Identification and Characterization of Glycine- and Arginine-Rich Motifs in Proteins by a Novel GAR Motif Finder Program
Abstract
1. Introduction
2. Materials and Methods
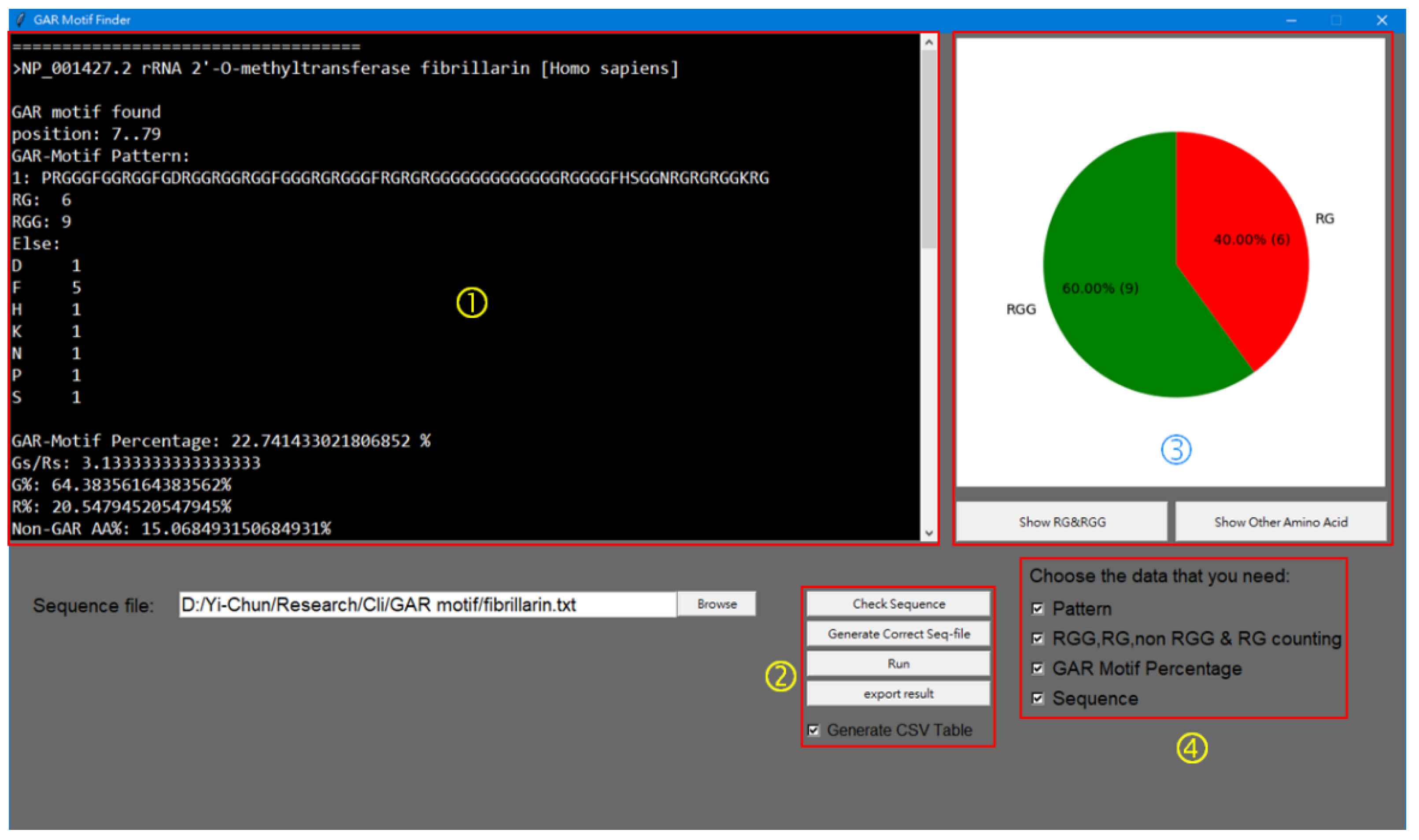
Program Coding of GAR Motif Finder (GMF)
3. Results
3.1. Analyses of the N-Terminal GAR Domain of Fibrillarin in Different Model Organisms
3.2. Development of the GAR MOTIF FINDER Program for Analyses of Long GAR Domain in FBL
3.3. Analyses of the GAR Domains in the Three Nucleolar Proteins by GMF
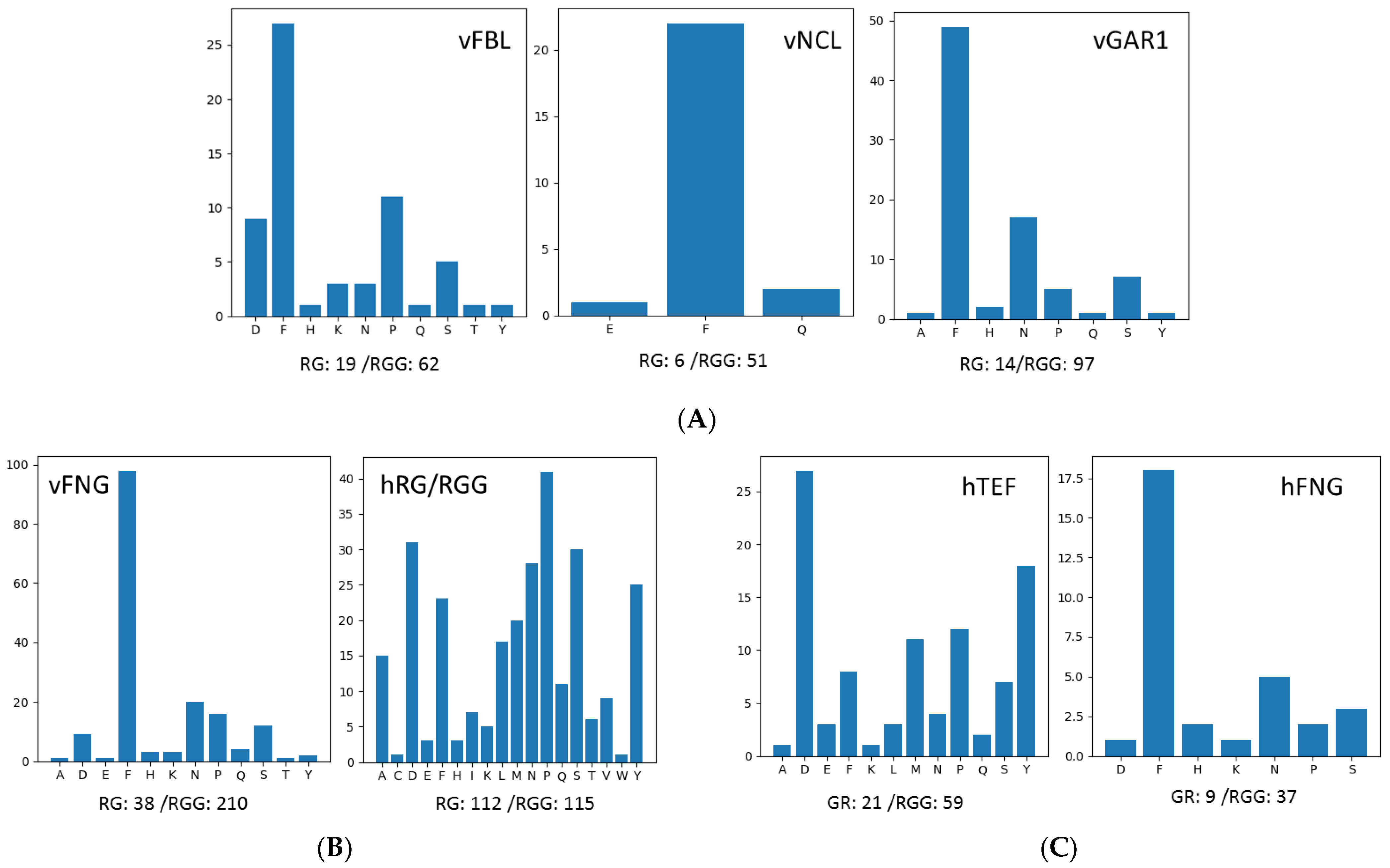
3.4. Analyses of Other High RG/RGG-Repeat-Containing Proteins by GMF
3.5. Analyses of Extra-Long GAR Motifs in Human and Other Proteomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shubina, M.Y.; Arifulin, E.A.; Sorokin, D.V.; Sosina, M.A.; Tikhomirova, M.A.; Serebryakova, M.V.; Smirnova, T.; Sokolov, S.S.; Musinova, Y.R.; Sheval, E.V. The GAR domain integrates functions that are necessary for the proper localization of fibrillarin (FBL) inside eukaryotic cells. PeerJ 2020, 8, e9029. [Google Scholar] [CrossRef] [PubMed]
- Tollervey, D.; Lehtonen, H.; Jansen, R.; Kern, H.; Hurt, E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 1993, 72, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Snaar, S.; Wiesmeijer, K.; Jochemsen, A.G.; Tanke, H.J.; Dirks, R.W. Mutational analysis of fibrillarin and its mobility in living human cells. J. Cell Biol. 2000, 151, 653–662. [Google Scholar] [CrossRef]
- El Hassouni, B.; Sarkisjan, D.; Vos, J.C.; Giovannetti, E.; Peters, G.J. Targeting the Ribosome Biogenesis Key Molecule Fibrillarin to Avoid Chemoresistance. Curr. Med. Chem. 2019, 26, 6020–6032. [Google Scholar] [CrossRef]
- Stamm, S.; Lodmell, J.S. C/D box snoRNAs in viral infections: RNA viruses use old dogs for new tricks. Non-Coding RNA Res. 2019, 4, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.W.; Xu, G.; Wang, Y.; Shan, L.; Luan, P.F.; Wang, Y.; Wu, M.; Yang, L.Z.; Xing, Y.H.; Yang, L.; et al. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Mol. Cell 2019. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; O’Connor, T.R.; Bailey, T.L.; Richard, S. Defining the RGG/RG motif. Mol. Cell 2013, 50, 613–623. [Google Scholar] [CrossRef]
- Dragon, F.; Pogacić, V.; Filipowicz, W. In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell Biol. 2000, 20, 3037–3048. [Google Scholar] [CrossRef]
- Lischwe, M.A.; Ochs, R.L.; Reddy, R.; Cook, R.G.; Yeoman, L.C.; Tan, E.M.; Reichlin, M.; Busch, H. Purification and partial characterization of a nucleolar scleroderma antigen (Mr = 34,000; pI, 8.5) rich in NG,NG-dimethylarginine. J. Biol. Chem. 1985, 260, 14304–14310. [Google Scholar] [CrossRef]
- Lischwe, M.A.; Roberts, K.D.; Yeoman, L.C.; Busch, H. Nucleolar specific acidic phosphoprotein C23 is highly methylated. J. Biol. Chem. 1982, 257, 14600–14602. [Google Scholar] [CrossRef]
- Frankel, A.; Clarke, S. RNase treatment of yeast and mammalian cell extracts affects in vitro substrate methylation by type I protein arginine N-methyltransferases. Biochem. Biophys. Res. Commun. 1999, 259, 391–400. [Google Scholar] [CrossRef]
- Whitehead, S.E.; Jones, K.W.; Zhang, X.; Cheng, X.; Terns, R.M.; Terns, M.P. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J. Biol. Chem. 2002, 277, 48087–48093. [Google Scholar] [CrossRef] [PubMed]
- Kiledjian, M.; Dreyfuss, G. Primary structure and binding activity of the hnRNP U protein: Binding RNA through RGG box. Embo. J. 1992, 11, 2655–2664. [Google Scholar] [CrossRef]
- Liu, Q.; Dreyfuss, G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell Biol. 1995, 15, 2800–2808. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Clarke, S.G. Protein Arginine Methylation in Mammals: Who, What, and Why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef]
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Erce, M.A.; Lai, Y.W.; Tomasetig, F.; Hart-Smith, G.; Hamey, J.J.; Wilkins, M.R. Crosstalk of Phosphorylation and Arginine Methylation in Disordered SRGG Repeats of Saccharomycescerevisiae Fibrillarin and Its Association with Nucleolar Localization. J. Mol. Biol. 2020, 432, 448–466. [Google Scholar] [CrossRef]
- Hofweber, M.; Dormann, D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019, 294, 7137–7150. [Google Scholar] [CrossRef]
- Ai, L.S.; Lin, C.H.; Hsieh, M.; Li, C. Arginine methylation of a glycine and arginine rich peptide derived from sequences of human FMRP and fibrillarin. Proc. Natl. Sci. Counc. Repub. China B 1999, 23, 175–180. [Google Scholar]
- Lin, C.H.; Huang, H.M.; Hsieh, M.; Pollard, K.M.; Li, C. Arginine methylation of recombinant murine fibrillarin by protein arginine methyltransferase. J. Protein Chem. 2002, 21, 447–453. [Google Scholar] [CrossRef]
- Lee, Y.J.; Hsieh, W.Y.; Chen, L.Y.; Li, C. Protein arginine methylation of SERBP1 by protein arginine methyltransferase 1 affects cytoplasmic/nuclear distribution. J. Cell Biochem. 2012, 113, 2721–2728. [Google Scholar] [CrossRef]
- Lee, Y.J.; Wei, H.M.; Chen, L.Y.; Li, C. Localization of SERBP1 in stress granules and nucleoli. FEBS J. 2014, 281, 352–364. [Google Scholar] [CrossRef]
- Wei, H.M.; Hu, H.H.; Chang, G.Y.; Lee, Y.J.; Li, Y.C.; Chang, H.H.; Li, C. Arginine methylation of the cellular nucleic acid binding protein does not affect its subcellular localization but impedes RNA binding. FEBS Lett. 2014, 588, 1542–1548. [Google Scholar] [CrossRef]
- Chong, P.A.; Vernon, R.M.; Forman-Kay, J.D. RGG/RG Motif Regions in RNA Binding and Phase Separation. J. Mol. Biol. 2018, 430, 4650–4665. [Google Scholar] [CrossRef] [PubMed]
- Li, K.K.C.; Chau, B.L.; Lee, K.A.W. Differential interaction of PRMT1 with RGG-boxes of the FET family proteins EWS and TAF15. Protein Sci. 2018, 27, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Bikkavilli, R.K.; Malbon, C.C. Arginine methylation of G3BP1 in response to Wnt3a regulates -catenin mRNA. J. Cell Sci. 2011, 124, 2310–2320. [Google Scholar] [CrossRef]
- Guo, Z.; Zheng, L.; Xu, H.; Dai, H.; Zhou, M.; Pascua, M.R.; Chen, Q.M.; Shen, B. Methylation of FEN1 suppresses nearby phosphorylation and facilitates PCNA binding. Nat. Chem. Biol. 2010, 6, 766–773. [Google Scholar] [CrossRef]
- Angrand, G.; Quillevere, A.; Loaec, N.; Dinh, V.T.; Le Senechal, R.; Chennoufi, R.; Duchambon, P.; Keruzore, M.; Martins, R.P.; Teulade-Fichou, M.P.; et al. Type I arginine methyltransferases are intervention points to unveil the oncogenic Epstein-Barr virus to the immune system. Nucleic Acids Res. 2022, 50, 11799–11819. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Chang, P.C.; Huerta, S.; Izumiya, C.; Davis, R.; Tepper, C.G.; Kim, K.Y.; Shevchenko, B.; Wang, D.H.; Jung, J.U.; et al. Protein arginine methyltransferase 1-directed methylation of Kaposi sarcoma-associated herpesvirus latency-associated nuclear antigen. J. Biol. Chem. 2012, 287, 5806–5818. [Google Scholar] [CrossRef]
- Mostaqul Huq, M.D.; Gupta, P.; Tsai, N.-P.; White, R.; Parker, M.G.; Wei, L.-N. Suppression of receptor interacting protein 140 repressive activity by protein arginine methylation. EMBO J. 2006, 25, 5094–5104. [Google Scholar] [CrossRef] [PubMed]
- Masuzawa, T.; Oyoshi, T. Roles of the RGG Domain and RNA Recognition Motif of Nucleolin in G-Quadruplex Stabilization. ACS Omega 2020, 5, 5202–5208. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Gu, H.; Zhou, J.; Mulhern, D.; Wang, Y.; Lee, K.A.; Yang, V.; Aguiar, M.; Kornhauser, J.; Jia, X.; et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell Proteom. 2014, 13, 372–387. [Google Scholar] [CrossRef]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.S.; Varela, J.A.; Lin, J.Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Strohl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-pi Interactions. Cell 2018, 173, 720–734.e15. [Google Scholar] [CrossRef] [PubMed]
- Kaneb, H.M.; Dion, P.A.; Rouleau, G.A. The FUS about arginine methylation in ALS and FTLD. Embo J. 2012, 31, 4249–4251. [Google Scholar] [CrossRef] [PubMed]
- Weimann, M.; Grossmann, A.; Woodsmith, J.; Özkan, Z.; Birth, P.; Meierhofer, D.; Benlasfer, N.; Valovka, T.; Timmermann, B.; Wanker, E.E.; et al. A Y2H-seq approach defines the human protein methyltransferase interactome. Nat. Methods 2013, 10, 339–342. [Google Scholar] [CrossRef]
- Gittings, L.M.; Foti, S.C.; Benson, B.C.; Gami-Patel, P.; Isaacs, A.M.; Lashley, T. Heterogeneous nuclear ribonucleoproteins R and Q accumulate in pathological inclusions in FTLD-FUS. Acta Neuropathol. Commun. 2019, 7, 18. [Google Scholar] [CrossRef]
- Estell, C.; Davidson, L.; Steketee, P.C.; Monier, A.; West, S. ZC3H4 restricts non-coding transcription in human cells. eLife 2021, 10. [Google Scholar] [CrossRef]
- Musiani, D.; Bok, J.; Massignani, E.; Wu, L.; Tabaglio, T.; Ippolito, M.R.; Cuomo, A.; Ozbek, U.; Zorgati, H.; Ghoshdastider, U.; et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal 2019, 12. [Google Scholar] [CrossRef]
- van Dijk, T.B.; Gillemans, N.; Stein, C.; Fanis, P.; Demmers, J.; van de Corput, M.; Essers, J.; Grosveld, F.; Bauer, U.M.; Philipsen, S. Friend of Prmt1, a novel chromatin target of protein arginine methyltransferases. Mol. Cell Biol. 2010, 30, 260–272. [Google Scholar] [CrossRef]
- Takai, H.; Masuda, K.; Sato, T.; Sakaguchi, Y.; Suzuki, T.; Suzuki, T.; Koyama-Nasu, R.; Nasu-Nishimura, Y.; Katou, Y.; Ogawa, H.; et al. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014, 9, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Ng, H.H.; Zhang, Y.; Hendrich, B.; Johnson, C.A.; Turner, B.M.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D.; Bird, A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999, 23, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; Sugimoto, K.; Fukushima, T.; Shinkai, Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001, 276, 25309–25317. [Google Scholar] [CrossRef] [PubMed]
- Rippe, K. Liquid-Liquid Phase Separation in Chromatin. Cold Spring Harb. Perspect. Biol. 2022, 14. [Google Scholar] [CrossRef]
- Greig, J.A.; Nguyen, T.A.; Lee, M.; Holehouse, A.S.; Posey, A.E.; Pappu, R.V.; Jedd, G. Arginine-Enriched Mixed-Charge Domains Provide Cohesion for Nuclear Speckle Condensation. Mol. Cell 2020, 77, 1237–1250.e1234. [Google Scholar] [CrossRef]
- Swiercz, R.; Person, M.D.; Bedford, M.T. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3). Biochem. J. 2005, 386, 85–91. [Google Scholar] [CrossRef]
- Swiercz, R.; Cheng, D.; Kim, D.; Bedford, M.T. Ribosomal protein rpS2 is hypomethylated in PRMT3-deficient mice. J. Biol. Chem. 2007, 282, 16917–16923. [Google Scholar] [CrossRef]
| Species | GAR/Full-Length | G/R Ratio | Non-GR Amino Acids | G% 1 | R% 1 | non-GR% 1 |
|---|---|---|---|---|---|---|
| S. cerevisiae | 76/327 | 42/17 2.47 | 3F, 8S, 6A | 55.2 | 22.3 | 22.3 |
| C. elegans | 107/352 | 62/22 2.82 | 7F, 5S, 5D, 3P, H, A, M | 57.9 | 20.6 | 21.5 |
| D. melanogaster | 92/344 | 71/15 4.73 | 4F, P, A | 77.2 | 16.3 | 6.5 |
| D. rerio | 72/317 | 43/16 2.68 | 6F, 2P, D, K, S, E, T | 59.7 | 22.2 | 18.1 |
| H. sapiens | 72/321 | 47/15 3.29 | 5F, D, H, S, N, K | 65.3 | 20.8 | 13.9 |
| M. musculus | 78/327 | 51/16 3.19 | 6F, Q, S, N, K | 65.4 | 20.5 | 12.8 |
| X. laevis | 76/325 | 43/17 2.53 | 7F, 4D, 2P, Y, K, S | 56.6 | 22.4 | 21.1 |
| A. carolinensis | 66/311 | 36/17 2.11 | 5F, 3P, 2D, 2S, N | 54.5 | 25.8 | 19.7 |
| Accession Number | Pattern | Position | RG | RGG | Else | Total% | G/R | G% | R% | Non- GR% |
|---|---|---|---|---|---|---|---|---|---|---|
| FBL | ||||||||||
| >NP_998167. [Danio rerio] | PRGGGGRGGFGGRGRGGGDRGGRGGFRGGRGG | 7..38 | 1 | 7 | ‘P’: 1, ‘F’: 2, ‘D’: 1 | 10.1 | 2.5 | 62.5 | 25.0 | 12.5 |
| GGFRGRGGGRGTPRGRGGGRGGGRGGFRGG | 50..79 | 3 | 5 | ‘F’: 2, ‘T’: 1, ‘P’: 1 | 9.5 | 2.3 | 60.0 | 26.7 | 13.3 | |
| >NP_989101.1 [Xenopus tropicalis] | PRGGRGGYGDRGGFGDRGGGRGRGGFRGRGGGGDRGGFGGRGGFGGRGGFGDRGGFRGGFKSPGRGGPRGGRGGRGG | 7..83 | 2 | 15 | ‘P’: 3, ‘Y’: 1, ‘D’: 4, ‘F’: 7, ‘K’: 1, ‘S’: 1 | 23.7 | 2.5 | 55.8 | 22.1 | 22.1 |
| >XP_003224982.1 [Anolis carolinensis] | PRGGRGDRGGRGGFGDRGRGGFRGGRGGGFNSPGRGGGPFRGGRGGSRGRGGPRGGGRGGRGGFRGG | 7..73 | 3 | 14 | ‘P’: 4, ‘D’: 2, ‘F’: 5, ‘N’: 1, ‘S’: 2 | 21.5 | 2.1 | 53.7 | 25.4 | 20.9 |
| >NP_032017.2 [Mus musculus] | PRGGGFGGRGGFGDRGGRGGGRGGRGGFGGGRGGFGGGGRGRGGGGGGFRGRGGGGGRGGGFQSGGNRGRGGGRGGKRG | 7..85 | 4 | 12 | ‘P’: 1, ‘F’: 6, ‘D’: 1, ‘Q’: 1, ‘S’: 1, ‘N’: 1, ‘K’: 1 | 24.2 | 3.2 | 64.6 | 20.3 | 15.2 |
| >NP_001427.2 [Homo sapiens] | PRGGGFGGRGGFGDRGGRGGRGGFGGGRGRGGGFRGRGRGGGGGGGGGGGGGRGGGGFHSGGNRGRGRGGKRG | 7..79 | 6 | 9 | ‘P’: 1, ‘F’: 5, ‘D’: 1, ‘H’: 1, ‘S’: 1, ‘N’: 1, ‘K’: 1 | 22.7 | 3.1 | 64.4 | 20.6 | 15.1 |
| NCL | ||||||||||
| >NP_001070120.2 [Danio rerio] | ERGGGGRG | 541..548 | 1 | 1 | ‘E’: 1 | 1.1 | 2.5 | 62.5 | 25.0 | 12.5 |
| GGRGGFGGGRGGFGGRGGGRGGFGGRGGGGRGGGFRGGRGGRGGGGGFRGGRGGGGRGG | 633..691 | 0 | 12 | ‘F’: 5 | 8.4 | 3.5 | 71.2 | 20.3 | 8.5 | |
| >XP_031758857.1 [Xenopus tropicalis] | QRGGRGGFGRGGGFRGGRGGRGGGGGRGGFGGRGGGRGRGGFGGRGGGGFRGG | 641..693 | 1 | 11 | ‘Q’: 1, ‘F’: 5 | 7.5 | 2.9 | 66.4 | 22.6 | 11.3 |
| >XP_003225545.1 [Anolis carolinensis] | GQRGGGGGGFGRGGRGGGGRGGGRGGFGRGGGRGFGGRGGGFRGGRGG | 634..681 | 1 | 9 | ‘Q’: 1, ‘F’: 4 | 6.9 | 3.3 | 68.8 | 20.8 | 10.4 |
| >NP_035010.3 [Mus musculus] | GGRGGGRGGFGGRGGGRGGRGGFGGRGRGGFGGRGGFRGGRGG | 651..693 | 1 | 9 | ‘F’: 4 | 6.1 | 2.9 | 67.4 | 23.3 | 9.3 |
| >NP_005372.2 [Homo sapiens] | GGRGGGRGGFGGRGGGRGGRGGFGGRGRGGFGGRGGFRGGRGG | 654..696 | 1 | 9 | ‘F’: 4 | 6.1 | 2.9 | 67.4 | 23.3 | 9.3 |
| GAR1 | ||||||||||
| >NP_957269.2 [Danio rerio] | FRGGGGGRGGGFNRGGGGGRGGGFGGGRGGGFGGGRGGGFGGGRGGRGG | 3..51 | 0 | 8 | ‘F’: 5, ‘N’: 1 | 21.8 | 4.4 | 71.4 | 16.3 | 12.2 |
| PRGGRGGGGRGGRGGGFRGGRGANGGGRGGFGGRGGGFGGRGGGGGGFRGGRGGGGGRGFRGG | 162..224 | 2 | 11 | ‘P’: 1, ‘F’: 5, ‘A’: 1, ‘N’: 1 | 28.0 | 3.2 | 66.7 | 20.6 | 12.7 | |
| >NP_001011252.1 [Xenopus tropicalis] | FRGRGGFNRGGGGGRGGGGFGGRGGGRGGYGQGGGRGGFGRGGGRGGFNRGG | 3..54 | 1 | 9 | ‘F’: 5, ‘N’: 2, ‘Y’: 1, ‘Q’: 1 | 23.9 | 3.3 | 63.5 | 19.2 | 17.3 |
| PRGGGRGGGRGGGRGRGGGRGGGGGFRGGRGGGFGGGGGFRGSRGGGFRGGRGFRGG | 161..217 | 3 | 10 | ‘P’: 1, ‘F’: 5, ‘S’: 1 | 26.2 | 2.9 | 64.9 | 22.8 | 12.3 | |
| >XP_008110322.1 [Anolis carolinensis] | FRGRGGGNRGGGFNRGGGFNRGGGGFNRGGFSRGGGRGGFGRGGGRGGFNRGG | 3..55 | 1 | 10 | ‘F’: 7, ‘N’: 5, ‘S’: 1 | 24.9 | 2.6 | 54.7 | 20.8 | 24.5 |
| PRGGRGGRGGRGGGRGGGGRGGGGFRGGRGGGGFRGGRGGGGGGRGFRGRG | 162..212 | 3 | 10 | ‘P’: 1, ‘F’: 3 | 23.9 | 2.6 | 66.7 | 25.5 | 7.8 | |
| >NP_080854.1 [Mus musculus] | FRGGGRGGFNRGGGGGGFNRGGGSNNHFRGGGGGGGGSFRGGGGGGGGSFRGGGRGGFGRGGGRGG | 3..68 | 0 | 10 | ‘F’: 7, ‘N’: 4, ‘S’: 3, ‘H’: 1 | 28.6 | 4.1 | 62.1 | 15.2 | 22.7 |
| PRGGGGGGRGGRGGGRGGGGRGGGRGGGFRGGRGGGGGFRGGRGGGGFRGRG | 179..230 | 2 | 10 | ‘P’: 1, ‘F’: 3 | 22.5 | 3.0 | 69.2 | 23.1 | 7.7 | |
| >NP_061856.1 [Homo sapiens] | FRGGGRGGFNRGGGGGGFNRGGSSNHFRGGGGGGGGGNFRGGGRGGFGRGGGRGG | 3..57 | 0 | 9 | ‘F’: 6, ‘N’: 4, ‘S’: 2, ‘H’: 1 | 25.4 | 3.7 | 60.0 | 16.4 | 23.6 |
| PRGGGRGGRGGGRGGGGRGGGRGGGFRGGRGGGGGGFRGGRGGGFRGRG | 168..216 | 2 | 10 | ‘P’: 1, ‘F’: 3 | 22.6 | 2.8 | 67.4 | 24.5 | 8.2 | |
| Accession Number/Protein | Pattern | Position | RG | RGG | Else | Total% | G/R | G% | R% | Non-GR % |
|---|---|---|---|---|---|---|---|---|---|---|
| >NP_004951.1 FUS 1 | DRGGRGRGG | 212..220 | 1 | 2 | ‘D’: 1 | 1.7 | 1.7 | 55.6 | 33.3 | 11.1 |
| PRGRGGGRGGRGGMGGSDRGG | 241..261 | 1 | 4 | ‘P’: 1, ‘M’: 1, ‘S’: 1, ‘D’: 1 | 4.0 | 2.4 | 57.1 | 23.8 | 19.1 | |
| NRGGGNGRGGRGRGGPMGRGG | 376..396 | 1 | 4 | ‘N’: 2, ‘P’: 1, ‘M’: 1 | 4.0 | 2.4 | 57.1 | 23.8 | 19.1 | |
| RRGGRGGYDRGGYRGRGGDRGGFRGGRGGGDRGG | 472..505 | 1 | 8 | ‘Y’: 2, ‘D’: 3, ‘F’: 1 | 6.5 | 1.8 | 52.9 | 29.4 | 17.7 | |
| >sp|Q01844.1 EWS | NRGRGRGGFDRGGMSRGGRGGGRGGMGSAGERGG | 299..332 | 2 | 6 | ‘N’: 1, ‘F’: 1, ‘D’: 1, ‘M’: 2, ‘S’: 2, ‘A’: 1, ‘E’: 1 | 5.2 | 2.1 | 50.0 | 23.5 | 26.5 |
| MRGGLPPREGRGMPPPLRGG | 454..473 | 1 | 2 | ‘M’: 2, ‘L’: 2, ‘P’: 5, ‘E’: 1 | 3.1 | 1.5 | 30.0 | 20.0 | 50.0 | |
| GGRGGDRGGFPPRGPRGSRG | 488..507 | 3 | 2 | ‘D’: 1, ‘F’: 1, ‘P’: 3, ‘S’: 1 | 3.1 | 1.8 | 45.0 | 25.0 | 30.0 | |
| GGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGG | 560..617 | 2 | 10 | ‘D’: 4, ‘P’: 2, ‘M’: 4, ‘L’: 1, ‘F’: 3 | 8.8 | 2.4 | 53.5 | 22.4 | 24.1 | |
| GGRRGGRGG | 630..638 | 0 | 2 | 1.4 | 2.0 | 66.7 | 33.3 | 0.0 | ||
| >NP_631961.1 TAF15 | NRGYGGSQGGGRGRGGYDKDGRG | 174..196 | 3 | 1 | ‘N’: 1, ‘Y’: 2, ‘S’: 1, ‘Q’: 1, ‘D’: 2, ‘K’: 1 | 3.9 | 2.8 | 47.8 | 17.4 | 34.8 |
| MRGGGSGGGRRGRGGYRGRGGFQGRGG | 325..351 | 2 | 4 | ‘M’: 1, ‘S’: 1, ‘Y’: 1, ‘F’: 1, ‘Q’: 1 | 4.6 | 2.1 | 55.6 | 25.9 | 18.5 | |
| FRGRGYGGERGYRGRGGRGGDRGG | 394..417 | 4 | 3 | ‘F’: 1, ‘Y’: 2, ‘E’: 1, ‘D’: 1 | 4.1 | 1.7 | 50.0 | 29.2 | 20.8 | |
| GGDRGGGYGGDRGGGYGGDRGGGYGGDRGGYGGDRGGGYGGDRGGYGGDRGGYGGDRGGYGGDRGGYGGDRSRGGYGGDRGG | 456..537 | 0 | 11 | ‘D’: 11, ‘Y’: 10, ‘S’: 1 | 13.9 | 4.0 | 58.5 | 14.6 | 26.8 | |
| GGDRGGGYGGDRGG | 559..572 | 0 | 2 | ‘D’: 2, ‘Y’: 1 | 2.4 | 4.5 | 64.3 | 14.3 | 21.4 | |
| >sp|Q8ND56.3 Lsm14a (RAP55A) | RRGRGGHRGGRG | 269..280 | 2 | 2 | ‘H’: 1 | 2.6 | 1.2 | 50.0 | 41.7 | 8.3 |
| NRGRGGYRGRGGLGFRGGRGRGGGRGGTFTAPRGFRGGFRGGRGG | 403..447 | 4 | 8 | ‘N’: 1, ‘Y’: 1, ‘L’: 1, ‘F’: 4, ‘T’: 2, ‘A’: 1, ‘P’: 1 | 9.7 | 1.8 | 48.9 | 26.7 | 24.4 | |
| >NP_001018077.1 SERBP1 | IRGRGGLGRGRGGRGRGMGRGDGFDSRG | 162..189 | 6 | 2 | ‘I’: 1, ‘L’: 1, ‘M’: 1, ‘D’: 2, ‘F’: 1, ‘S’: 1 | 6.9 | 1.6 | 46.4 | 28.6 | 25.0 |
| GRGGRGGRGGRGRGGRPNRG | 366..385 | 2 | 4 | ‘P’: 1, ‘N’: 1 | 4.9 | 1.6 | 55.0 | 35.0 | 10.0 | |
| >NP_112420.1 hnRNPA1 | DRGSGKKRG | 139..147 | 2 | 0 | ‘D’: 1, ‘S’: 1, ‘K’: 2 | 2.4 | 1.5 | 33.3 | 22.2 | 44.4 |
| GRGGNFSGRGGFGGSRGG | 217..234 | 0 | 3 | ‘N’: 1, ‘F’: 2, ‘S’: 2 | 4.8 | 3.3 | 55.6 | 16.7 | 27.8 | |
| >NP_114032.2 hnRNPU | NRGGGHRGRGGFNMRGGNFRGGAPGNRGGYNRRGNMPQRGG | 701..741 | 2 | 6 | ‘N’: 6, ‘H’: 1, ‘F’: 2, ‘M’: 2, ‘A’: 1, ‘P’: 2, ‘Y’: 1, ‘Q’: 1 | 5.0 | 1.8 | 39.0 | 22.0 | 39.0 |
| GRGSYSNRGNYNRGGMPNRGNYNQNFRGRGNNRG | 761..794 | 6 | 1 | ‘S’: 2, ‘Y’: 3, ‘N’: 9, ‘M’: 1, ‘P’: 1, ‘Q’: 1, ‘F’: 1 | 4.1 | 1.3 | 26.5 | 20.6 | 52.9 | |
| >NP_077726.1 DDX4 | NRGFSKRGG | 124..132 | 1 | 1 | ‘N’: 1, ‘F’: 1, ‘S’: 1, ‘K’: 1 | 1.2 | 1.5 | 33.3 | 22.2 | 44.4 |
| RRGGRGSFRGCRGG | 146..159 | 2 | 2 | ‘S’: 1, ‘F’: 1, ‘C’: 1 | 1.9 | 1.2 | 42.9 | 35.7 | 21.4 | |
| >NP_005745.1 G3BP1 | LRGPGGPRGGLGGGMRGPPRGG | 428..449 | 2 | 2 | ‘L’: 2, ‘P’: 4, ‘M’: 1 | 4.7 | 2.8 | 50.0 | 18.2 | 31.8 |
| >NP_002015.1 FMRP | GRGSRPYRNRGHGRRG | 470..485 | 3 | 0 | ‘S’: 1, ‘P’: 1, ‘Y’: 1, ‘N’: 1, ‘H’: 1 | 2.5 | 0.8 | 31.3 | 37.5 | 31.3 |
| RRGDGRRRGGGGRGQGGRGRGG | 527..548 | 3 | 2 | ‘D’: 1, ‘Q’: 1 | 3.5 | 1.5 | 54.6 | 36.4 | 9.1 | |
| >NP_005078.2 FXR1 | GRGRGRRG | 385..392 | 3 | 0 | 1.3 | 1.0 | 50.0 | 50.0 | 0.0 | |
| GGRGRSVSGGRGRGGPRGG | 443..461 | 2 | 2 | ‘S’: 2, ‘V’: 1, ‘P’: 1 | 3.1 | 2.0 | 52.6 | 26.3 | 21.1 | |
| >NP_004851.2 FXR2 | GGRGRG | 430..435 | 2 | 0 | 0.9 | 2.0 | 66.7 | 33.3 | 0.0 | |
| GGRGRG | 486..491 | 2 | 0 | 0.9 | 2.0 | 66.7 | 33.3 | 0.0 | ||
| >NP_006550.1 KHDR1 (SAM68) | SRGGGGGSRGG | 44..54 | 0 | 2 | ‘S’: 2 | 2.5 | 3.5 | 63.6 | 18.2 | 18.2 |
| SRGRGVPVRGRG | 281..292 | 4 | 0 | ‘S’: 1, ‘V’: 2, ‘P’: 1 | 2.7 | 1.0 | 33.3 | 33.3 | 33.3 | |
| PRGRGVGPPRGALVRGTPVRGAITRGATVTRG | 301..332 | 7 | 0 | ‘P’: 4, ‘V’: 4, ‘A’: 3, ‘L’: 1, ‘T’: 4, ‘I’: 1 | 7.2 | 1.1 | 25.0 | 21.9 | 53.1 | |
| >NP_005889.3 Caprin-1 | SRGVSRGGSRGARGLMNGYRGPANGFRGG | 607..635 | 4 | 2 | ‘S’: 3, ‘V’: 1, ‘A’: 2, ‘L’: 1, ‘M’: 1, ‘N’: 2, ‘Y’: 1, ‘P’: 1, ‘F’: 1 | 4.1 | 1.7 | 34.5 | 20.7 | 44.8 |
| KRGSGQSGPRGAPRGRGGPPRPNRG | 675..699 | 4 | 1 | ‘K’: 1, ‘S’: 2, ‘Q’: 1, ‘P’: 5, ‘A’: 1, ‘N’: 1 | 3.5 | 1.3 | 32.0 | 24.0 | 44.0 | |
| >NP_056422.2 CHTOP | ARGAIGGRGLPIIQRGLPRGGLRGG | 96..120 | 3 | 2 | ‘A’: 2, ‘I’: 3, ‘L’: 3, ‘P’: 2, ‘Q’: 1 | 10.1 | 1.8 | 36.0 | 20.0 | 44.0 |
| LRGGMSLRGQNLLRGG | 127..142 | 1 | 2 | ‘L’: 4, ‘M’: 1, ‘S’: 1, ‘Q’: 1, ‘N’: 1 | 6.5 | 1.7 | 31.3 | 18.8 | 50.0 | |
| RRGGVRGRGGPGRGGLGRGAMGRGGIGGRGRGMIGRGRGGFGGRGRGRGRGRG | 152..204 | 10 | 5 | ‘V’: 1, ‘P’: 1, ‘L’: 1, ‘A’: 1, ‘M’: 2, ‘I’: 2, ‘F’: 1 | 21.4 | 1.8 | 52.8 | 30.2 | 17.0 | |
| >NP_055542.1 kmt2b (MML4) | ARGRFPGRPRGAGGGGGRGGRG | 16..37 | 3 | 1 | ‘A’: 2, ‘F’: 1, ‘P’: 2 | 0.8 | 1.8 | 50.0 | 27.3 | 22.7 |
| QRGRGRGRGRGWGPSRG | 90..106 | 6 | 0 | ‘Q’: 1, ‘W’: 1, ‘P’: 1, ‘S’: 1 | 0.6 | 1.2 | 41.2 | 35.3 | 23.5 | |
| QRGRAPRGRG | 144..153 | 3 | 0 | ‘Q’: 1, ‘A’: 1, ‘P’: 1 | 0.4 | 0.8 | 30.0 | 40.0 | 30.0 | |
| RRGGQSSRGGRGGRGRGRGG | 280..299 | 2 | 4 | ‘Q’: 1, ‘S’: 2 | 0.7 | 1.4 | 50.0 | 35.0 | 15.0 |
| Accession Number/Name 1 | Pattern | Position | RG | RGG | Else | % | G/R | G% | R% | Non-GR % | Isoforms |
|---|---|---|---|---|---|---|---|---|---|---|---|
| >NP_055983.1/ ZC3H4 | SRGRGSRGRGRGYRGRGSRGGSRGRGMGRGSRGRGRG | 235..271 | 13 | 1 | ‘S’: 5, ‘Y’: 1, ‘M’: 1 | 2.8 | 1.1 | 43.2 | 37.8 | 18.9 | 11 |
| SRGRGLSRGRGRGSRGRGKGMGRGRGRGGSRGG | 319..351 | 9 | 2 | ‘S’: 4, ‘L’: 1, ‘K’: 1, ‘M’: 1 | 2.5 | 1.4 | 45.5 | 33.3 | 21.2 | 11 | |
| >NP_003918.1/ MBD2 | GARGGGRGRGRWKQAGRGGGVCGRGRGRGRGRGRGRGRGRGRG | 53..95 | 12 | 2 | ‘A’: 2, ‘W’: 1, ‘K’: 1, ‘Q’: 1, ‘V’: 1, ‘C’: 1 | 10.5 | 1.4 | 48.8 | 34.9 | 16.3 | 2 |
| >NP_001095868.1/ hnRNP R | VRGRGGGRGGRGAPPPPRGRGAPPPRGRAGYSQRGAPLGPPRGSRGGRGGPAQQQRGRGSRGSRGNRGG | 502..570 | 11 | 5 | ‘V’: 1, ‘A’: 5, ‘P’: 11, ‘Y’: 1, ‘S’: 4, ‘Q’: 4, ‘L’: 1, ‘N’: 1 | 10.8 | 1.4 | 34.8 | 24.6 | 40.6 | 17 |
| >NP_006363.4 /hnRNP Q isoform 1 | GARGRGGRGARGAAPSRGRGAAPPRGRAGYSQRGGPGSARGVRGARGGAQQQRGRGVRGARGGRGG | 494..559 | 11 | 5 | ‘A’: 11, ‘P’: 4, ‘S’: 3, ‘Y’: 1, ‘Q’: 4, ‘V’: 2 | 10.6 | 1.4 | 36.4 | 25.8 | 37.9 | 6 |
| >NP_001153145.1/ hnRNP Q isoform 2 | GARGRGGRGARGAAPSRGRGAAPPRGRAGYSQRGGPGSARGVRGARGGAQQQRGRG | 396..451 | 10 | 3 | ‘A’: 10, ‘P’: 4, ‘S’: 3, ‘Y’: 1, ‘Q’: 4, ‘V’: 1 | 12.1 | 1.4 | 33.9 | 25.0 | 41.1 | 5 |
| >NP_001153148.1/ hnRNP Q isoform 5 | GARGRGGRGARGAAPSRGRGAAPPRGRAGYSQRGGPGSARGVRGARGGAQQQRGRGG | 494..550 | 9 | 4 | ‘A’: 10, ‘P’: 4, ‘S’: 3, ‘Y’: 1, ‘Q’: 4, ‘V’: 1 | 10.2 | 1.4 | 35.1 | 24.6 | 40.4 | 4 |
| >NP_003741.1/ eIF-3A | DRGPRRGLDDDRGPRRGMDDDRGPRRGMDDDRGPRRGMDDDRGPRRGLDDDRG | 1066..1118 | 11 | 0 | ‘D’: 16, ‘P’: 5, ‘L’: 2, ‘M’: 3 | 3.8 | 0.7 | 20.8 | 30.2 | 49.1 | 1 |
| >NP_001296171.1/myosin XVB | GRGHGRGSKGRGRGKADEGRGHERGDEGRGRGKADEGRGHERGYEGRG | 414..461 | 11 | 0 | ‘H’: 3, ‘S’: 1, ‘K’: 3, ‘A’: 2, ‘D’: 3, ‘E’: 6, ‘Y’: 1 | 1.6 | 1.6 | 37.5 | 22.9 | 39.6 | 1 |
| >NP_056422.2/ CHTOP | RRGGVRGRGGPGRGGLGRGAMGRGGIGGRGRGMIGRGRGGFGGRGRGRGRGRG | 152..204 | 10 | 5 | ‘V’: 1, ‘P’: 1, ‘L’: 1, ‘A’: 1, ‘M’: 2, ‘I’: 2, ‘F’: 1 | 21.4 | 1.8 | 52.8 | 30.2 | 17.0 | 2 |
| >NP_001273560.1/RBM26 | KRGILSSGRGRGIHSRGRGAVHGRGRGRGRGRG | 849..881 | 10 | 0 | ‘K’: 1, ‘I’: 2, ‘L’: 1, ‘S’: 3, ‘H’: 2, ‘A’: 1, ‘V’: 1 | 3.3 | 1.2 | 36.4 | 30.3 | 33.3 | 40 |
| >NP_694984.5/ BRWD3 | SRGGRGRGGRGRGSRGRGGGGTRGRGRGRGGRGASRG | 1683..1719 | 9 | 4 | ‘S’: 3, ‘T’: 1, ‘A’: 1 | 2.1 | 1.5 | 51.4 | 35.1 | 13.5 | 3 |
| >NP_001276342.1/EHMT2 | MRGLPRGRGLMRARGRGRAAPPGSRGRGRGGPHRGRG | 1..37 | 9 | 1 | ‘M’: 2, ‘L’: 2, ‘P’: 4, ‘A’: 3, ‘S’: 1, ‘H’: 1 | 3.0 | 1.0 | 32.4 | 32.4 | 35.1 | 4 |
| >NP_689813.2/ ZNF579 | HRGRGRGRGRGRGRGRGRGRGG | 15..36 | 9 | 1 | ‘H’: 1 | 3.9 | 1.1 | 50.0 | 45.5 | 4.5 | 4 |
| >NP_002943.2/ RPS2 | GNRGGFRGGFGSGIRGRGRGRGRGRGRGRGARGG | 20..53 | 8 | 3 | ‘N’: 1, ‘F’: 2, ‘S’: 1, ‘I’: 1, ‘A’: 1 | 11.6 | 1.5 | 50.0 | 32.4 | 17.6 | 1 |
| >NP_003478.1/ TAF15 | GGDRGGGYGGDRGGGYGGDRGGGYGGDRGGYGGDRGGGYGGDRGGYGGDRGGYGGDRGGYGGDRGGYGGDRSRGGYGGDRGG | 453..534 | 0 | 11 | ‘D’: 11, ‘Y’: 10, ‘S’: 1 | 13.9 | 4.0 | 58.5 | 14.6 | 26.8 | 3 |
| >NP_053733.2/ EWS | GGDRGRGGPGGMRGGRGGLMDRGGPGGMFRGGRGGDRGGFRGGRGMDRGGFGGGRRGG | 565..622 | 2 | 10 | ‘D’: 4, ‘P’: 2, ‘M’: 4, ‘L’: 1, ‘F’: 3 | 8.8 | 2.4 | 53.4 | 22.4 | 24.1 | 28 |
| >NP_061856.1/ GAR1 | PRGGGRGGRGGGRGGGGRGGGRGGGFRGGRGGGGGGFRGGRGGGFRGRG | 168..216 | 2 | 10 | ‘P’: 1, ‘F’: 3 | 22.6 | 2.8 | 67.3 | 24.5 | 8.2 | 2 |
| >NP_001427.2/ fibrillarin | PRGGGFGGRGGFGDRGGRGGRGGFGGGRGRGGGFRGRGRGGGGGGGGGGGGGRGGGGFHSGGNRGRGRGGKRG | 7..79 | 6 | 9 | ‘P’: 1, ‘F’: 5, ‘D’: 1, ‘H’: 1, ‘S’: 1, ‘N’: 1, ‘K’: 1 | 22.7 | 3.1 | 64.4 | 20.5 | 15.1 | 2 |
| >NP_005372.2/ nucleolin | GGRGGGRGGFGGRGGGRGGRGGFGGRGRGGFGGRGGFRGGRGG | 654..696 | 1 | 9 | ‘F’: 4 | 6.1 | 2.9 | 67.4 | 23.3 | 9.3 | 1 |
| >NP_001107565.1/LSM14 | NRGRGGYRGRGGLGFRGGRGRGGGRGGTFTAPRGFRGGFRGGRGG | 403..447 | 4 | 8 | ‘N’: 1, ‘Y’: 1, ‘L’: 1, ‘F’: 4, ‘T’: 2, ‘A’: 1, ‘P’: 1 | 9.7 | 1.8 | 48.9 | 26.7 | 24.4 | 24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-C.; Huang, S.-H.; Chang, C.-P.; Li, C. Identification and Characterization of Glycine- and Arginine-Rich Motifs in Proteins by a Novel GAR Motif Finder Program. Genes 2023, 14, 330. https://doi.org/10.3390/genes14020330
Wang Y-C, Huang S-H, Chang C-P, Li C. Identification and Characterization of Glycine- and Arginine-Rich Motifs in Proteins by a Novel GAR Motif Finder Program. Genes. 2023; 14(2):330. https://doi.org/10.3390/genes14020330
Chicago/Turabian StyleWang, Yi-Chun, Shang-Hsuan Huang, Chien-Ping Chang, and Chuan Li. 2023. "Identification and Characterization of Glycine- and Arginine-Rich Motifs in Proteins by a Novel GAR Motif Finder Program" Genes 14, no. 2: 330. https://doi.org/10.3390/genes14020330
APA StyleWang, Y.-C., Huang, S.-H., Chang, C.-P., & Li, C. (2023). Identification and Characterization of Glycine- and Arginine-Rich Motifs in Proteins by a Novel GAR Motif Finder Program. Genes, 14(2), 330. https://doi.org/10.3390/genes14020330





