MIR149 rs2292832 and MIR499 rs3746444 Genetic Variants Associated with the Risk of Rheumatoid Arthritis
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval of the Study
2.2. Study Population
2.3. Genomic DNA Extraction and Genotyping of Selected SNPs Using TaqMan Assay
2.4. Statistical Analysis
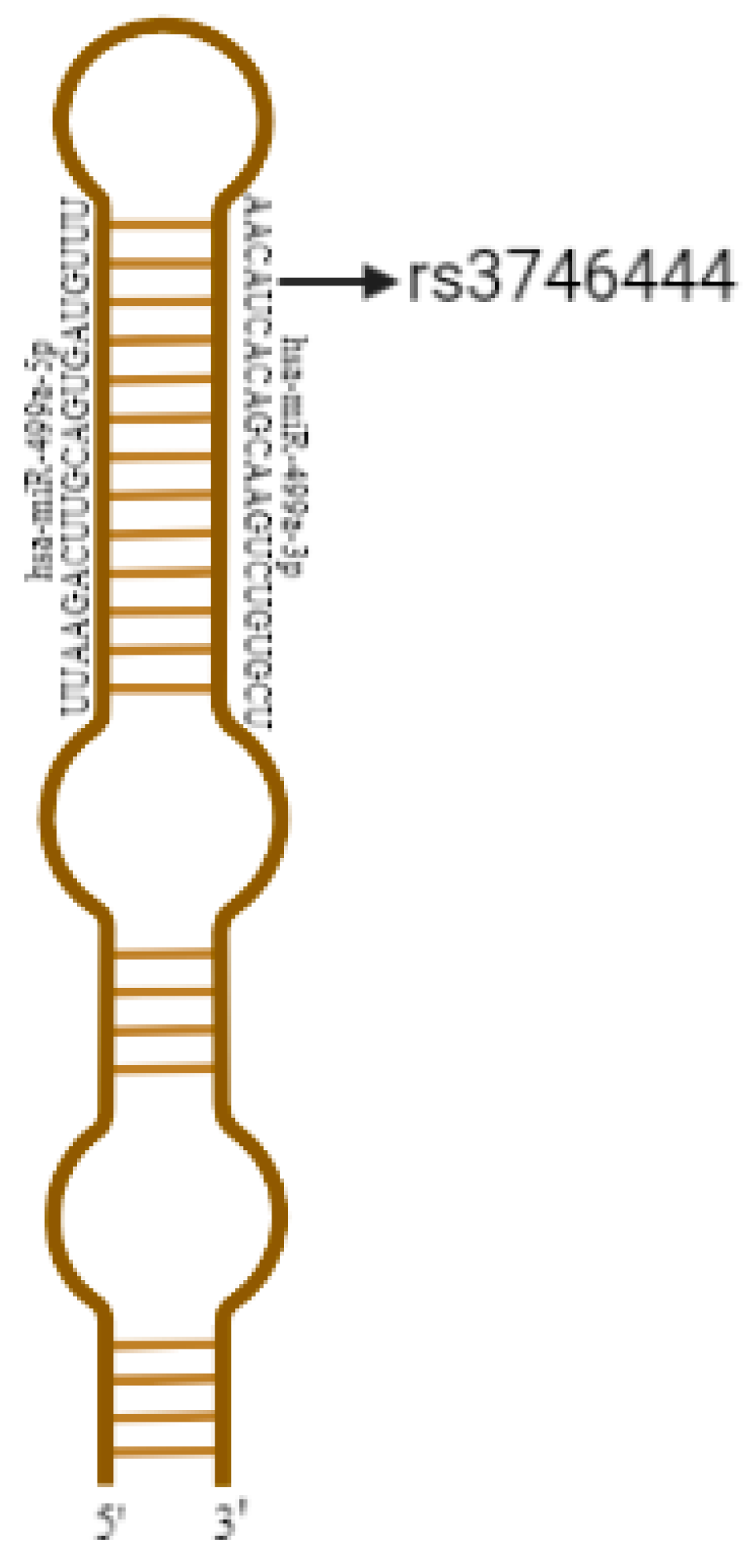
2.5. In Silico Analysis of miRNAs’ Primary Structures
3. Results
3.1. Genetics Analysis
3.2. In Silco Analysis of miRNAs
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Symmons, D.P.M. Epidemiology of rheumatoid arthritis: Determinants of onset, persistence and outcome. Best Pract. Res. Clin. Rheumatol. 2002, 16, 707–722. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.E. Accelerated immunosenescence in rheumatoid arthritis: Impact on clinical progression. Immun. Ageing 2020, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Maiuolo, J.; Muscoli, C.; Gliozzi, M.; Musolino, V.; Carresi, C.; Paone, S.; Ilari, S.; Mollace, R.; Palma, E.; Mollace, V. Endothelial dysfunction and extra-articular neurological manifestations in rheumatoid arthritis. Biomolecules 2021, 11, 81. [Google Scholar] [CrossRef] [PubMed]
- Deane, K.D.; Demoruelle, M.K.; Kelmenson, L.B.; Kuhn, K.A.; Norris, J.M.; Holers, V.M. Genetic and environmental risk factors for rheumatoid arthritis. Best Pract. Res. Clin. Rheumatol. 2017, 31, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, Y.; Zhu, H.; Zhao, M.; Lu, Q. The pathogenic role of dysregulated epigenetic modifications in autoimmune diseases. Front. Immunol. 2019, 10, 2305. [Google Scholar] [CrossRef]
- Patil, K.; Kuttikrishnan, S.; Khan, A.Q.; Ahmad, F.; Alam, M.; Buddenkotte, J.; Ahmad, A.; Steinhoff, M.; Uddin, S. Molecular pathogenesis of Cutaneous T cell Lymphoma: Role of chemokines, cytokines, and dysregulated signaling pathways. Semin. Cancer Biol. 2021, 86, 382–399. [Google Scholar] [CrossRef]
- Stahl, E.A.; Raychaudhuri, S.; Remmers, E.F.; Xie, G.; Eyre, S.; Thomson, B.P.; Li, Y.; Kurreeman, F.A.; Zhernakova, A.; Hinks, A. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 2010, 42, 508. [Google Scholar] [CrossRef]
- Kim, K.; Bang, S.-Y.; Lee, H.-S.; Bae, S.-C. Update on the genetic architecture of rheumatoid arthritis. Nat. Rev. Rheumatol. 2017, 13, 13–24. [Google Scholar] [CrossRef]
- Akhtar, M.; Ali, Y.; Islam, Z.-U.; Arshad, M.; Rauf, M.; Ali, M.; Maodaa, S.N.; Al-Farraj, S.A.; El-Serehy, H.A.; Jalil, F. Characterization of Rheumatoid Arthritis Risk-Associated SNPs and Identification of Novel Therapeutic Sites Using an In-Silico Approach. Biology 2021, 10, 501. [Google Scholar] [CrossRef]
- Ali, Y.; Khan, S.; Chen, Y.; Farooqi, N.; Islam, Z.-U.; Akhtar, M.; Aman, A.; Shah, A.A.; Jamal, M.; Jalil, F. Association of AFF3 Gene Polymorphism rs10865035 with Rheumatoid Arthritis: A Population-Based Case-Control Study on a Pakistani Cohort. Genet. Res. 2021, 2021, 5544198. [Google Scholar] [CrossRef]
- Akhtar, M.; Khan, S.; Ali, Y.; Haider, S.; ud Din, J.; Islam, Z.-U.; Jalil, F. Association study of CCR6 rs3093024 with Rheumatoid Arthritis in a Pakistani cohort. Saudi J. Biol. Sci. 2020, 27, 3354–3358. [Google Scholar] [CrossRef]
- Chen, X.-M.; Huang, Q.-C.; Yang, S.-L.; Chu, Y.-L.; Yan, Y.-H.; Han, L.; Huang, Y.; Huang, R.-Y. Role of micro RNAs in the pathogenesis of rheumatoid arthritis: Novel perspectives based on review of the literature. Medicine 2015, 94, e1326. [Google Scholar] [CrossRef]
- Moran-Moguel, M.C.; Petarra-del Rio, S.; Mayorquin-Galvan, E.E.; Zavala-Cerna, M.G. Rheumatoid arthritis and miRNAs: A critical review through a functional view. J. Immunol. Res. 2018, 2018, 2474529. [Google Scholar] [CrossRef]
- Manoharan, A.P. The Landscape of Caenorhabditis elegans 3’UTRs. Ph.D. Thesis, University of Michigan, Ann Arbor, MI, USA, 2011. [Google Scholar]
- Gurtan, A.M.; Sharp, P.A. The role of miRNAs in regulating gene expression networks. J. Mol. Biol. 2013, 425, 3582–3600. [Google Scholar] [CrossRef]
- Imamura, K.; Akimitsu, N. Long non-coding RNAs involved in immune responses. Front. Immunol. 2014, 5, 573. [Google Scholar] [CrossRef]
- Gam, R.; Shah, P.; Crossland, R.E.; Norden, J.; Dickinson, A.M.; Dressel, R. Genetic association of hematopoietic stem cell transplantation outcome beyond histocompatibility genes. Front. Immunol. 2017, 8, 380. [Google Scholar] [CrossRef]
- Cammaerts, S.; Strazisar, M.; De Rijk, P.; Del Favero, J. Genetic variants in microRNA genes: Impact on microRNA expression, function, and disease. Front. Genet. 2015, 6, 186. [Google Scholar] [CrossRef]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Li, L.-J.; Gao, L.-B.; Lv, M.-L.; Dong, W.; Su, X.-W.; Liang, W.-B.; Zhang, L. Association between SNPs in pre-miRNA and risk of chronic obstructive pulmonary disease. Clin. Biochem. 2011, 44, 813–816. [Google Scholar] [CrossRef]
- Xiao, M.; Ma, Y.; Chen, X.; Kuang, B. Single nucleotide polymorphism of miR-149 and susceptibility of rheumatoid arthritis. Zhong Nan Da Xue Xue Bao. Yi Xue Ban J. Cent. South Univ. Med. Sci. 2015, 40, 495–498. [Google Scholar]
- Yang, B.; Chen, J.; Li, Y.; Zhang, J.; Li, D.; Huang, Z.; Cai, B.; Li, L.; Shi, Y.; Ying, B.; et al. Association of polymorphisms in pre-miRNA with inflammatory biomarkers in rheumatoid arthritis in the Chinese Han population. Hum. Immunol. 2012, 73, 101–106. [Google Scholar] [CrossRef]
- Hashemi, M.; Eskandari-Nasab, E.; Zakeri, Z.; Atabaki, M.; Bahari, G.; Jahantigh, M.; Taheri, M.; Ghavami, S. Association of pre-miRNA-146a rs2910164 and pre-miRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Mol. Med. Rep. 2013, 7, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Amal, S.; Aly, N.M.; Galil, S.M.A.; Moustafa, M.A.; Kandel, W.A. Association of microRNAs genes polymorphisms with rheumatoid arthritis in Egyptian female patients. Jt. Bone Spine 2013, 80, 626–631. [Google Scholar]
- Fattah, S.A.; Ghattas, M.H.; Saleh, S.M.; Abo-Elmatty, D.M. Pre-micro RNA-499 gene polymorphism rs3746444 T/C is associated with susceptibility to rheumatoid arthritis in Egyptian population. Indian J. Clin. Biochem. 2018, 33, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.A.; Ismail, N.M.; Toraih, A.A.; Hussein, M.H.; Fawzy, M.S. Precursor miR-499a variant but not miR-196a2 is associated with rheumatoid arthritis susceptibility in an Egyptian population. Mol. Diagn. Ther. 2016, 20, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Alemán-Ávila, I.; Jiménez-Morales, M.; Beltrán-Ramírez, O.; Barbosa-Cobos, R.E.; Jiménez-Morales, S.; Sánchez-Muñoz, F.; Valencia-Pacheco, G.; Amezcua-Guerra, L.M.; Juárez-Vicuña, Y.; Hernández, D.M.R.-B.; et al. Functional polymorphisms in pre-miR146a and pre-miR499 are associated with systemic lupus erythematosus but not with rheumatoid arthritis or Graves’ disease in Mexican patients. Oncotarget 2017, 8, 91876. [Google Scholar] [CrossRef]
- Shaker, O.G.; Abdelaleem, O.O.; Fouad, N.A.; Ali, A.M.E.A.; Ahmed, T.I.; Ibrahem, E.G.; Abdelghaffar, N.K. Association between miR-155, its polymorphism and ischemia-modified albumin in patients with rheumatoid arthritis. J. Interferon Cytokine Res. 2019, 39, 428–437. [Google Scholar] [CrossRef]
- Smolen, J.S.; Landewé, R.; Bijlsma, J.; Burmester, G.; Chatzidionysiou, K.; Dougados, M.; Nam, J.; Ramiro, S.; Voshaar, M.; Van Vollenhoven, R.; et al. EULAR recommendations for the management of rheumatoid arthritis with synthetic and biological disease-modifying antirheumatic drugs: 2016 update. Ann. Rheum. Dis. 2017, 76, 960–977. [Google Scholar] [CrossRef]
- Smolen, J.S.; Aletaha, D.; Koeller, M.; Weisman, M.H.; Emery, P. New therapies for treatment of rheumatoid arthritis. Lancet 2007, 370, 1861–1874. [Google Scholar] [CrossRef]
- Van Rooij, E. The art of microRNA research. Circ. Res. 2011, 108, 219–234. [Google Scholar] [CrossRef]
- Filková, M.; Jüngel, A.; Gay, R.E.; Gay, S. MicroRNAs in rheumatoid arthritis. BioDrugs 2012, 26, 131–141. [Google Scholar] [CrossRef]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., III; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- McInnes, I.B.; Schett, G. The pathogenesis of rheumatoid arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef]
- Shen, L.; Zhang, H.; Yan, T.; Zhou, G.; Liu, R. Association between interleukin 17A polymorphisms and susceptibility to rheumatoid arthritis in a Chinese population. Gene 2015, 566, 18–22. [Google Scholar] [CrossRef]
- Karami, J.; Aslani, S.; Jamshidi, A.; Garshasbi, M.; Mahmoudi, M. Genetic implications in the pathogenesis of rheumatoid arthritis; an updated review. Gene 2019, 702, 8–16. [Google Scholar] [CrossRef]
- Kochi, Y.; Okada, Y.; Suzuki, A.; Ikari, K.; Terao, C.; Takahashi, A.; Yamazaki, K.; Hosono, N.; Myouzen, K.; Tsunoda, T. A regulatory variant in CCR6 is associated with rheumatoid arthritis susceptibility. Nat. Genet. 2010, 42, 515. [Google Scholar] [CrossRef]
- Mohammadi, F.S.; Aslani, S.; Mostafaei, S.; Jamshidi, A.; Riahi, P.; Mahmoudi, M. Are genetic variations in IL-21–IL-23R–IL-17A cytokine axis involved in a pathogenic pathway of rheumatoid arthritis? Bayesian hierarchical meta-analysis. J. Cell. Physiol. 2019, 234, 17159–17171. [Google Scholar] [CrossRef]
- Salvi, V.; Gianello, V.; Tiberio, L.; Sozzani, S.; Bosisio, D. Cytokine targeting by miRNAs in autoimmune diseases. Front. Immunol. 2019, 10, 15. [Google Scholar] [CrossRef]
- Latini, A.; Ciccacci, C.; Novelli, G.; Borgiani, P. Polymorphisms in miRNA genes and their involvement in autoimmune diseases susceptibility. Immunol. Res. 2017, 65, 811–827. [Google Scholar] [CrossRef]
- Li, J.; Wan, Y.; Guo, Q.; Zou, L.; Zhang, J.; Fang, Y.; Zhang, J.; Zhang, J.; Fu, X.; Liu, H. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res. Ther. 2010, 12, 1–12. [Google Scholar] [CrossRef]
- Song, G.; Bae, S.-C.; Seo, Y.; Kim, J.-H.; Choi, S.; Ji, J.; Lee, Y.H. The association between susceptibility to inflammatory arthritis and miR-146a, miR-499 and IRAK1 polymorphisms. Z. Für Rheumatol. 2015, 74, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Schellekens, G.A.; Visser, H.; De Jong, B.A.; Van Den Hoogen, F.H.; Hazes, J.M.; Breedveld, F.C.; Van Venrooij, W.J. The diagnostic properties of rheumatoid arthritis antibodies recognizing a cyclic citrullinated peptide. Arthritis Rheum. 2000, 43, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Chavanas, S.; Méchin, M.-C.; Takahara, H.; Kawada, A.; Nachat, R.; Serre, G.; Simon, M. Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene 2004, 330, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Ayeldeen, G.; Nassar, Y.; Ahmed, H.; Shaker, O.; Gheita, T. Possible use of miRNAs-146a and-499 expression and their polymorphisms as diagnostic markers for rheumatoid arthritis. Mol. Cell. Biochem. 2018, 449, 145–156. [Google Scholar] [CrossRef]
- Ul Islam, Z.; Baneen, U.; Khaliq, T.; Nurulain, S.M.; Muneer, Z.; Hussain, S. Association analysis of miRNA-146a and miRNA-499 polymorphisms with rheumatoid arthritis: A case–control and trio-family study. Clin. Exp. Med. 2022, 1–9. [Google Scholar] [CrossRef]
- Filkova, M.; Trenkmann, M.; Stanczyk, J.; Frank, M.; Kolling, C.; Huber, L.C.; Michel, B.A.; Gay, R.E.; Senolt, L.; Gay, S. MiR-196a Is An Important Regulator of Synovial Fibroblasts In the Pathogenesis of Rheumatoid Arthritis. In Arthritis and Rheumatism; Wiley: Hoboken, NJ, USA, 2011; p. S986. [Google Scholar]
- Mahesh, G.; Biswas, R. MicroRNA-155: A master regulator of inflammation. J. Interferon Cytokine Res. 2019, 39, 321–330. [Google Scholar] [CrossRef]
| Variable | Cases (n = 300) | Controls (n = 300) | p-Value |
|---|---|---|---|
| Gender (male/female) | 113/187 | 119/181 | 0.6121 |
| Mean age in years (±SD) | 43.5 (±14.5) | 42 (±12.6) | 0.217 |
| Mean disease duration in years (±SD) | 4.1 (±3.7) | - | - |
| Sero-positive antibody (Me) | 100% (RF-positive) | - | - |
| Mean ESR (±SD) | 40.60 (±15.8) | - | - |
| SNP ID | miRNA | Names of Mature miRNA Sequences | Chromosome No. | miRNA Location (Coordinates) | Coded Allele | Other Allele | MAF |
|---|---|---|---|---|---|---|---|
| rs2292832 | MIR149 | hsa-miR-149-5p | 2 | 240456001-240456089 | T | C | 0.38 |
| hsa-miR-149-3p | |||||||
| rs374644 | MIR499A | hsa-miR-499-5p | 20 | 34990376-34990497 [+] | A | G | 0.12 |
| hsa-miR-499-3p | |||||||
| rs11614913 | MIR196a2 | hsa-miR-196a-5p | 12 | 53991738-53991847 [+] | C | T | 0.49 |
| hsa-miR-196a-3p | |||||||
| rs1044165 | MIR223 | hsa-miR-223-5p | X | 66018870-66018979 | G | A | 0.00 |
| hsa-miR-223-3p | |||||||
| rs767649 | MIR155 | hsa-miR-155-5p | 21 | 25573980-255740444 | T | C | 0.29 |
| miRNA SNPs | Models | Genotypes | Cases (300) | Controls (300) | Odds Ratio | χ2-Value | p-Value |
|---|---|---|---|---|---|---|---|
| MIR149 rs2292832 | Co-dominant | TT CT CC | 55 (18.4%) 136 (45.4%) 109 (36.3%) | 112 (37.4%) 123 (41.0%) 65 (21.6%) | - | 31.23 | <0.0001 |
| Dominant | CC TT + CT | 109 (36.3%) 291 (63.7%) | 65 (21.6%) 235 (78.4%) | 2.063 1.437–2.962 | - | 0.0001 | |
| Recessive | TT CC + CT | 55 (18.4%) 245 (81.6%) | 112 (37.4%) 188 (62.6%) | 0.376 0.259–0.548 | - | <0.0001 | |
| Heterozygous | CT CC + TT | 136 (45.4%) 164 (54.6%) | 123 (41.0%) 177 (59%) | 1.267 0.919–1.748 | - | 0.16 | |
| Additive | C T | 264 (44.0%) 354 (56.0%) | 347 (57.8%) 253 (42.2%) | 0.506 0.402–0.637 | - | <0.0001 | |
| MIR499 rs3746444 | Co-dominant | AA AG GG | 72 (24.0%) 110 (36.7%) 118 (39.3%) | 129 (43.0%) 138 (46.0%) 33 (11.0%) | - | 59.19 | 0.0001 |
| Dominant | GG AA + AG | 118 (39.3%) 182 (60.7%) | 33 (11.0%) 267 (89.0%) | 5.246 3.414–8.061 | - | <0.0001 | |
| Recessive | AA GG + AG | 72 (24.0%) 228 (76.0%) | 129 (43.0%) 171 (57.0%) | 0.653 0.466–0.916 | - | 0.014 | |
| Heterozygous | AG AA + GG | 110 (36.7%) 190 (62.3%) | 138 (46.0%) 162 (54.0%) | 0.679 0.490–0.942 | - | 0.025 | |
| Additive | A G | 254 (42.4%) 346 (57.6%) | 291 (58.5%) 309 41.5%) | 0.779 0.620–0.978 | - | 0.03 | |
| MIR196a2 rs11614913 | Co-dominant | CC CT TT | 107 (35.7%) 132 (44.0%) 61 (20.3%) | 112 (37.3%) 115 (38.3%) 73 (24.4%) | - | 2.35 | 0.30 |
| Dominant | CC TT + CT | 107 (35.7%) 193 (64.3%) | 112 (37.3%) 188 (42.7%) | 0.930 0.667–1.298 | - | 0.73 | |
| Recessive | TT CC + CT | 61 (20.3%) 239 (79.6%) | 73 (24.4%) 227 (75.6%) | 0.826 0.563–1.213 | - | 0.38 | |
| Heterozygous | CT CC + TT | 132 (44.0%) 168 (56%) | 115 (38.3%) 185 (61.7%) | 1.264 0.912–1.751 | - | 0.18 | |
| Additive | C T | 346 (57.7%) 254 (42.3%) | 339 (56.5%) 261 (43.5%) | 1.049 0.834–1.318 | - | 0.72 | |
| MIR223 rs1044165 | Co-dominant | AA AG GG | 3 (1.0%) 26 (8.7%) 271 (90.3%) | 8 (2.7%) 24 (8.0%) 268 (89.3%) | - | 2.369 | 0.30 |
| Dominant | GG AA + AG | 271 (90.3%) 29 (9.7%) | 268 (89.3%) 32 (10.7%) | 1.116 0.656–1.896 | - | 0.78 | |
| Recessive | AA GG + AG | 3 (1.0%) 297 (99.0%) | 8 (2.7%) 292 (97.3%) | 0.368 0.096–1.404 | - | 0.22 | |
| Heterozygous | AG AA + GG | 26 (8.7%) 274 (91.3%) | 24 (8.0%) 276 (92.0%) | 1.091 0.611–1.948 | - | 0.88 | |
| Additive | A G | 32 (5.3%) 568 (94.7%) | 40 (6.7%) 560 (93.3%) | 0.788 0.488–1.274 | - | 0.39 | |
| MIR155 rs767649 | Co-dominant | CC CT TT | 6 (2.0%) 72 (24.0%) 222 (74.0%) | 3 (1.0%) 92(30.7%) 205 (68.3%) | - | 4.116 | 0.12 |
| Dominant | TT CC + CT | 222 (74.0%) 78 (26.0%) | 205 (68.3%) 95 (31.7%) | 1.319 0.925–1.88 | 0.14 | ||
| Recessive | CC TT + CT | 6 (2.0%) 294 (98.0%) | 3 (1.0%) 297 (99.0%) | 2.02 0.50–8.157 | - | 0.50 | |
| Heterozygous | CT CC + TT | 72 (24.0%) 228 (76%) | 92 (30.7%) 208 (69.3%) | 0.714 0.497–1.025 | - | 0.08 | |
| Additive | C T | 84 (14.0%) 516 (86.0%) | 92 (16.3%) 502 (83.7%) | 0.833 0.607–1.144 | - | 0.29 |
| Parameters | MIR149 rs2292832 | MIR499A rs3746444 | MIR196A2 rs11614913 | |||
|---|---|---|---|---|---|---|
| Sequences | Reference Sequence | Mutated Sequence | Reference Sequence | Mutated Sequence | Reference Sequence | Mutated Sequence |
| Ensemble thermodynamic free energy | −54.29 kcal/mol | −56.49 kcal/mol | −64.92 kcal/mol | −59.55 kcal/mol | −52.02 kcal/mol | −42.82 kcal/mol |
| Ensemble diversity | 7.21 | 7.20 | 9.65 | 13.42 | 7.18 | 4.86 |
| Optimal secondary structure with the lowest free energy | −52.70 kcal/mol | −54.90 kcal/mol | −63.20 kcal/mol | −57.70 kcal/mol | −49.90 kcal/mol | −41.60 kcal/mol |
| Secondary structure of the centroid | −52.70 kcal/mol | 54.90 kcal/mol | −63.00 kcal/mol | −54.80 kcal/mol | −41.30 kcal/mol | −44.30 kcal/mol |
| Ensemble MFE structure frequency | 7.56% | 7.57% | 6.14% | 4.96% | 6.14% | 13.85% |
| Author | Year | Country | Disease | Control Source | Genotype Method | Cases | Controls | HWE |
|---|---|---|---|---|---|---|---|---|
| MIR149 rs22928323 | ||||||||
| Xiao | 2015 | China | RA | HB | PCR-RFLP | 200 | 120 | No |
| MIR499 rs3743444 | ||||||||
| Yang | 2012 | China | RA | PB | PCR-RFLP | 208 | 240 | Yes |
| Hashemi | 2013 | Iran | RA | PB | T-ARMS-PCR | 104 | 110 | Yes |
| Amal | 2013 | Egypt | RA | PB | PCR-RFLP | 217 | 245 | Yes |
| Zhang | 2013 | China | RA | HB | MALDI-TOP MS | 206 | 466 | Yes |
| Yang | 2016 | China | RA | PB | TaqMan | 386 | 576 | Yes |
| Toraih | 2016 | Egypt | RA | PB | Real-time PCR | 95 | 200 | No |
| Aleman-avila | 2017 | Mexico | RA | PB | TaqMan | 412 | 486 | Yes |
| Ayeldeen | 2018 | Egypt | RA | PB | Real-time PCR | 52 | 56 | Yes |
| Fataah | 2018 | Egypt | RA | PB | PCR-RFLP | 100 | 100 | Yes |
| MIR196a rs 11614913 | ||||||||
| Toraih | 2016 | Egypt | RA | PB | Real-time PCR | 95 | 200 | Yes |
| Aleman-avila | 2017 | Mexico | RA | PB | TaqMan | 412 | 486 | No |
| MIR155 rs7676649 | ||||||||
| Shaker | 2019 | Egypt | RA | PB | TaqMan | 79 | 78 | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, Y.; Chen, Y.; Islam, Z.U.; Aman, A.; Almutairi, M.M.; Alouffi, A.; Mohammed, A.; Shah, A.A.; Rehman, Z.U.; Hussain, I.; et al. MIR149 rs2292832 and MIR499 rs3746444 Genetic Variants Associated with the Risk of Rheumatoid Arthritis. Genes 2023, 14, 431. https://doi.org/10.3390/genes14020431
Ali Y, Chen Y, Islam ZU, Aman A, Almutairi MM, Alouffi A, Mohammed A, Shah AA, Rehman ZU, Hussain I, et al. MIR149 rs2292832 and MIR499 rs3746444 Genetic Variants Associated with the Risk of Rheumatoid Arthritis. Genes. 2023; 14(2):431. https://doi.org/10.3390/genes14020431
Chicago/Turabian StyleAli, Yasir, Yangchao Chen, Zia Ul Islam, Aisha Aman, Mashal M. Almutairi, Abdulaziz Alouffi, Aymen Mohammed, Aftab Ali Shah, Zia Ur Rehman, Ibrar Hussain, and et al. 2023. "MIR149 rs2292832 and MIR499 rs3746444 Genetic Variants Associated with the Risk of Rheumatoid Arthritis" Genes 14, no. 2: 431. https://doi.org/10.3390/genes14020431
APA StyleAli, Y., Chen, Y., Islam, Z. U., Aman, A., Almutairi, M. M., Alouffi, A., Mohammed, A., Shah, A. A., Rehman, Z. U., Hussain, I., Ali, A., & Jalil, F. (2023). MIR149 rs2292832 and MIR499 rs3746444 Genetic Variants Associated with the Risk of Rheumatoid Arthritis. Genes, 14(2), 431. https://doi.org/10.3390/genes14020431








