Sex-Specific Effects of the Genetic Variant rs10487505 Upstream of leptin in the Development of Obesity
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Leptin Gene Expression Analyses in AT
2.3. Measurement of Leptin Serum Concentrations
2.4. Genotyping
2.5. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Subjects
3.2. Characteristics of Leptin Gene Expression in a Cohort with Obesity
3.3. rs10487505 Is Associated with Circulating Leptin in Women but not with LEP mRNA in SCAT
3.4. rs10487505 C-allele Is Associated with BMI and Metabolic Parameters
3.5. The BMI Decreasing Effect Is Specific to Postmenopausal Women
4. Discussion
4.1. rs10487505 Is Associated with Circulating Leptin Levels, but the Association Is not Mediated by LEP mRNA Expression
4.2. rs10487505 C Allele Is Associated with Lower BMI in a Cohort with Obesity
4.3. rs10487505 Correlates with Obesity-Related and Inflammatory Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- van Gaal, L.F.; Mertens, I.L.; de Block, C.E. Mechanisms linking obesity with cardiovascular disease. Nature 2006, 444, 875–880. [Google Scholar] [CrossRef]
- de Pergola, G.; Silvestris, F. Obesity as a major risk factor for cancer. J. Obes. 2013, 2013, 291546. [Google Scholar] [CrossRef]
- Zhang, Y.; Proenca, R.; Maffei, M.; Barone, M.; Leopold, L.; Friedman, J.M. Positional cloning of the mouse obese gene and its human homologue. Nature 1994, 372, 425–432. [Google Scholar] [CrossRef]
- Ingalls, A.M.; Dickie, M.M.; Shell, G.D. Obese, a new mutation in the house mouse. J. Hered. 1950, 41, 317–318. [Google Scholar] [CrossRef]
- Maffei, M.; Halaas, J.; Ravussin, E.; Pratley, R.E.; Lee, G.H.; Zhang, Y.; Fei, H.; Kim, S.; Lallone, R.; Ranganathan, S. Leptin levels in human and rodent: Measurement of plasma leptin and ob RNA in obese and weight-reduced subjects. Nat. Med. 1995, 1, 1155–1161. [Google Scholar] [CrossRef] [PubMed]
- Cowley, M.A.; Smart, J.L.; Rubinstein, M.; Cerdán, M.G.; Diano, S.; Horvath, T.L.; Cone, R.D.; Low, M.J. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 2001, 411, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Park, H.-K.; Ahima, R.S. Physiology of leptin: Energy homeostasis, neuroendocrine function and metabolism. Metab. Clin. Exp. 2015, 64, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Ahima, R.S.; Kelly, J.; Elmquist, J.K.; Flier, J.S. Distinct physiologic and neuronal responses to decreased leptin and mild hyperleptinemia. Endocrinology 1999, 140, 4923–4931. [Google Scholar] [CrossRef]
- Blüher, S.; Shah, S.; Mantzoros, C.S. Leptin deficiency: Clinical implications and opportunities for therapeutic interventions. J. Investig. Med. 2009, 57, 784–788. [Google Scholar] [CrossRef]
- Myers, M.G.; Cowley, M.A.; Münzberg, H. Mechanisms of leptin action and leptin resistance. Annu. Rev. Physiol. 2008, 70, 537–556. [Google Scholar] [CrossRef]
- Saad, M.F.; Damani, S.; Gingerich, R.L.; Riad-Gabriel, M.G.; Khan, A.; Boyadjian, R.; Jinagouda, S.D.; el-Tawil, K.; Rude, R.K.; Kamdar, V. Sexual dimorphism in plasma leptin concentration. J. Clin. Endocrinol. Metab. 1997, 82, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Enzi, G.; Gasparo, M.; Biondetti, P.R.; Fiore, D.; Semisa, M.; Zurlo, F. Subcutaneous and visceral fat distribution according to sex, age, and overweight, evaluated by computed tomography. Am. J. Clin. Nutr. 1986, 44, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Kilpeläinen, T.O.; Carli, J.F.M.; Skowronski, A.A.; Sun, Q.; Kriebel, J.; Feitosa, M.F.; Hedman, Å.K.; Drong, A.W.; Hayes, J.E.; Zhao, J.; et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat. Commun. 2016, 7, 10494. [Google Scholar] [CrossRef] [PubMed]
- Dallner, O.S.; Marinis, J.M.; Lu, Y.-H.; Birsoy, K.; Werner, E.; Fayzikhodjaeva, G.; Dill, B.D.; Molina, H.; Moscati, A.; Kutalik, Z.; et al. Dysregulation of a long noncoding RNA reduces leptin leading to a leptin-responsive form of obesity. Nat. Med. 2019, 25, 507–516. [Google Scholar] [CrossRef]
- Klöting, N.; Fasshauer, M.; Dietrich, A.; Kovacs, P.; Schön, M.R.; Kern, M.; Stumvoll, M.; Blüher, M. Insulin-sensitive obesity. Am. J. Physiol. Endocrinol. Metab. 2010, 299, E506–E515. [Google Scholar] [CrossRef]
- American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37 (Suppl. 1), S81–S90. [Google Scholar] [CrossRef]
- Mardinoglu, A.; Heiker, J.T.; Gärtner, D.; Björnson, E.; Schön, M.R.; Flehmig, G.; Klöting, N.; Krohn, K.; Fasshauer, M.; Stumvoll, M.; et al. Extensive weight loss reveals distinct gene expression changes in human subcutaneous and visceral adipose tissue. Sci. Rep. 2015, 5, 14841. [Google Scholar] [CrossRef]
- Maurano, M.T.; Humbert, R.; Rynes, E.; Thurman, R.E.; Haugen, E.; Wang, H.; Reynolds, A.P.; Sandstrom, R.; Qu, H.; Brody, J.; et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012, 337, 1190–1195. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Pelletier, J. Exploring the Impact of Single-Nucleotide Polymorphisms on Translation. Front. Genet. 2018, 9, 507. [Google Scholar] [CrossRef]
- Milewicz, A.; Demissie, M. Metabolic and endocrine changes in climacteric women. Int. Congr. Ser. 2002, 1229, 3–7. [Google Scholar] [CrossRef]
- ter Horst, K.W.; Gilijamse, P.W.; Koopman, K.E.; de Weijer, B.A.; Brands, M.; Kootte, R.S.; Romijn, J.A.; Ackermans, M.T.; Nieuwdorp, M.; Soeters, M.R.; et al. Insulin resistance in obesity can be reliably identified from fasting plasma insulin. Int. J. Obes. 2015, 39, 1703–1709. [Google Scholar] [CrossRef] [PubMed]
- Díez, J.J.; Iglesias, P. The role of the novel adipocyte-derived hormone adiponectin in human disease. Eur. J. Endocrinol. 2003, 148, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, S.; Blüher, M.; Stengler, K. Geschlechtsspezifische Analyse der Inanspruchnahme bariatrischer Chirurgie. Adipositas -Ursachen Folgeerkrankungen Ther. 2012, 6, 164–167. [Google Scholar] [CrossRef]
- Tony Burdett, Emma Hastings Dani Welter. GWAS Catalog. Available online: https://www.ebi.ac.uk/gwas/variants/rs10487505 (accessed on 16 January 2023).
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP—Database for Single Nucleotide Polymorphisms and Other Classes of Minor Genetic Variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef] [PubMed]
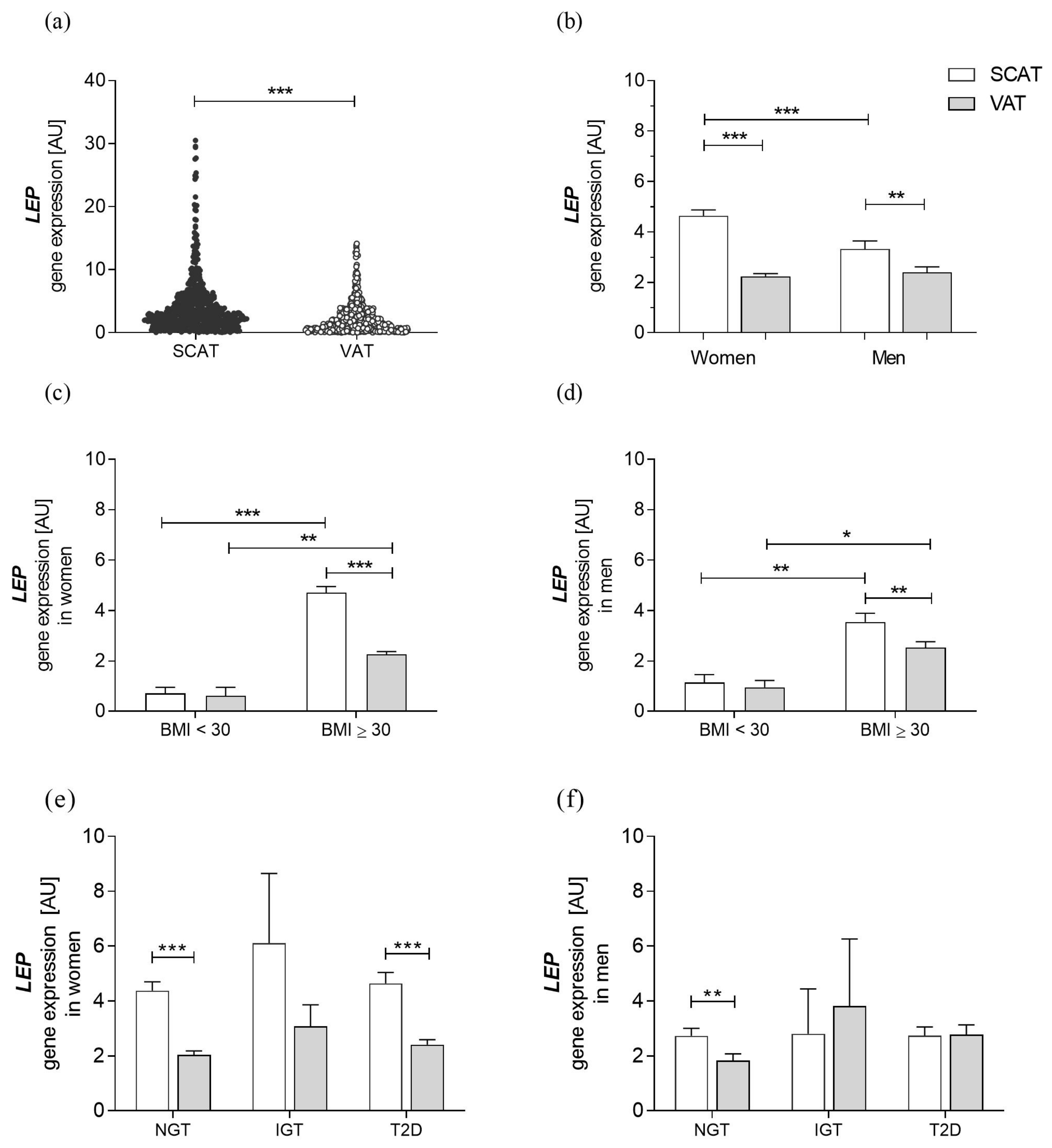
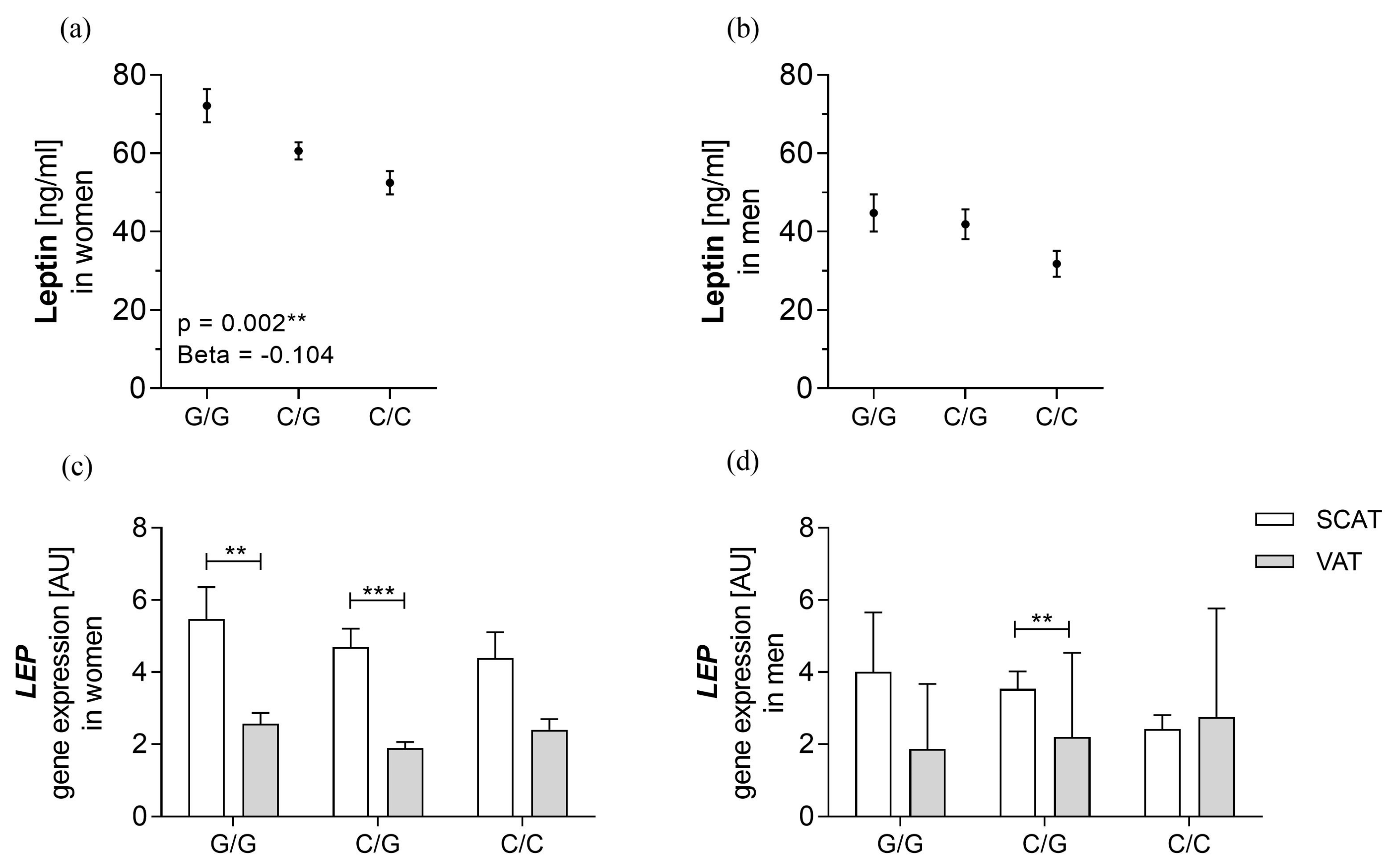

| BMI < 30 kg/m2 | BMI ≥ 30 kg/m2 | p-Value | |
|---|---|---|---|
| (N = 138) | (N = 1497) | ||
| Age (years) | 61.77 ± 15.57 | 47.16 ± 12.03 | ≤0.001 |
| Women/Men (N) | 71/67 | 1042/455 | |
| T2D (N) | 23 | 622 | |
| Body weight (kg) | 69.74 ± 12.13 | 143.94 ± 31.13 | ≤0.001 |
| Height (m) | 1.69 ± 0.09 | 1.70 ± 0.10 | 0.194 |
| BMI (kg/m2) | 24.34 ± 3.19 | 49.18 ± 8.92 | ≤0.001 |
| Waist circumference (cm) | 84.33 ± 17.32 | 138.61 ± 21.05 | ≤0.001 |
| Hip circumference (cm) | 91.81 ± 13.52 | 145.41 ± 18.72 | ≤0.001 |
| WHR | 0.91 ± 0.11 | 0.96 ± 0.11 | 0.001 |
| Body fat (%) | 20.87 ± 4.50 | 47.12 ± 8.79 | ≤0.001 |
| CRP (mg/l) | 13.48 ± 25.87 | 11.35 ± 13.76 | 0.358 |
| FPG (mmol/l) | 5.85 ± 1.37 | 6.49 ± 2.67 | ≤0.001 |
| FPI (pmol/l) | 57.41 ± 77.03 | 148.58 ± 142.14 | ≤0.001 |
| HbA1c (%) | 5.60 ± 0.72 | 6.19 ± 1.31 | ≤0.001 |
| Total cholesterol (mmol/l) | 5.08 ± 1.12 | 4.74 ± 1.06 | 0.036 |
| HDL-C (mmol/l) | 1.64 ± 0.51 | 1.15 ± 0.44 | ≤0.001 |
| LDL-C (mmol/l) | 3.00 ± 1.07 | 3.03 ± 0.95 | 0.781 |
| Triglycerides (mmol/l) | 1.22 ± 0.52 | 2.26 ± 15.78 | 0.629 |
| ALAT [µkat/l] | 0.45 ± 0.30 | 0.77 ± 1.69 | 0.028 |
| ASAT [µkat/l] | 0.49 ± 0.26 | 0.81 ± 4.02 | 0.368 |
| gGT [µkat/l] | 1.03 ± 1.52 | 0.85 ± 2.83 | 0.472 |
| Leptin serum levels [ng/mL] | 8.25 ± 6.87 | 57.77 ± 35.42 | ≤0.001 |
| IL-6 serum levels [pg/mL] | 2.69 ± 3.24 | 6.74 ± 5.80 | ≤0.001 |
| Adiponectin serum levels [μg/mL] | 12.71 ± 6.19 | 6.54 ± 3.49 | ≤0.001 |
| Subjects with leptin mRNA expression in AT (N) | 4 | 306 |
| Genetic Variants | NGT N (%) | T2D N (%) | MAF NGT/T2D | p-Value (OR [95% CI]) Adj. for Age, Sex, BMI |
|---|---|---|---|---|
| rs10487505 (C/G) | ||||
| Entire cohort | ||||
| CC | 186 (23.8%) | 131 (20.2%) | 0.4943/0.4645 | 0.433(1.069 [0.905–1.262]) |
| CG | 402 (51.3%) | 340 (52.5%) | ||
| GG | 195 (24.9%) | 177 (27.3%) | ||
| Women | ||||
| CC | 148 (26.2%) | 77 (14.6%) | 0.5133/0.3510 | 0.205(1.142 [0.930–1.403]) |
| CG | 283 (50.2%) | 216 (41.0%) | ||
| GG | 133 (23.6%) | 234 (44.4%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Molder, J.; Guiu-Jurado, E.; Moulla, Y.; Stroh, C.; Dietrich, A.; Schön, M.R.; Gärtner, D.; Lohmann, T.; Dressler, M.; Stumvoll, M.; et al. Sex-Specific Effects of the Genetic Variant rs10487505 Upstream of leptin in the Development of Obesity. Genes 2023, 14, 378. https://doi.org/10.3390/genes14020378
Molder J, Guiu-Jurado E, Moulla Y, Stroh C, Dietrich A, Schön MR, Gärtner D, Lohmann T, Dressler M, Stumvoll M, et al. Sex-Specific Effects of the Genetic Variant rs10487505 Upstream of leptin in the Development of Obesity. Genes. 2023; 14(2):378. https://doi.org/10.3390/genes14020378
Chicago/Turabian StyleMolder, Janine, Esther Guiu-Jurado, Yusef Moulla, Christine Stroh, Arne Dietrich, Michael R. Schön, Daniel Gärtner, Tobias Lohmann, Miriam Dressler, Michael Stumvoll, and et al. 2023. "Sex-Specific Effects of the Genetic Variant rs10487505 Upstream of leptin in the Development of Obesity" Genes 14, no. 2: 378. https://doi.org/10.3390/genes14020378
APA StyleMolder, J., Guiu-Jurado, E., Moulla, Y., Stroh, C., Dietrich, A., Schön, M. R., Gärtner, D., Lohmann, T., Dressler, M., Stumvoll, M., Kovacs, P., Blüher, M., & Klöting, N. (2023). Sex-Specific Effects of the Genetic Variant rs10487505 Upstream of leptin in the Development of Obesity. Genes, 14(2), 378. https://doi.org/10.3390/genes14020378






