Abstract
Despite high diversity in the Oriental region, ticks of the genus Haemaphysalis have been neglected regarding their genetic data and vector potential. This study aimed to genetically characterize three species of the genus Haemaphysalis: Haemaphysalis cornupunctata, Haemaphysalis kashmirensis, and Haemaphysalis montgomeryi infesting goats and sheep, and Rickettsia spp. associated with these tick species in the Hindu Kush Himalayan range of Pakistan. Altogether, 834 ticks were collected by examining 120 hosts including goats (64/120, 53.3%) and sheep (56/120, 46.6%), in which 86 (71.6%) hosts were found to be tick-infested. The morphologically identified ticks were subjected to DNA extraction and PCR for the amplification of partial 16S rDNA and cox fragments. Rickettsia spp. associated with the collected ticks were detected through the amplification of gltA, ompA and ompB partial fragments. The 16S rDNA of H. cornupunctata and H. montgomeryi showed a maximum identity of 100% with the sequences of the same species, whereas the 16S rDNA of H. kashmirensis showed the highest identity of 93–95% with Haemaphysalis sulcata. The cox sequence of H. montgomeryi displayed 100% identity with the same species. In comparison, the cox sequences of H. cornupunctata and H. kashmirensis showed maximum identities of 87.65–89.22% with Haemaphysalis punctata and 89.34% with H. sulcata, respectively. The gltA sequence of Rickettsia sp. from H. kashmirensis showed the highest identity of 97.89% with Rickettsia conorii subsp. raoultii, while the ompA and ompB fragments from the same DNA samples revealed 100% and 98.16% identity with Rickettsia sp. and “Candidatus Rickettsia longicornii”, respectively. Another gltA sequence amplified from H. montgomeryi ticks showed 100% identity with Rickettsia hoogstraalii, while the attempts to amplify ompA and ompB for R. hoogstraalii were unsuccessful. In the phylogenetic tree, the 16S rDNA of H. cornupunctata clustered with the corresponding species while its cox clustered with H. punctata. Both 16S rDNA and cox sequences of H. kashmirensis clustered with H. sulcata. The gltA sequence of Rickettsia sp. was clustered individually in the spotted fever (SF) group of Rickettsia, while the gltA sequence of R. hoogstraalii was clustered with the same species in the transition group of Rickettsia. In the SF group, the rickettsial ompA and ompB sequence clustered with undetermined Rickettsia sp. and “Candidatus Rickettsia longicornii”, respectively. This is the earliest study regarding the genetic characterization of H. kashmirensis. This study indicated that ticks belong to the genus Haemaphysalis have the potential of harboring and/or transmitting Rickettsia spp. in the region.
1. Introduction
Ticks are notorious ectoparasites infesting the majority of terrestrial and semiterrestrial vertebrates [1]. Ticks are cosmopolitan in distribution, particularly prevalent in tropical and subtropical regions [1]. In addition to anemia, a reduction in dairy products and meat, ticks can also transmit numerous infectious agents including viruses, bacteria, protozoa, and helminths to vertebrate hosts [2].
Haemaphysalis (Acari: Ixodidae) is the second largest genus of ixodid ticks comprising approximately 176 species [1]. Haemaphysalis ticks inhabit various landscapes mainly in the Oriental, Afrotropical, and Palearctic regions, while some of its members have been recorded in Australasia (Australia, New Zealand, and New Guinea), infesting a variety of free-roaming and domestic animals [1,3]. Some species of Haemaphysalis have been identified as potential vectors for various protozoan, viral, and bacterial pathogens to humans, domestic, and wild animals [4,5]. Haemaphysalis ticks such as Haemaphysalis longicornis, Haemaphysalis concinna, Haemaphysalis qinghaiensis, and Haemaphysalis flava have been associated with various Rickettsia spp. including Rickettsia japonica, R. conorii, Rickettsia monacensis, Rickettsia heilongjiangensis, and several undetermined Rickettsia spp. [6,7,8,9]. Moreover, the number of Rickettsia spp. associated with Haemaphysalis ticks are continuously increasing because of the advancement in molecular approaches [10].
Important Haemaphysalis ticks such as H. kashmirensis, H. cornupunctata, and H. montgomeryi belong to the subgenus Herpetobia, Aboimisalis, and Segalia, respectively [11]. These tick species have been described by Hoogstraal and Varma (1962). Additionally, the H. kashmirensis and H. cornupunctata ticks were collected from Kashmir, while H. montgomeryi from India [12]. Later on, life stages such as larva and nymph of these ticks were also described [12,13]. Haemaphysalis kashmirensis, H. cornupunctata, and H. montgomeryi inhabit in the Oriental and Palearctic regions [1]. In the Indian subcontinent, the H. cornupunctata, H. kashmirensis, and H. montgomeryi have been reported from different regions of India, Kashmir, Nepal, and Pakistan, particularly located in the Hindu Kush Himalayan (HKH) range [14,15,16,17,18].
The livestock hosts in Pakistan are known to be infested by a variety of Haemaphysalis ticks [16,19,20]. Genetic characterization of closely related Haemaphysalis ticks such as H. kashmirensis and H. cornupunctata is important for the surveillance and control of these ticks in the Oriental region including Pakistan, which is an epidemic hotspot for these parasites [14,16,18]. The capabilities of Haemaphysalis ticks as a vector for Rickettsia spp. have been neglected. These ticks need to be molecularly screened for associated Rickettsia spp. This study was aimed to the molecular characterization of H. kashmirensis and H. cornupunctata ticks infesting goats and sheep, and associated Rickettsia spp. in the HKH range of Pakistan.
2. Materials and Methods
2.1. Study Sites
The present study was conducted in four districts, namely: Dir Lower (34°52′12.1” N, 71°49′00.8” E), Dir Upper (35°12′30.06” N, 71°52′31.22” E), Bajaur (34°44′4.95” N, 71°30′47.80” E), and Swat (34°45′0.8634” N, 72°21′26.42” E) of the HKH ranges of Khyber Pakhtunkhwa (KP), Pakistan. The geographical coordinates of the tick collection sites were noted by Global Positioning System (GPS) and used for the designing of a map through ArcGIS 10.3.1 (ESRI, Redlands, CA, USA) (Figure 1).
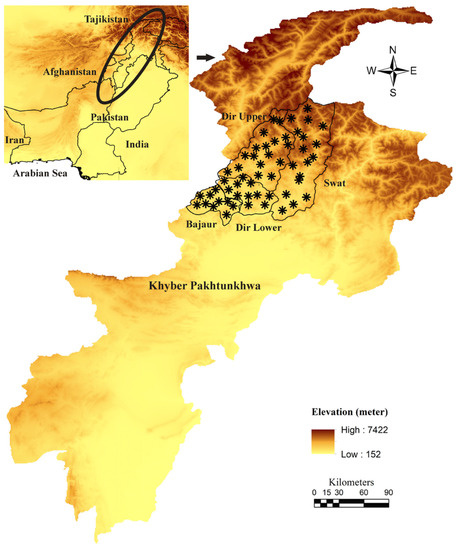
Figure 1.
Elevation-based map showing tick collection sites (black asterisk).
2.2. Ethical Approval and Consent
The design of the present study was approved by the Advanced Study and Research Board (Dir/A&R/AWKUM/2022/9396) and the Ethical Committee of the Faculty of Chemical and Life Sciences, Abdul Wali Khan University Mardan, Pakistan. All animal owners were informed verbally, and approval was taken before observing their hosts.
2.3. Tick Collection and Morphological Identification
Villages of the four selected districts were randomly visited between March 2021 to February 2022 for tick collection. Small ruminants including goats and sheep were examined for ticks. When found, ticks from each host were separately collected in labeled tubes using a fine tweezer. The necessary information regarding collection sites, host type and gender, collection date, and environmental conditions (temperature and humidity) were noted. Collected ticks were rinsed with distilled water followed by 70% ethanol to remove contaminants. Subsequently, standard identification keys [12,13,14,21] were used to identify tick species based on observation of morphological features such as shape of capitulum, cornua and palp articles, cervical and lateral grooves, size and shape of coxa spurs, number of festoons, and shape of genital aperture. These morphological observations were performed under a stereomicroscope (SZ61, Olympus Corporation, Tokyo, Japan).
2.4. Statistical Analyses
Necessary tick data recorded from the four districts were compiled and arranged in spreadsheets using Microsoft Excel V. 2016 (Microsoft Office 365®). Prevalence (no. of infested hosts×100/total no. of examined hosts), mean abundance (total no. of collected ticks/total no. of examined hosts), and mean intensity (total no. of collected ticks/no. of infested hosts) was determined. The necessary climate data for each month or seasonal were obtained via mean temperature (°C), relative humidity (%), and total rainfall (mm) (climate-data.org; accessed on 15 November 2022).
2.5. DNA Extraction and PCR Amplification
Altogether, 108 tick specimens (three males, three females, and three nymphs per species per district) were randomly selected for molecular analysis. The morphologically identified ticks were individually dissected with a sterile blade and ground with a hygienic pestle in 1.5 mL Eppendorf tubes. The ground samples were individually subjected to the phenol–chloroform protocol [22] for DNA extraction. The extracted genomic DNA was quantified by using NanoDrop (Nano-Q, Optizen, South Korea).
The extracted DNA was subjected to conventional PCR to amplify 16S rDNA (460 bp) and cox (710 bp) fragments of tick species and citrate synthase (gltA), outer membrane protein subunit A (ompA), and outer membrane protein subunit B (ompB) fragment for any Rickettsia spp. Each PCR reaction comprised 25 µL volume: 8.5 µL of PCR water “nuclease free”, 1 µL of each primer at a concentration of 10 pmol/µL, 2 µL of extracted DNA (50–100 ng/µL), and 12.5 µL DreamTaq green MasterMix (2X) (Table 1). In each PCR reaction, PCR water was used as a negative control while Rhipicephalus microplus and Rickettsia massiliae DNA were taken as positive control for ticks and Rickettsia spp., respectively. PCR products were analyzed by horizontal electrophoresis in 2% agarose gel and examined under ultraviolet light of a Gel Documentation System (BioDoc-It™ Imaging Systems, Upland, CA, USA).

Table 1.
Primers and PCR conditions used in the current study.
2.6. Sequencing and Phylogenetic Analysis
Amplicons were purified with GeneClean II Kit (Qbiogene, Illkirch, France) following the manufacturer’s protocol and sequenced bi-directionally (Macrogen, Inc., Seoul, South Korea) via the Sanger-based sequencing method. The obtained sequences were trimmed by removing the poor-quality regions followed by obtaining a consensus sequence in SeqMan v 5.00 (DNASTAR, Inc., Madison, WI, USA). Maximum identity sequences were retrieved from GenBank using the Basic Local Alignment Search Tool (BLAST) [28] on the National Center for Biotechnology Information (NCBI) user interface. They were aligned with the obtained sequences in BioEdit Sequence Alignment Editor v. 7.0.5 [29] using CLUSTAL W multiple alignments [30]. The Neighbor-Joining method with the Kimura 2-parameter model was applied to construct phylogenetic trees with 1000 bootstrap replicates in Molecular Evolutionary Genetic Analysis (MEGA-X) software [31]. The coding (cox, gltA, ompA, and ompB) nucleotide sequences were aligned by MUSCLE [32]. The final positions in the dataset comprised the obtained sequence of each fragment.
3. Results
3.1. Ticks, Hosts, and Seasonal Data
Altogether, 834 tick specimens belonging to three tick species including H. cornupunctata (258, 30.9%: 145 females, 86 males, 27 nymphs), H. kashmirensis (191, 22.8%: 102 females, 67 males, 22 nymphs), and H. montgomeryi (385, 46.2%: 202 females, 148 males, 35 nymphs) were morphologically identified. The details about the ticks’ prevalence and their life stages in each district are provided in Table 2.

Table 2.
Data on the number of ticks and hosts, and detection of Rickettsia spp. by polymerase chain reaction.
Ticks were collected from 86 out of 120 examined hosts (goats and sheep) with an overall 71.3% prevalence. Goats were observed with a high prevalence (49/64, 76.6%) compared to sheep (37/56, 66.1%). An overall mean abundance of 7.0 ticks/examined host was noted, while the mean intensity was 9.7 ticks/infested host. Goats were found with higher mean abundance and mean intensity (8.1 and 10.6, respectively) compared to sheep (5.6 and 8.6, respectively). Seasonal parameters, including temperature, humidity, and rainfall with tick parameters such as the prevalence of infestation, mean abundance, and mean intensity are shown in Figure 2.
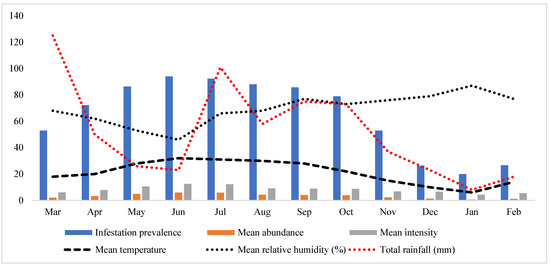
Figure 2.
The graph shows the variation in prevalence, mean abundance, and mean intensity related to mean temperature, mean relative humidity, and the total rainfall during this study.
3.2. Molecular Analysis
Altogether, 72 sequences were obtained (one 16S rDNA and one cox for one female, one male, and one nymph of each tick species) for ticks. Trimmed sequences for H. cornupunctata, H. kashmirensis, and H. montgomeryi were 16S rDNA (395 bp) and cox (654 bp). The 16S rDNA of H. cornupunctata showed 100% identity with the same species from Pakistan. In contrast, its cox showed 87.65–89.22% identity with the same subgenus species: H. (Aboimisalis) punctata from Portugal, Iran, France, China, Hungary, and Romania. The BLAST outcome for the 16S rDNA of H. kashmirensis revealed 93–95% identity with the same subgenus species: H. (Herpetobia) sulcata reported from France, Turkey, China, and Pakistan, compared to cox which showed 89.34% identity with H. sulcata reported from China and Iran. The sequences obtained for H. montgomeryi were 100% identical to the sequences of the same species reported from Pakistan [16]. Thus, such sequences were excluded from further analysis.
For Rickettsia spp., bidirectional sequences were obtained for each amplified fragment. The consensus sequences of gltA (380 bp), ompA (504 bp), and ompB (449 bp) were amplified from H. kashmirensis, while gltA (340 bp) was amplified from H. montgomeryi. Rickettsial gltA sequence (380 bp) amplified from H. kashmirensis showed 97.89% identity with R. conorii subsp. raoultii from Brazil, China, and France, while another rickettsial gltA consensus sequence from H. montgomeryi displayed 100% identity with R. hoogstraalii reported from Italy. The ompA amplified from the same samples of H. kashmirensis showed 100% identity with Rickettsia sp. from China and Algeria. The ompB sequence of the corresponding sample showed 98.16% identity with “Candidatus Rickettsia longicornii” reported from China. The rate of infection was 15.74% (17/108) recorded for Rickettsia sp. (based on gltA, ompA, and ompB) followed by 5.56% (6/108) for R. hoogstraalii (based only on gltA). The details regarding the infection rate and the number of sequences obtained for each Rickettsia spp. are shown in Table 2.
The obtained 16S rDNA sequences were submitted to GenBank under the accession numbers: OQ024373 (H. cornupunctata) and OQ024650 (H. kashmirensis); cox sequences under the accession numbers: OQ096502 (H. cornupunctata) and OQ096625 (H. kashmirensis); rickettsial gltA sequences under the accession numbers: OQ160793 (Rickettsia sp.) and OQ160792 (R. hoogstraalii); rickettsial ompA sequence under the accession number: OQ108505 (Rickettsia sp.), while rickettsial ompB sequence under the accession number: OQ055189 (Rickettsia sp.).
3.3. Phylogenetic Analysis
In a phylogenetic tree based on 16S rDNA, the H. cornupunctata sequence clustered with the same species reported from Pakistan (ON911369), whereas this species grouped in a sister clade with the species of the same subgenus: H. punctata reported from China (NC062064, MF002566, and MG021187) and Turkey (KR870978) (Figure 3). The 16S rDNA sequence of H. kashmirensis clustered with the species of the same subgenus: H. (Herpetobia) sulcata reported from France (KX576650), Turkey (MZ463298 and KR870979), China (NC062063), and Pakistan (MT799947) (Figure 3). In a phylogenetic tree based on cox, the H. cornupunctata sequence clustered with the species of the same subgenus: H. punctata reported from Portugal (LC508354), Iran (MH532298), France (ON387756), China (NC062064 and MZ596002), Hungary (MW193894-MW193895), and Romania (JX394187) (Figure 4). Whereas the obtained cox sequence of H. kashmirensis sequence clustered with the species of the same subgenus: H. (Herpetobia) sulcata reported from China (MN836696-MN836698 and NC062063) and Iran (MH532303) (Figure 4).
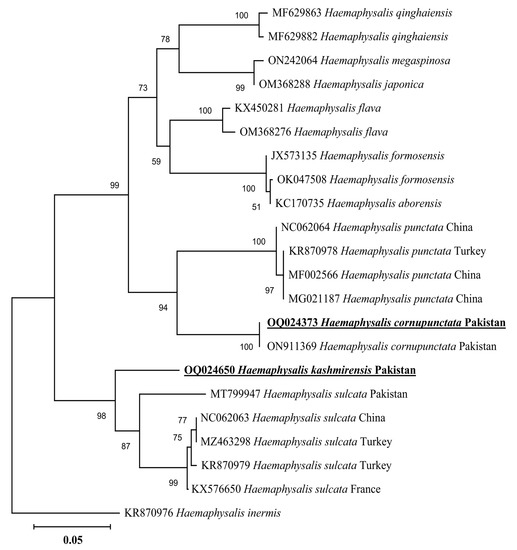
Figure 3.
Neighbor-Joining phylogenetic tree based on 16S rDNA of H. cornupunctata and H. kashmirensis. The 16S rDNA sequence of Haemaphysalis inermis was employed as an outgroup. All sequences have been denoted by their GenBank accession numbers, followed by species name and country name. The bootstrap values (1000-replications) are shown at each node. The sequences (OQ024373 and OQ024650) of the present study have been marked with bold and underlined fonts.
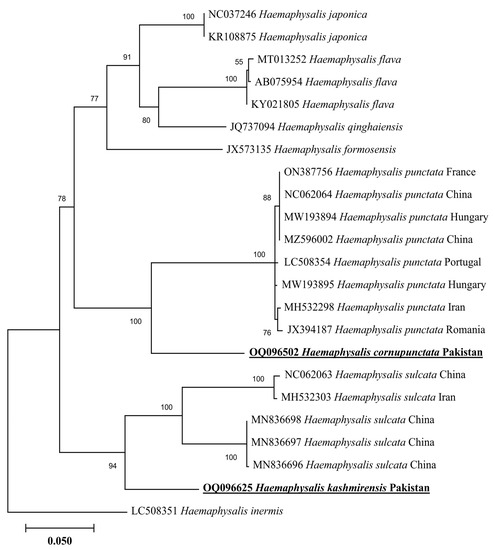
Figure 4.
Neighbor-Joining phylogenetic tree based on cox of H. cornupunctata and H. kashmirensis. The cox sequence of H. inermis was employed as an outgroup. All sequences have been denoted by their GenBank accession numbers, followed by species name and country name. The bootstrap values (1000-replications) are shown at each node. The sequences (OQ096502 and OQ096625) of the present study have been marked with bold and underlined fonts.
Phylogenetic tree based on rickettsial gltA, the Rickettsia sp. detected in H. kashmirensis clustered individually with the sequence of Rickettsia spp. of the spotted fever group. In contrast, the R. hoogstraalii detected in H. montgomeryi was clustered with the same species reported from Italy (KY418024-KY418025) (Figure 5). The rickettsial ompA fragment detected in H. kashmirensis clustered with the Rickettsia sp. reported from China (MT361020, MG228270, and MN644903) and Algeria (MZ064523-MZ064524 and JN943296) (Figure 6) and grouped in a sister clade with “Candidatus Rickettsia longicornii” and “Candidatus Rickettsia jingxinensis”, while the rickettsial ompB fragment clustered with “Candidatus Rickettsia longicornii” reported from China (MN026546, MK620854, MG906675, and MT511089) (Figure 7).
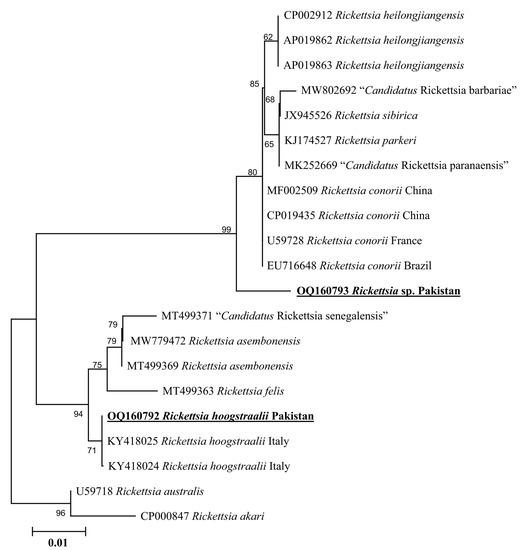
Figure 5.
Neighbor-Joining phylogenetic tree based on gltA sequences of Rickettsia sp. and R. hoogstraalii. The gltA sequences of Rickettsia australis and Rickettsia akari were employed as an outgroup. All sequences have been denoted by their GenBank accession numbers, followed by species name and country name. The bootstrap values (1000-replications) are shown at each node. The sequences (OQ160793 and OQ160792) of the present study have been marked with bold and underlined fonts.
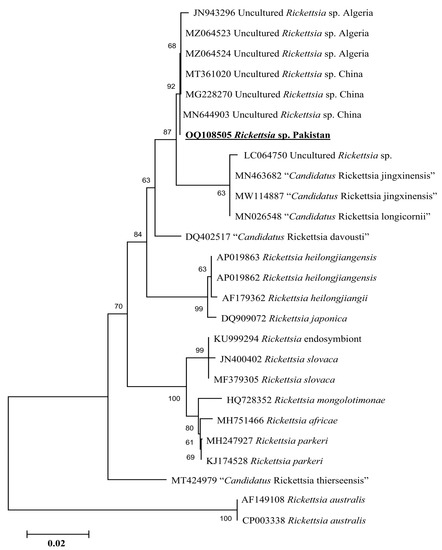
Figure 6.
Neighbor-Joining phylogenetic tree based on ompA sequences of a Rickettsia sp. The ompA sequences of R. australis were employed as an outgroup. All sequences have been denoted by their GenBank accession numbers, followed by species name and country name. The bootstrap values (1000-replications) are shown at each node. The sequence (OQ108505) of the present study has been marked with bold and underlined fonts.
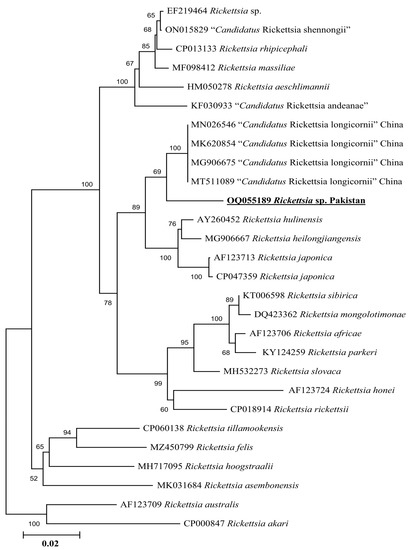
Figure 7.
Neighbor-Joining phylogenetic tree based on ompB sequences of a Rickettsia sp. The ompB sequences of R. australis and R. akari were employed as an outgroup. All sequences have been denoted by their GenBank accession numbers, followed by species name and country name. The bootstrap values (1000-replications) are shown at each node. The sequence (OQ055189) of the present study has been marked with bold and underlined fonts.
4. Discussion
As the geo-climatic conditions of the Oriental region including Pakistan suit the flourishment of Haemaphysalis ticks, the largest diversity of these ticks has been reported in this region [16]. The second most diverse genus of hard ticks (Ixodidae), Haemaphysalis comprises ~173 tick species globally. In Pakistan, 13 Haemaphysalis spp. have been reported; however, genetic data of these ticks are limited [16]. Despite the huge diversity, genetic data regarding the genus Haemaphysalis ticks and associated Rickettsia spp. have been largely neglected. For this purpose, Haemaphysalis ticks were collected from goats and sheep in northern Pakistan, where several species of this genus are considered endemic [16,18,19]. Herein, the collected ticks were morpho-molecularly identified as H. cornupunctata, H. kashmirensis, and H. montgomeryi. The genetic characterization based on 16S rDNA and cox partial sequences of H. kashmirensis, and cox sequence for H. cornupunctata was achieved for the first time. An undetermined Rickettsia sp. based on gltA, ompA, and ompB sequence was molecularly characterized in H. kashmirensis, whereas R. hoogstraalii based on only gltA sequence was detected in H. montgomeryi.
The surveyed areas are part of the HKH mountain range, which has been considered as one of the most important biodiversity hotspots [18,33]. Previously, H. kashmirensis, H. cornupunctata, and H. montgomeryi have been reported from many locations of the HKH range, which spans different territories of the Indian subcontinent, including Pakistan [15,17,18], Kashmir and India [14], and Afghanistan [34]. These findings suggest that HKH mountain regions have a great diversity of the Haemaphysalis ticks owing to the abundance of suitable hosts and conducive climate conditions.
Haemaphysalis kashmirensis was found less in number than H. cornupunctata and H. montgomeryi, which could be due to the association of the later species with hosts other than goats and sheep in the family Bovidae [1]. Moreover, the adult ticks (female and male) were outnumbered by the immature ticks (nymphs), while no larval stage of any tick species was found on goats and sheep. Previous studies suggested that Agama tuberculata (Kashmir Rock Agama) is the main host of the nymphal and larval stages of H. kashmirensis, which is found in the HKH range [21]. Similarly, animals belonging to the Muridae, Herpestidae, Erinaceidae, Cricetidae, and Soricidae families have been recorded as the main hosts for the nymphal and larval ticks of H. cornupunctata and H. montgomeryi [1].
Haemaphysalis ticks have been reported as vectors for Rickettsia spp. including R. hoogstraalii and Rickettsia rhipicephali [5,35]. Herein, Rickettsia sp. was detected through gltA, ompA, and ompB, whereas R. hoogstraalii was detected only through gltA. The genetic characterization of R. hoogstraalii was also attempted through ompA and ompB; however, the amplifications of these fragments were unsuccessful. Amplification failures are common in the case of ompA, ompB that might be the lack of targeted genes, as shown in the transition group Rickettsia or due to primer mismatching [36,37,38]. Rickettsia hoogstraalii has been detected in ticks of the genus Haemaphysalis such as H. sulcata (Cyprus and Italy), H. punctata (Italy), and Haemaphysalis parva (Turkey) [39,40,41]. The pathogenicity of R. hoogstraalii is poorly known [42].
Genetic analyses based on molecular markers such as 16S rDNA and cox genes are extremely helpful in unveiling the systematics of ticks and phylogenetic positioning [43,44,45,46,47,48,49]. In the phylogenetic tree based on 16S rDNA and cox of H. cornupunctata, this species appeared in a monophyletic branch with H. punctata from Turkey, France, China, Iran, Romania, Portugal, and Hungary. Previously, these two tick species (H. cornupunctata and H. punctata) have been assigned to the same subgenus (Aboimisalis) on a morphological basis [11]. In a phylogenetic tree based on 16S rDNA and cox of H. kashmirensis, this specie clustered in a monophyletic clade with H. sulcata previously reported from Pakistan, China, Turkey, France, and Iran. On a morphological basis, H. sulcata and H. kashmirensis have been placed in the same subgenus, Herpetobia [11]. A phylogenetic clustering of these species with different closest species of the same subgenus could be associated with the missing genetic data of corresponding species in the GenBank. The phylogenetic analysis based on rickettsial gltA showed that the Rickettsia sp. detected in H. kashmirensis belonged to the SF group, whereas R. hoogstraalii detected in H. montgomeryi belonged to the transition group. The phylogenetic tree based on ompA and ompB, obtained from the same sample in which Rickettsia sp. was detected, validated the gltA-based phylogenetic analysis for Rickettsia sp.
5. Conclusions
This study contributes to the missing information regarding the genetic data of some Haemaphysalis ticks, especially H. kashmirensis, which was genetically characterized for the first time. The relationship of H. cornupunctata with subgenus Aboimisalis and H. kashmirensis with subgenus Herpetobia established on a morphological basis was confirmed through molecular-based phylogenetic analysis. Furthermore, a Rickettsia sp. was molecularly assessed in H. kashmirensis, whereas R. hoogstraalii was detected in H. montgomeryi. This study may assist in understanding the identification, evolutionary history, and molecular epidemiology of Haemaphysalis ticks and associated Rickettsia spp. Further studies should genetically characterize and evaluate the pathogenicity of these Rickettsia spp.
Author Contributions
A.A. (Abdulaziz Alouffi) designed the study. S.M.K., Z.U.I., A.A. (Abdulaziz Alouffi), M.M.A., T.T. and H.A., performed the experimental designing of the study. A.A. (Abid Ali), S.M.K., M.K., S.U., M.N. and M.K.O. collected the ticks. A.A. (Abid Ali), S.M.K., M.K., S.U., M.N., A.A. (Abdulaziz Alouffi), M.M.A. and M.K.O. carried out experiments, phylogenetic, and statistical analysis. All authors have read and agreed to the published version of the manuscript.
Funding
The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia.
Institutional Review Board Statement
The design of the present study was approved by the Advanced Study and Research Board (Dir/A&R/AWKUM/2022/9396) and the Ethical Committee of the Faculty of Chemical and Life Sciences, Abdul Wali Khan University Mardan, Pakistan.
Informed Consent Statement
Not applicable.
Data Availability Statement
All the relevant data are within the manuscript.
Acknowledgments
Authors highly acknowledge the financial support provided by Pakistan Science Foundation and Higher Education Commission to drive this research. The researchers supporting project number (RSP2023R494), King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guglielmone, A.A.; Petney, T.N.; Robbins, R.G. Ixodidae (Acari: Ixodoidea): Descriptions and redescriptions of all known species from 1758 to December 31, 2019. Zootaxa 2020, 4871, 1–322. [Google Scholar] [CrossRef] [PubMed]
- Jongejan, F.; Uilenberg, G. The global importance of ticks. Parasitology 2004, 129, S3–S14. [Google Scholar] [CrossRef] [PubMed]
- Apanaskevich, D.A.; Tomlinson, J.A. Description of four new species of Haemaphysalis Koch, 1844 (Acari: Ixodidae) from the H. (Rhipistoma) spinulosa subgroup, parasites of carnivores and rodents in Africa. Syst. Parasitol. 2019, 96, 625–657. [Google Scholar] [CrossRef] [PubMed]
- Guan, G.; Moreau, E.; Liu, J.; Hao, X.; Ma, M.; Luo, J.; Chauvin, A.; Yin, H. Babesia sp. BQ1 (Lintan): Molecular evidence of experimental transmission to sheep by Haemaphysalis qinghaiensis and Haemaphysalis longicornis. Parasitol. Int. 2010, 59, 265–267. [Google Scholar] [CrossRef]
- Jiang, J.; An, H.; Lee, J.S.; O’Guinn, M.L.; Kim, H.C.; Chong, S.T.; Zhang, Y.; Song, D.; Burrus, R.G.; Bao, Y.; et al. Molecular characterization of Haemaphysalis longicornis-borne rickettsiae, Republic of Korea and China. Ticks Tick-Borne Dis. 2018, 9, 1606–1613. [Google Scholar] [CrossRef]
- Lee, J.H.; Park, H.S.; Jung, K.D.; Jang, W.J.; Koh, S.E.; Kang, S.S.; Lee, I.Y.; Lee, W.J.; Kim, B.J.; Kook, Y.H.; et al. Identification of the spotted fever group rickettsiae detected from Haemaphysalis longicornis in Korea. Microbiol. Immunol. 2003, 47, 301–304. [Google Scholar] [CrossRef]
- Cheng, C.; Fu, W.; Ju, W.; Yang, L.; Xu, N.; Wang, Y.M.; Li, H.; Wang, Y.L.; Hu, M.X.; Wen, J.; et al. Diversity of spotted fever group Rickettsia infection in hard ticks from Suifenhe, Chinese–Russian border. Ticks Tick-Borne Dis. 2016, 7, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Noh, Y.; Lee, Y.S.; Kim, H.C.; Chong, S.T.; Klein, T.A.; Jiang, J.; Richards, A.L.; Lee, H.K.; Kim, S.Y. Molecular detection of Rickettsia species in ticks collected from the southwestern provinces of the Republic of Korea. Parasit. Vectors 2017, 10, 1–10. [Google Scholar] [CrossRef]
- Igolkina, Y.; Rar, V.; Vysochina, N.; Ivanov, L.; Tikunov, A.; Pukhovskaya, N.; Epikhina, T.; Golovljova, I.; Tikunova, N. Genetic variability of Rickettsia spp. in Dermacentor and Haemaphysalis ticks from the Russian Far East. Ticks Tick-Borne Dis. 2018, 9, 1594–1603. [Google Scholar] [CrossRef]
- Qin, X.R.; Han, H.J.; Han, F.J.; Zhao, F.M.; Zhang, Z.T.; Xue, Z.F.; Ma, D.Q.; Qi, R.; Zhao, M.; Wang, L.J.; et al. Rickettsia japonica and novel Rickettsia species in ticks, China. Emerg. Infect. Dis. 2019, 25, 992. [Google Scholar] [CrossRef]
- Hoogstraal, H.; Kim, K.C. Tick and mammal coevolution, with emphasis on Haemaphysalis. In Coevolution of Parasitic Arthropods and Mammals; Kim, K.C., Ed.; Wiley International Science: New York, NY, USA, 1985; pp. 505–568. [Google Scholar]
- Hoogstraal, H.; Trapido, H.; Kohls, G.M. Studies on southeast Asian Haemaphysalis ticks (Ixodoidea, Ixodidae). Speciation in the H. (Kaiseriana) obesa group: H. semermis Neumann, H. obesa Larrousse, H. roubaudi Toumanoff, H. montgomeryi Nuttall, and H. hirsuta sp. n. J. Parasitol. 1966, 52, 169–191. [Google Scholar] [CrossRef]
- Dhanda, V.; Kulkarni, S.M. Immature stages of Haemaphysalis cornupunctata Hoogstraal and Varma, 1962 (Acarina: Ixodidae) with new host and locality records, and notes on its ecology. Orient. Insects 1969, 3, 15–21. [Google Scholar] [CrossRef]
- Hoogstraal, H.; Varma, M.G.R. Haemaphysalis cornupunctata sp. n. and H. kashmirensis sp. n. from Kashmir, with Notes on H. sundrai Sharif and H. sewelli Sharif of India and Pakistan (Ixodoidea, Ixodidae). J. Parasitol. 1962, 48, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.; Budachetri, K.; Mukherjee, N.; Williams, J.; Kausar, A.; Hassan, M.J.; Adamson, S.; Dowd, S.E.; Apanskevich, D.; Arijo, A.; et al. A study of ticks and tick-borne livestock pathogens in Pakistan. PLoS Negl. Trop. Dis. 2017, 11, e0005681. [Google Scholar] [CrossRef]
- Ali, A.; Numan, M.; Ullah, S.; Khan, M.; Kamran, K. Genetic Characterization of Haemaphysalis (Rhipistoma) indica and Haemaphysalis (Segalia) montgomeryi Ticks (Ixodoidea: Ixodidae). Ticks Tick-Borne Dis. 2022, 14, 102105. [Google Scholar] [CrossRef]
- Alam, S.; Khan, M.; Alouffi, A.; Almutairi, M.M.; Ullah, S.; Numan, M.; Islam, N.; Khan, Z.; Aiman, O.; Safi, S.Z.; et al. Spatio-Temporal Patterns of Ticks and Molecular Survey of Anaplasma marginale, with Notes on Their Phylogeny. Microorganisms 2022, 10, 1663. [Google Scholar] [CrossRef]
- Khan, Z.; Shehla, S.; Alouffi, A.; Obaid, M.K.; Khan, A.Z.; Almutairi, M.M.; Numan, M.; Aiman, O.; Alam, S.; Ullah, S.; et al. Molecular Survey and Genetic Characterization of Anaplasma marginale in Ticks Collected from Livestock Hosts in Pakistan. Animals 2022, 12, 1708. [Google Scholar] [CrossRef]
- Ali, A.; Khan, M.A.; Zahid, H.; Yaseen, P.M.; Qayash Khan, M.; Nawab, J.; Ur Rehman, Z.; Ateeq, M.; Khan, S.; Ibrahim, M. Seasonal dynamics, record of ticks infesting humans, wild and domestic animals and molecular phylogeny of Rhipicephalus microplus in Khyber Pakhtunkhwa Pakistan. Front. Physiol. 2019, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Ahmad., I.; Ullah, S.; Alouffi, A.; Almutairi, M.M.; Khan, M.; Numan, M.; Safi, S.Z.; Chitimia-Dobler, L.; Tanaka, T.; Ali, A. Description of Male, Redescription of Female, Host Record, and Phylogenetic Position of Haemaphysalis danieli. Pathogens 2022, 11, 1495. [Google Scholar] [CrossRef]
- Hoogstraal, H.; McCarthy, V.C. Hosts and distribution of Haemaphysalis kashmirensis with descriptions of immature stages and definition of the subgenus Herpetobia Canestrini (resurrected). J. Parasitol. 1965, 51, 674–679. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Folmer, O.; Hoeh, W.R.; Black, M.B.; Vrijenhoek, R.C. Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mol. Marine Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Mangold, A.J.; Bargues, M.D.; Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol. Res. 1998, 84, 478–484. [Google Scholar] [CrossRef]
- Labruna, M.B.; Whitworth, T.; Bouyer, D.H.; McBride, J.; Camargo, L.M.A.; Camargo, E.P.; Popov, V.; Walker, D.H. Rickettsia bellii and Rickettsia amblyommii in Amblyomma ticks from the state of Rondônia, Western Amazon, Brazil. J. Med. Entomol. 2004, 41, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Roux, V.; Fournier, P.E.; Raoult, D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein rOmpA. J. Clin. Microbiol. 1996, 34, 2058–2065. [Google Scholar] [CrossRef] [PubMed]
- Roux, V.; Raoult, D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB). Int. J. Syst. Evol. Microbiol. 2000, 50, 1449–1455. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, T.; Biosciences, I.; Carlsbad, C.J.G.B.B. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Aiman, O.; Ullah, S.; Chitimia-Dobler, L.; Nijhof, A.M.; Ali, A. First report of Nosomma monstrosum ticks infesting Asian water buffaloes (Bubalus bubalis) in Pakistan. Ticks Tick-Borne Dis. 2022, 13, 101899. [Google Scholar] [CrossRef] [PubMed]
- Hoogstraal, H. Biological patterns in the Afghanistan tick fauna. In Proceedings of the 3rd International Congress of Acarology; Springer: Dordrecht, The Netherlands, 1973; pp. 511–514. [Google Scholar]
- Zhao, L.; Li, J.; Cui, X.; Jia, N.; Wei, J.; Xia, L.; Wang, H.; Zhou, Y.; Wang, Q.; Liu, X.; et al. Distribution of Haemaphysalis longicornis and associated pathogens: Analysis of pooled data from a China field survey and global published data. Lancet Planet. Health 2020, 4, e320–e329. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; La Scola, B.; Audic, S.; Renesto, P.; Blanc, G.; Robert, C.; Fournier, P.E.; Claverie, J.M.; Raoult, D. Genome sequence of Rickettsia bellii illuminates the role of amoebae in gene exchanges between intracellular pathogens. PLoS Genet. 2006, 2, e76. [Google Scholar] [CrossRef]
- Thu, M.J.; Qiu, Y.; Matsuno, K.; Kajihara, M.; Mori-Kajihara, A.; Omori, R.; Monma, N.; Chiba, K.; Seto, J.; Gokuden, M.; et al. Diversity of spotted fever group rickettsiae and their association with host ticks in Japan. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Reeves, W.K.; Mans, B.J.; Durden, L.A.; Miller, M.M.; Gratton, E.M.; Laverty, T.M. Rickettsia hoogstraalii and a Rickettsiella from the Bat tick Argas transgariepinus, in Namibia. J. Parasitol. 2020, 106, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Chochlakis, D.; Ioannou, I.; Sandalakis, V.; Dimitriou, T.; Kassinis, N.; Papadopoulos, B.; Tselentis, Y.; Psaroulaki, A. Spotted fever group Rickettsiae in ticks in Cyprus. Microb. Ecol. 2012, 63, 314–323. [Google Scholar] [CrossRef]
- Orkun, Ö.; Karaer, Z.; Çakmak, A.; Nalbantoğlu, S. Spotted fever group rickettsiae in ticks in Turkey. Ticks Tick-Borne Dis. 2014, 5, 213–218. [Google Scholar] [CrossRef] [PubMed]
- Chisu, V.; Leulmi, H.; Masala, G.; Piredda, M.; Foxi, C.; Parola, P. Detection of Rickettsia hoogstraalii, Rickettsia helvetica, Rickettsia massiliae, Rickettsia slovaca and Rickettsia aeschlimannii in ticks from Sardinia, Italy. Ticks Tick-Borne Dis. 2017, 8, 347–352. [Google Scholar] [CrossRef]
- Parola, P.; Paddock, C.D.; Socolovschi, C.; Labruna, M.B.; Mediannikov, O.; Kernif, T.; Abdad, M.Y.; Stenos, J.; Bitam, I.; Fournier, P.E.; et al. Update on tick-borne rickettsioses around the world: A geographic approach. Clin. Microbiol. Rev. 2013, 26, 657–702. [Google Scholar] [CrossRef]
- Norris, D.E.; Klompen, J.S.H.; Black, W.C. Comparison of the mitochondrial 12S and 16S ribosomal DNA genes in resolving phylogenetic relationships among hard ticks (Acari: Ixodidae). Ann. Entomolo. Soc. Am. 1999, 92, 117–129. [Google Scholar] [CrossRef]
- Latrofa, M.S.; Dantas-Torres, F.; Annoscia, G.; Cantacessi, C.; Otranto, D. Comparative analyses of mitochondrial and nuclear genetic markers for the molecular identification of Rhipicephalus spp. Infect. Genet. Evol. 2013, 20, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Numan, M.; Islam, N.; Adnan, M.; Safi, S.Z.; Chitimia-Dobler, L.; Labruna, M.B.; Ali, A. First genetic report of Ixodes kashmiricus and associated Rickettsia sp. Parasit. Vectors 2022, 15, 1–12. [Google Scholar] [CrossRef]
- Ali, A.; Numan, M.; Khan, M.; Aiman, O.; Muñoz-Leal, S.; Chitimia-Dobler, L.; Labruna, M.B.; Nijhof, A. Ornithodoros (Pavlovskyella) ticks associated with a Rickettsia sp. in Pakistan. Parasit. Vectors 2022, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Zahid, H.; Zeb, I.; Tufail, M.; Khan, S.; Haroon, M.; Bilal, M.; Hussain, M.; Alouffi, A.S.; Muñoz-Leal, S.; et al. Risk factors associated with tick infestations on equids in Khyber Pakhtunkhwa, Pakistan, with notes on Rickettsia massiliae detection. Parasit. Vectors 2021, 14, 1–12. [Google Scholar] [CrossRef]
- Zahid, H.; Muñoz-Leal, S.; Khan, M.Q.; Alouffi, A.S.; Labruna, M.B.; Ali, A. Life cycle and genetic identification of Argas persicus infesting domestic fowl in Khyber Pakhtunkhwa, Pakistan. Front. Vet. Sci. 2021, 8, 302. [Google Scholar] [CrossRef]
- Khan, M.; Islam, N.; Khan, A.; Islam, Z.U.; Muñoz-Leal, S.; Labruna, M.B.; Ali, A. New records of Amblyomma gervaisi from Pakistan, with detection of a reptile-associated Borrelia sp. Ticks Tick-Borne Dis. 2022, 13, 102047. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).