supplementary Figure S1), and two species (Deinococcus radiodurans and Helicobacter pylor) with function at the TnpB-containing mobile elements previously defined by He [22,23] and Pasternak [11,14] were used for further TCME mining.
The genomes of a high assembly level (chromosome and complete) of Bacillus cereus (152), Clostridioides difficile (133), Deinococcus radiodurans (12), Escherichia coli (2467), Helicobacter pylor (335), and Salmonella enterica (1495) were downloaded from NCBI and used for TCMEs mining. These genomes were translated into proteins with the transeq program embedded in emboss software [24]. The TnpB homology proteins were then searched against the translated proteins by using the hmmsearch program of HMM3 with the HMM profile. Target hits with sequence lengths shorter than 50 aa were discarded, and the remaining target hits (genomic coordinates) were used to extract the DNA sequences with flank extensions of 1.7 kb upstream and 1.7 kb downstream of hits in genomes. The extracted DNA sequences were clustered by the USEARCH program with a 70% identity, and representative sequences from each cluster from each species were aligned by MAFFT. The boundaries of complete elements and structure organizations were manually checked and defined for each cluster. The elements with detectable LE and RE boundaries (identifiable cleavage site sequences) were designated full TCME elements. The translated proteins from the truncated elements with undetectable LE or RE or missing both LE and RE boundaries were used as queries to BLASTP against the ISfinder database to define their classification, and the truncated elements were discarded in cases of no homology hit to the ORFBs of IS605, IS607, or IS1341 by BLASTP searching.
Finally, all the full TCME elements obtained were clustered again using the USEARCH program with a 90% identity. The consensus sequences were derived for each cluster and BLASTN to the ISfinder database, and the obtained IS elements were designated as the known IS elements; that is, if they shared over 90% sequence identity with any known sequences from IS605, IS607, or IS1341 group with highly conserved LE and RE sequences and the cleavage sites. If otherwise, they were designated as new IS elements and named according to the nomenclature rules of ISfinder [25,26,27].
The upstream and downstream sequences of the TnpB ORFs of all the identified IS1341 elements were aligned to the Y1 or SR ORF coding sequences (forward or reverse) of IS605 and IS607. If the IS1341 elements contained Y1 or SR-derived sequences (more than 20 bp match by alignment), they were designated as the decays of IS605 or IS607 and removed from the IS1341 group.
2.2. Sequence Analysis and Phylogenetic Tree Construction
The potential ORFs of the obtained TCMEs were predicted by the BioEdit software. The protein domains were defined using the profile hidden Markov models by the online hmmscan web server (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan accessed on 6 October 2022). The obtained TnpA and TnpB sequences and the reference sequences from ISfinder (https://www-is.biotoul.fr/index.php accessed on 6 October 2022) were submitted for alignment using the E-INS-I method from the MAFFT software [28], and the final alignments were used for the phylogenetic tree construction. The phylogenetic tree was inferred [29], and the ultrafast bootstrap approach with 1000 replicates was applied. The best-suited aa substitution model was selected by ModelFinder.
2.3. Hairpin Structures Prediction
The repeat sequences at the subterminal were predicted by the Novopro web tool. The hairpin structures of LE and RE were predicted by the Mfold (DNA Folding Form (mfold.org, accessed on 6 October 2022) and RNAfold web servers (univie.ac.at, accessed on 6 October 2022).
3. Results
3.1. Distribution of TnpB-Containing Mobile Elements (TCMEs) in the Six Species of Bacteria
High-quality assembled genomes of six species, including 152 B. cereus, 133 C. difficile, 2467 E. coli, 12 D. radiodurans, 335 H. pylori, and 1495 S. enterica genomes, were downloaded and submitted for TCME annotation. Overall, 687 copies in B. cereus, 1875 copies in C. difficile, 44 in D. radiodurans, 4825 copies in E. coli, 734 copies in H. pylori, and 1831 copies in S. enterica were obtained and designated as TCME-derived sequences after filtering the sequences without any homology to the TnpBs of IS605, IS605, or IS1341 from ISfinder, as previously described in the methodology.
The expansion of TCMEs across these species varied significantly: over 90% of C. difficile, D. radiodurans, and E. coli contained TCMEs, and 83% of the detected genomes of B. cereus harbored TCME copies. In comparison, only 64% and 44% of the detected genomes of H. pylori and S. enterica contained TCME copies, respectively. In addition, for TCME-detected genomes, the average copy number of TCMEs varied significantly across the six species (Table 1). The highest average TCME copy number (over 14) was observed for each genome of C. difficile, followed by B. cereus with an average of 5.28 TCME copies in each genome, and D. radiodurans and H. pylori with approximately 3.5 TCME copies in each genome. In contrast, the average copy numbers of TCMEs in E. coli (2.05 ± 0.94) and S. enterica (1.25 ± 0.61) were less than three in each genome. Most identified TCME copies (over 76%) were presented by full elements with detectable LE and RE boundaries (cleavage sites) in these species except for S. enterica, in which only 41% of TCME copies were full (Table 1).

Table 1.
Distribution of TnpB-containing mobile elements (TCMEs) in six species.
3.2. Protein Type and Structure Organization of TCMEs in Six Species
Overall, 39 IS elements (including 27 new IS elements and 12 IS elements overlapping with ISfinder) were defined for all obtained TCMEs sequences in these genomes based on the structural organization and sequence identity summarized in Supplementary Table S1. The consensus sequences were derived for each IS element. Based on the structure organization, the 39 IS elements were classified into three groups (IS605, IS607, and IS1341), including 15 IS605 elements, 6 IS607 elements and 18 IS1341 elements (Supplementary Table S1 and Figure 1). Furthermore, different genetic structural organizations were observed for IS605 (Figure 1A) and IS607 (Figure 1B). The Y1, TnpB ORFs of IS605a and IS605b were oriented in the same direction with Y1 upstream of TnpB. However, the ORFs of IS605a partially overlapped. In contrast, IS605b was separated (Figure 1A). Similar structures were observed for IS607a and IS607b (Figure 1B), but the ORFA of IS607 encodes aserine recombinase. In addition, the ORFs of IS605c were separated and oriented in the reverse direction (Figure 1A). Only one structure was observed for 18 IS1341 elements (Figure 1C), and we found that more than half (10 elements) of the detected IS1341 elements were shorter than IS605 and IS607, ranging from 1252 bp to 1393 bp. However, 8 IS1341 elements had relatively long lengths ranging from 1512 bp to 2382 bp. The lengths of IS1341 TnpBs ranged from 362 aa to 489 aa (Supplementary Table S1).
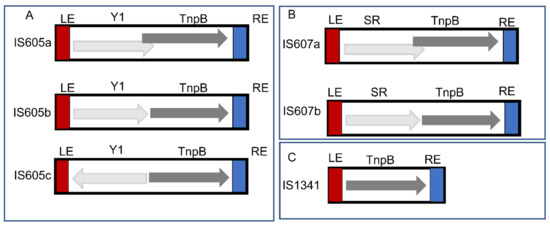
Figure 1.
Structure organization of TCME elements. ORFs are presented as boxes with arrowheads showing the direction of transcription: LE and RE are red and blue boxes, respectively. (A) Three genetic structures of IS605 elements. (B) Two genetic structures of IS607 elements. (C) Genetic structure of IS1341 elements.
Most IS605 elements have a total length of 1.74~1.95 kilobases (kb), two ORFs that encode Y1 transposase of about 140 (102–164 aa), and TnpB nucleuses of about 400 (340–489) aa which are flanked by LE and RE and their cleavage sequences (Supplementary Table S1). Most IS607 elements have a total length of 1.62~2.40 kb and harbor two ORFs that encode serine recombinases of about 200 (150–217 aa) and TnpB nucleuses of about 400 (361–447) aa, respectively, which are flanked by LE and RE and their cleavage sequences (Supplementary Table S1). Both left and right cleavage sites of these IS elements are relatively divergent across 39 IS elements (Supplementary Table S1). However, some cleavage motifs are more frequently detected, such as TCAA (16 IS elements) and TTCA (8 IS elements) for RE cleavage sites and TTAT (10 IS elements) for LE cleavage sites (Supplementary Table S1).
All elements of the IS607 carry several flawed, short, directly repeated sequences at the left end (LE) and right end (RE) and show a lack of inverted repeat sequences at the subterminal (Supplementary Figure S2) which were the putative binding sites of serine recombinases. The sub-terminal palindromic structures were detected for all LEs and REs of IS605 elements (Supplementary File S1), which were the putative binding sites of Y1 transposases, and the structures of hairpins were detectable for the LEs and REs of most IS1341 elements. The alignment of the right flanks (the downstream of the coding sequences of TnpB to RE) of these TCMEs revealed that half of the identified IS elements (seventeen) contain two conserved (GC and G enriched) motifs in the right end of REs (Supplementary Figure S5). In contrast, some TCMEs tend to increase AT and TG nucleotides in the right subterminal of REs (Supplementary Figure S5), which may function as RNA guides.
3.3. Domains and Phylogenetic Analysis of Y1, SR, and TnpB
The overall sequence identity of 39 TnpBs was very low, representing about 24% and ranging from 7.2% to 98.7% (Figure 2A). The phylogenetic tree revealed that the mined TnpBs, together with the TnpB from ISfinder (233 proteins), were classified into two main branches (TnpB-A and TnpB-B) and two minor branches (TnpB-C and TnpB-D) with high bootstrap supports (>70%). IscBs, which were defined previously [19], were used as an outgroup (Figure 3A). In addition, the phylogeny of TnpB seems to be independent of the phylogeny of the TnpB-containing IS elements (Figure 3A). Here, we found that three RuvC (I, II, III) segments, two Zinc-fingers, an arginine/lysine-rich region (RK-rich) and a helix-turn-helix motif (HTH) embedded in the WED domain in the N-terminal, were identified in the alignments of TnpB (Figure 4A,B). In addition, all TnpB proteins, which are highly conserved (Supplementary Figure S4), were defined by Karvelis et al. [16]. Their domain organizations differed from that of IscB, which includes three RuvC I, II, and III motifs, one arginine-rich region (R-rich), one HNH motif, one CXXC zinc finger, and an additional PLMP in the N-terminal IscB [17].
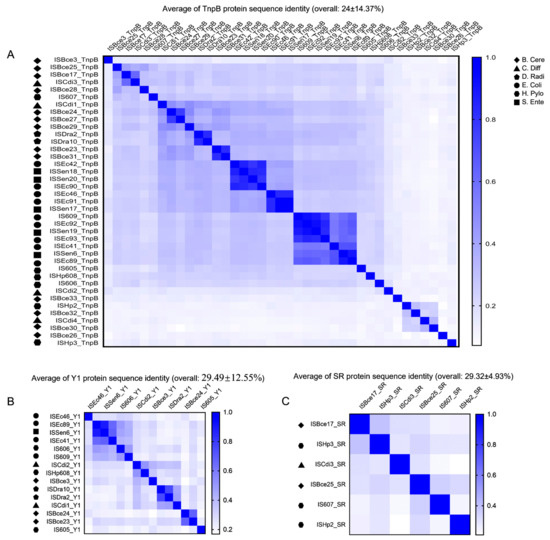
Figure 2.
The overall identity of Y1, SR, and TnpB. (A) The identity of 39 mined TnpB proteins in six species. (B) The identity of 15 mined Y1 proteins in six species. (C) The identity of 6 mined SR proteins in six species.
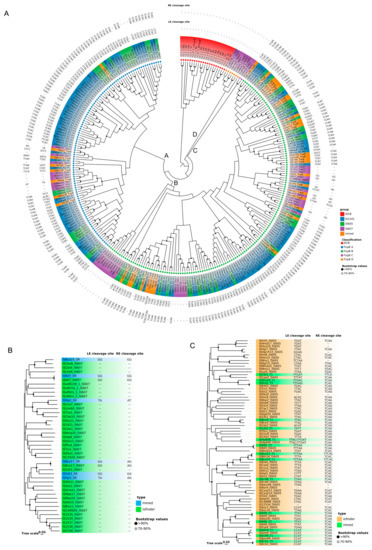
Figure 3.
The phylogenetic tree of SR, Y1, and TnpB. The bootstrap values are indicated with a black circle (>90%) and a grey circle (70–90%). (A) The phylogenetic tree of TnpB and IscB. The TnpB proteins are classified into two main branches (TnpB-A and TnpB-B) and two minor branches (TnpB-C and TnpB-D), are indicated with blue, green, purple, and orange circles, respectively. The IscB formed a single branch shown with red circles. The mined TnpBs from six species are also shown in orange background. The IS1341 subgroup is shown in blue background. The IS605 subgroup is shown in green background. The IS607 family is shown with purple background. (B) The phylogenetic tree of SR. The SR proteins from ISfinder are shown in green background. The mined SR proteins are shown in blue background. (C) The phylogenetic tree of Y1. The Y1 proteins from ISfinder are shown in orange. The mined-Y1 proteins are shown in green.

Figure 4.
The alignments of TnpB, Y1, and SR protein sequences. The alignment was performed with MAFFT and drawn by Jalview Version 2. (A) Schematic of TnpB. The key domains include HTH—helix turn helix (blue), WED—wedge domain (grey), RuvC I, II, III (green), HH—arginine rich helix (yellow), ZF—zinc finger (red). (B) The alignment of several sequences was selected from TnpB-B, TnpB-B, TnpB-C and TnpB-D subgroups according to the phylogenetic tree. The HTH, RKrich HH, ZF, and RuvC I, II, and III domains are marked using a red box. (C) The alignments of the Y1 sequence. The HuH, Y and Q residues are indicated. (D) The alignments of SR. The DNA banding, catalytic, and Helix domains are also shown. The active site seine is marked using a black arrow.
The overall sequence identity (29.49 ± 12.55%, ranging from 16.9% to 89.5%) of mined Y1s was also detected to be low (Figure 2). The alignment of the fifteen mined Y1 proteins (102–164 aa) of the IS605 family revealed five signature sequence motifs, including the His-hydrophobic-His (HuH) motif required for metal ion binding [29]. The HuH triad, catalytic tyrosine (Y), and glutamine (Q) in the C terminal, which are important residues for the transposition [4,30], were highly conserved across these elements (Figure 4B). Most mined Y1s of IS605 seem to be intact; however, two (ISEc46 and ISBce24) are truncated in the N terminal.
Three key domains of SR, including the N-terminal DNA binding domain (DBD), the long central catalytic domain harboring the catalytic serine, and the C-terminal HTH domain were identified based on the alignment (Figure 4C). The average of SR protein sequence identity is approximately 29.32 ± 4.93%, ranging from 20.9% to 39.7% (Figure 2C). All mined SRs of IS607 seem to be complete except ISBce17, with a truncated N terminal, and the whole DBD missing. Phylogenetic trees of Y1s and SRs revealed that these Y1 and SR proteins appear to fall into separate clades. Their associated left and right cleavage sites were generally conserved in the same or close clades (Figure 3), which was also observed for the TnpB tree (Figure 3A).
3.4. Differential Distribution of TCMEs in the Genomes of Six Species
Our data revealed significant differential invasions of TCMEs across six species. Two groups of TCMEs were detected in three species, E. coli (IS605 and IS1341), H. pylori (IS605 and IS607), and S. enterica (IS605 and IS1341); all three groups of TCMEs were detected in B. cereus and C. difficile, while only one group (IS605) was detected in D. radiodurans. In addition, overlapping invasions of IS605 and IS1341 elements were observed for 229 E. coli and 63 S. enterica genomes, respectively. Co-invasions of IS605 and IS607 were observed for 44 H. pylori genomes, and overlapping invasions of IS605, IS607, and IS1341 were observed for 26 B. cereus and 15 C. difficile genomes, respectively (Figure 5). IS605 displayed the most extensive invasions in the genomes of six bacteria species: over 53.95%, 99.25%, 100%, 90.88%, 52.84%, and 20.27% of the total detected genomes of B. cereus, C. difficile, D. radiodurans, E. coli, H. pylori, and S. enterica contained IS605 copies, respectively, while IS607 elements represent a relatively narrow distribution and were only detected in three species (B. cereus, C. difficile, and H. pylori). They also invaded into low proportions of these genomes, representing 31.58% of B. cereus, 11.28% of C. difficile, and 24.18% of H. pylori, respectively. IS1341 was not detected in D. radiodurans and H. pylori, but presented in 112 B. cereus, 106 C. difficile, 312 E. coli, and 418 S. enterica genomes, accounting for 73.68%, 79.70%, 12.65%, and 27.96% of the total detected genomes of these species, respectively. Differential distributions of IS605 and IS607 elements were also observed across the investigated species (Table 2). The differential invasions of TCMEs in genomes may be related to the transposition activity of these elements; thus, the wide distribution of IS605 elements indicates that they may possess a higher transposition activity than that of IS607 and IS1341 elements.
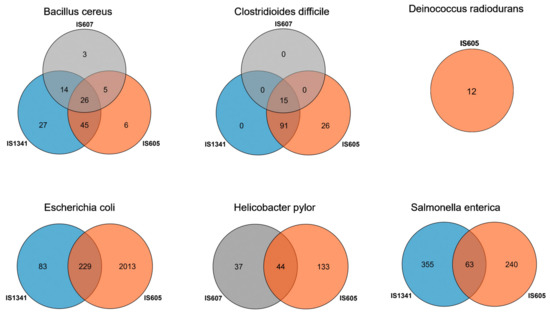
Figure 5.
The genome number of different TCME groups in six species. The IS607 is shown in grey color, IS605 is shown in orange color, and the color blue represents IS1341.

Table 2.
Distribution of TCMEs in the genomes of six species.
Substantial differential copy number variations were observed for the different groups and subgroups of TCMEs across these species. The average copy numbers of most TCME subgroups, including IS605b, IS607a, IS607b in B. cereus, IS605a, IS607b, and IS1341 in C. Diff, IS605c and IS1341 in E. Coli, IS605a in H. pylori, IS605c, and IS1341 in S. enterica, range from one to two in each detected genome. Similarly, for each detected genome, approximately three copies of IS1341 in B. cereus, IS605a in D. radiodurans, and IS605c and IS607a in H. pylori were observed. On average, over 12 copies were only observed for IS605b in C. difficile for each detected genome (Figure 6).
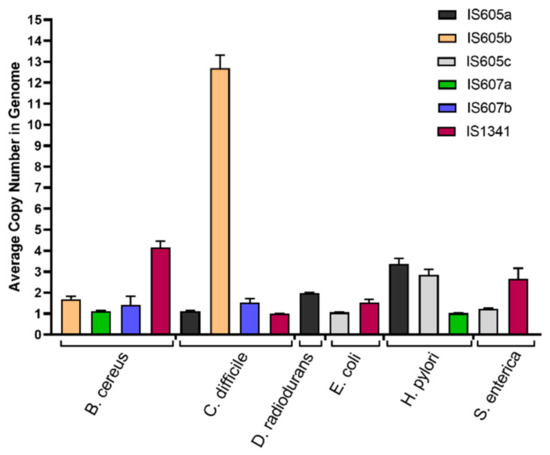
Figure 6.
The average copies number of different subgroups of TCMEs in genomes of six studied bacterial species. The IS605a, IS605b, IS605c, IS607a, IS607b, and IS1341 subgroups were shown in black, orange, grey, green, blue, and red, respectively.
4. Discussion
4.1. Co-Evolution Profiles of TnpB, an RNA-Guided Endonuclease, with Y1 and SR
IS605 was found by DNA sequencing as a putative, transposable element in a very early study [18], which was further confirmed in an H. pylori strain by hybridization and PCR [19]. Differential distribution of IS605 in H. pylori isolates from different parts of the world was observed [30]. Later, the sequence diversity and copy numbers in five strains of H. pylori were estimated in Southern blots of genomic DNAs; furthermore, PCR and hybridization showed a differential distribution of ISHp608 in H. pylori from different human populations were observed [15]. The invasion of IS200/IS605 was recently characterized in Halanaerobium hydrogeniformans [31].
Recent research revealed that the RNA-guided endonucleases of IscB and TnpB were identified as the ancestral proteins of the Cas9 and Cas12 nucleases, respectively, and have adopted CRISPR–Cas systems [1,4,32]. Similarly, they display cleavage DNA activities in both prokaryotes and eukaryotes. TnpB is guided by an RNA derived from the right-end element of a transposon [16] to cleave DNA next to the transposon-associated motif (named TAM). In contrast, IscB is guided by a noncoding RNA (called OMEGA RNA) derived from the upstream of IscB ORF [17]. In the current study, by mining TnpB-containing mobile elements (TCMEs), we characterized the co-evolution profiles of TnpB endonuclease with Y1 and serine recombinases in six species of bacteria. In total, 9996 TCMEs were identified in 4594 genomes from six species, and they were classified into 39 IS elements based on their structure organization and identity, belonging to the three designated groups of IS605, IS607, and IS1341 [4]. Our data mining revealed significant variations of TCME expansion across the genomes of these species and strains, and differential invasions of IS605, IS607, and IS1341, and their subgroups across these species and even across different strains. Furthermore, our data revealed that the phylogeny of TnpB is not related to the phylogeny of these mobile elements harboring TnpB proteins, which may indicate the existence of horizontal TnpB gene transfer among these groups of IS elements.
Surprisingly, we found most genomes of C. difficile (99%), D. radiodurans (100%), E. coli (94%), and B. cereus (83%) were invaded by TCMEs, while only about half of the H. pylori (64%) and S. enterica (44%) genomes contained TCME copies. Across the groups (IS605, IS607, and IS1341) of TCMEs, we found that the IS605 represented the widest distribution in the detected species and invaded all six species. However, dramatic differences in IS605 amplification in strain genomes between species were detected. IS605 invaded only about 20% of S. enterica strain genomes, and about 50% of B. cereus and H. pylori strain genomes harbored IS605 copies. In addition, most C. difficile-, D. radiodurans-, and E. coli-detected genomes (>90%) contained IS605 elements. IS607 and IS1341 displayed a relatively narrow distribution and invaded into few species and had fewer detected strain genomes of these species (<30%) compared to IS605. The exception to this is IS1341 in B. cereus and C. difficile, which invaded into above 70% of the detected strain genomes. These data indicate that TCMEs and their families experienced dramatically differential evolutionary histories across these bacteria species. They played roles in shaping the genome evolution of hosts and may contribute to bacterial genome plasticity [3].
Our data analysis also revealed that the copy numbers of TCME subgroups varied significantly in the detected genomes. Most subgroups in the detected genomes of most species were represented by one to two copies, while three to four copies in the detected genomes were observed for IS1341 in B. cereus, IS605a in D. radiodurans, IS605c and IS607a in H. pylori. Approximately 13 copies in the detected genomes of C. difficile were observed for IS605b (Figure 6). The expansion difference of TCMEs, their groups (IS605, IS607, and IS1341) and subgroups (IS605a, IS605b, IS605c, IS607a, IS607b, and IS1341) between species and strain genomes may be dependent on both hosts and mobile elements. The autoregulation of mobile elements has been observed in both prokaryotes and eukaryotes [33,34,35]. It has been proved that although Y1 recombinase is sufficient to carry out the cleavage and joining steps of IS605, TnpB is not necessary for the transposition based on the two members of the IS605 (IS608 and ISDra2) functional analysis [33,34]. On the other hand, it may inhibit transposition activity [11]. Extensive invasion of IS605 suggests that, compared with IS607 and IS1341, IS605 may have higher transposition activity, mainly contributed by Y1 recombinase. Further comparative evaluation of the transposition activities between Y1 and serine recombinases will help interpret these results.
4.2. Structure Organization of TCMEs
Based on the TCME mining, three types of structures were identified for IS605 which were summarized in the previous review [4]: two genetic structure types (SR and TnpB genes oriented in the same direction with encoding regions overlapping or separating) were identified for IS607 (Figure 1). However, more genetic structures of IS607 (SR and TnpB genes oriented in the reverse direction) may also exist in prokaryotes; this requires further analysis. IS1341 represents one genetic structure and only encodes TnpB flanked with LE and RE. We applied a stringent standard to exclude the putative decays of the IS605 and IS607 elements from IS1341 elements, where the upstream or downstream TnpB of IS1341s containing Y1- or SR-derived sequences (more than 20 bp) were designated as the decays of IS605 or IS607 elements and excluded from IS1341 group. We found that IS1341 displayed a distinct invasion profile from IS605 and IS607, such as the IS1341 elements in S. enterica, where 418 genomes (about 28%) contain IS1341 copies, while 303 genomes (about 20%) contain IS605 elements and IS1341 and IS605 only co-exist in 63 genomes (about 4%). According to the present findings, the identified IS1341 may evolve independently and may not be decayed from IS605 or IS607. Systematic IS1341 mining in prokaryote genomes can provide insight into their evolutionary history and origin. The transposition activity of IS1341 is worth further verification and would provide more substantial evidence for their origins.
Our data initially revealed four distinct branches of TnpB. All motifs of TnpB, including three RuvC (I, II, III) segments, two ZFs, one RK-rich, and one HTH, have been previously defined [16] and were highly conserved across species. However, variations of the C-terminal were observed, and their function remains unknown. Diverse Cas12 subfamilies, including miniature Cas12 proteins (Cas12f), were reported [36,37]; however, their phylogenetic and association analyses with TnpB, which was designated as the ancestor of Cas12 [16], remain largely unknown. In addition, it has been suggested that TnpB may evolve from IscB [17]. Systematically, mining TnpB and Cas12 homology proteins in prokaryotes and viruses and extensively analyzing their evolutionary relationships with IscB will provide a broader insight into the evolutionary history of these RNA-guided nucleases.
5. Conclusions
The current study identified 39 TnpB-containing mobile elements (TCMEs) from the total 4594 genomes of six bacteria species. TCME groups and subgroups displayed significant differential evolution profiles (copy number in genome and structural organization) across the six bacterial species. Overall, more than 80% of the genomes of B. cereus, C. difficile, and D. radiodurans, E. coli harbor TCMEs, while only 64% of H. pylori and 44% of S. enterica genomes contain TCMEs. IS605 displays the most extensive invasions in these genomes, while IS607 and IS1341 elements represent a relatively narrow distribution. Co-invasions of IS605, IS607, or IS1341 elements were observed for many genomes. Significant copy number variations were detected for the subgroups of TCMEs across these species. Additionally, although the key domains and motifs of TnpBs and their associated Y1 or serine recombinases are highly conserved, their overall sequence identities are low.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14020523/s1, Figure S1: The number of TnpB hits in 19 species, Figure S2: The LE and RE structures of IS607 group, Figure S3: The various copy number of IS elements in different genomes, Figure S4: Alignment of 274 TnpB sequences includes represented sequences from ISfinder database and 39-mined sequence from six species, Figure S5: The character of RE sequences, Table S1: The information of 39 TCMEs is six species, File S1: The structure of LE and RE of IS605 group.
Author Contributions
Y.W. created the figures and edited the table. Y.W. and M.G. analyzed the sequences and collected data. N.Y., Z.G. and S.S. collected the sequences. H.W., E.A. and N.U. revised the manuscript. B.G. and C.S. conceived the project and helped write the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded with grants from the National Natural Science Foundation of China (32271508 and 31671313) and the High-end Talent Support Program of Yangzhou University to Chengyi Song.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank all the authors for their suggestions and critical comments on the manuscript. We also thank the reviewers for their insightful comments on this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bao, W.; Jurka, J. Homologues of Bacterial TnpB-IS605 Are Widespread in Diverse Eukaryotic Transposable Elements. Mob. DNA 2013, 4, 1–16. [Google Scholar] [CrossRef]
- Gilbert, C.; Cordaux, R. Horizontal Transfer and Evolution of Prokaryote Transposable Elements in Eukaryotes. Genome Biol. Evol. 2013, 5, 822–832. [Google Scholar] [CrossRef]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; van Houdt, R. The Impact of Insertion Sequences on Bacterial Genome Plasticity and Adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- He, S.; Lavatine, L.; Dyda, F.; Siguier, P.; Caumont-Sarcos, A.; Chandler, M.; Corneloup, A.; Guynet, C.; Marty, B.; Ton Hoang, B. The IS200/IS605 Family and “Peel and Paste” Single-Strand Transposition Mechanism. Microbiol. Spectr. 2015, 3, 1–21. [Google Scholar] [CrossRef]
- Boocock, M.R.; Rice, P.A. A Proposed Mechanism for IS607-Family Serine Transposases. Mob. DNA 2013, 4, 24. [Google Scholar] [CrossRef]
- Johnson, R.C. Site-Specific DNA Inversion by Serine Recombinases. Microbiol. Spectr. 2015, 3, 1–36. [Google Scholar] [CrossRef]
- Guynet, C.; Hickman, A.B.; Barabas, O.; Dyda, F.; Chandler, M.; Ton-Hoang, B. In Vitro Reconstitution of a Single-Stranded Transposition Mechanism of IS608. Mol. Cell 2008, 29, 302–312. [Google Scholar] [CrossRef]
- Morero, N.R.; Zuliani, C.; Kumar, B.; Bebel, A.; Okamoto, S.; Guynet, C.; Hickman, A.B.; Chandler, M.; Dyda, F.; Barabas, O. Targeting IS608 Transposon Integration to Highly Specific Sequences by Structure-Based Transposon Engineering. Nucleic Acids Res. 2018, 46, 4152–4163. [Google Scholar] [CrossRef]
- Ton-Hoang, B.; Guynet, C.; Ronning, D.R.; Cointin-Marty, B.; Dyda, F.; Chandler, M. Transposition of ISHp608, Member of an Unusual Family of Bacterial Insertion Sequences. EMBO J. 2005, 24, 3325–3338. [Google Scholar] [CrossRef]
- Lavatine, L.; He, S.; Caumont-Sarcos, A.; Guynet, C.; Marty, B.; Chandler, M.; Ton-Hoang, B. Single Strand Transposition at the Host Replication Fork. Nucleic Acids Res. 2016, 44, 7866–7883. [Google Scholar] [CrossRef]
- Pasternak, C.; Dulermo, R.; Ton-Hoang, B.; Debuchy, R.; Siguier, P.; Coste, G.; Chandler, M.; Sommer, S. ISDra2 Transposition in Deinococcus Radiodurans Is Downregulated by TnpB. Mol. Microbiol. 2013, 88, 443–455. [Google Scholar] [CrossRef]
- Hickman, A.B.; James, J.A.; Barabas, O.; Pasternak, C.; Ton-Hoang, B.; Chandler, M.; Sommer, S.; Dyda, F. DNA Recognition and the Precleavage State during Single-Stranded DNA Transposition in D. Radiodurans. EMBO J. 2010, 29, 3840–3852. [Google Scholar] [CrossRef]
- Kersulyte, D.; Mukhopadhyay, A.K.; Shirai, M.; Nakazawa, T.; Berg, D.E. Functional Organization and Insertion Specificity of IS607, a Chimeric Element of Helicobacter Pylori. J. Bacteriol. 2000, 182, 5300–5308. [Google Scholar] [CrossRef]
- Pasternak, C.; Ton-Hoang, B.; Coste, G.; Bailone, A.; Chandler, M.; Sommer, S. Irradiation-Induced Deinococcus Radiodurans Genome Fragmentation Triggers Transposition of a Single Resident Insertion Sequence. PLoS Genet. 2010, 6, e1000799. [Google Scholar] [CrossRef]
- Kersulyte, D.; Velapatiño, B.; Dailide, G.; Mukhopadhyay, A.K.; Ito, Y.; Cahuayme, L.; Parkinson, A.J.; Gilman, R.H.; Berg, D.E. Transposable Element ISHp608 of Helicobacter Pylori: Nonrandom Geographic Distribution, Functional Organization, and Insertion Specificity. J. Bacteriol. 2002, 184, 992–1002. [Google Scholar] [CrossRef]
- Karvelis, T.; Druteika, G.; Bigelyte, G.; Budre, K.; Zedaveinyte, R.; Silanskas, A.; Kazlauskas, D.; Venclovas, Č.; Siksnys, V. Transposon-Associated TnpB Is a Programmable RNA-Guided DNA Endonuclease. Nature 2021, 599, 692–696. [Google Scholar] [CrossRef]
- Altae-Tran, H.; Kannan, S.; Demircioglu, F.E.; Oshiro, R.; Nety, S.P.; McKay, L.J.; Dlakić, M.; Inskeep, W.P.; Makarova, K.S.; Macrae, R.K.; et al. The Widespread IS200/IS605 Transposon Family Encodes Diverse Programmable RNA-Guided Endonucleases. Science 2021, 374, 57–65. [Google Scholar] [CrossRef]
- Censini, S.; Lange, C.; Xiang, Z.; Crabtree, J.E.; Ghiara, P.; Borodovsky, M.; Rappuoli, R.; Covacci, A. Cag, a Pathogenicity Island of Helicobacter Pylori, Encodes Type I-Specific and Disease-Associated Virulence Factors. Proc. Natl. Acad. Sci. USA 1996, 93, 14648–14653. [Google Scholar] [CrossRef]
- Kersulyte, D.; Akopyants, N.S.; Clifton, S.W.; Roe, B.A.; Berg, D.E. Novel Sequence Organization and Insertion Specificity of IS605 and IS606: Chimaeric Transposable Elements of Helicobacter Pylori. Gene 1998, 223, 175–186. [Google Scholar] [CrossRef]
- Kersulyte, D.; Kalia, A.; Zhang, M.J.; Lee, H.K.; Subramaniam, D.; Kiuduliene, L.; Chalkauskas, H.; Berg, D.E. Sequence Organization and Insertion Specificity of the Novel Chimeric ISHp609 Transposable Element of Helicobacter Pylori. J. Bacteriol. 2004, 186, 7521–7528. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile Hidden Markov Models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- He, S.; Guynet, C.; Siguier, P.; Hickman, A.B.; Dyda, F.; Chandler, M.; Ton-Hoang, B. IS200/IS605 Family Single-Strand Transposition: Mechanism of IS608 Strand Transfer. Nucleic Acids Res. 2013, 41, 3302–3313. [Google Scholar] [CrossRef]
- He, S.; Hickman, A.B.; Dyda, F.; Johnson, N.P.; Chandler, M.; Ton-Hoang, B. Reconstitution of a Functional IS608 Single-Strand Transpososome: Role of Non-Canonical Base Pairing. Nucleic Acids Res. 2011, 39, 8503–8512. [Google Scholar] [CrossRef]
- Rice, P.; Longden, L.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Siguier, P.; Varani, A.; Perochon, J.; Chandler, M. Exploring Bacterial Insertion Sequences with ISfinder: Objectives, Uses, and Future Developments. Methods Mol. Biol. 2012, 859, 91–103. [Google Scholar] [CrossRef]
- Kichenaradja, P.; Siguier, P.; Pérochon, J.; Chandler, M. ISbrowser: An Extension of ISfinder for Visualizing Insertion Sequences in Prokaryotic Genomes. Nucleic Acids Res. 2009, 38, D62–D68. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The Reference Centre for Bacterial Insertion Sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Hyung, H.L.; Ji, Y.Y.; Hyoun, S.K.; Ji, Y.K.; Kyoung, H.K.; Do, J.K.; Jun, Y.H.; Mikami, B.; Hye, J.Y.; Se, W.S. Crystal Structure of a Metal Ion-Bound IS200 Transposase. J. Biol. Chem. 2006, 281, 4261–4266. [Google Scholar] [CrossRef]
- Höök-Nikanne, J.; Berg, D.E.; Peek, R.M.; Kersulyte, D.; Tummuru, M.K.R.; Blaser, M.J. DNA Sequence Conservation and Diversity in Transposable Element IS605 of Helicobacter Pylori. Helicobacter 1998, 3, 79–85. [Google Scholar] [CrossRef]
- Sadler, M.; Mormile, M.R.; Frank, R.L. Characterization of the IS200/IS605 Insertion Sequence Family in Halanaerobium Hydrogeniformans. Genes 2020, 11, 484. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Makarova, K.S.; Koonin, E. v ISC, a Novel Group of Bacterial and Archaeal DNA Transposons That Encode Cas9 Homologs. J. Bacteriol. 2016, 198, 797–807. [Google Scholar] [CrossRef]
- Beuzón, C.R.; Marqués, S.; Casadesús, J. Repression of IS200 Transposase Synthesis by RNA Secondary Structures. Nucleic Acids Res. 1999, 27, 3690–3695. [Google Scholar] [CrossRef]
- Sittka, A.; Lucchini, S.; Papenfort, K.; Sharma, C.M.; Rolle, K.; Binnewies, T.T.; Hinton, J.C.D.; Vogel, J. Deep Sequencing Analysis of Small Noncoding RNA and MRNA Targets of the Global Post-Transcriptional Regulator, Hfq. PLoS Genet. 2008, 4, e1000163. [Google Scholar] [CrossRef]
- Bouuaert, C.C.; Lipkow, K.; Andrews, S.S.; Liu, D.; Chalmers, R. The Autoregulation of a Eukaryotic DNA Transposon. Elife 2013, 2, e00668. [Google Scholar] [CrossRef]
- Gao, P.; Yang, H.; Rajashankar, K.R.; Huang, Z.; Patel, D.J. Type v CRISPR-Cas Cpf1 Endonuclease Employs a Unique Mechanism for CrRNA-Mediated Target DNA Recognition. Cell. Res. 2016, 26, 901–913. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary Classification of CRISPR–Cas Systems: A Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).