EGFRvIII Promotes the Proneural–Mesenchymal Transition of Glioblastoma Multiforme and Reduces Its Sensitivity to Temozolomide by Regulating the NF-κB/ALDH1A3 Axis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Cell Culture
2.3. Cell Proliferation and Transwell Migration Assays
2.4. Real-Time Quantitative Reverse Transcription Polymerase Chain Reaction (RT-qPCR)
2.5. Western Blotting
2.6. Chromatin Immunoprecipitation (ChIP)
2.7. Immunohistochemistry and Immunofluorescence
2.8. In Vivo Xenograft Growth
2.9. Bioluminescence Imaging
2.10. Transcriptome Sequencing
2.11. Statistical Analysis
3. Results
3.1. EGFRvIII Affected the Gene Expression Profile of GBM Patients, and GBM Patients with EGFRvIII (+) Had Low Sensitivity to TMZ
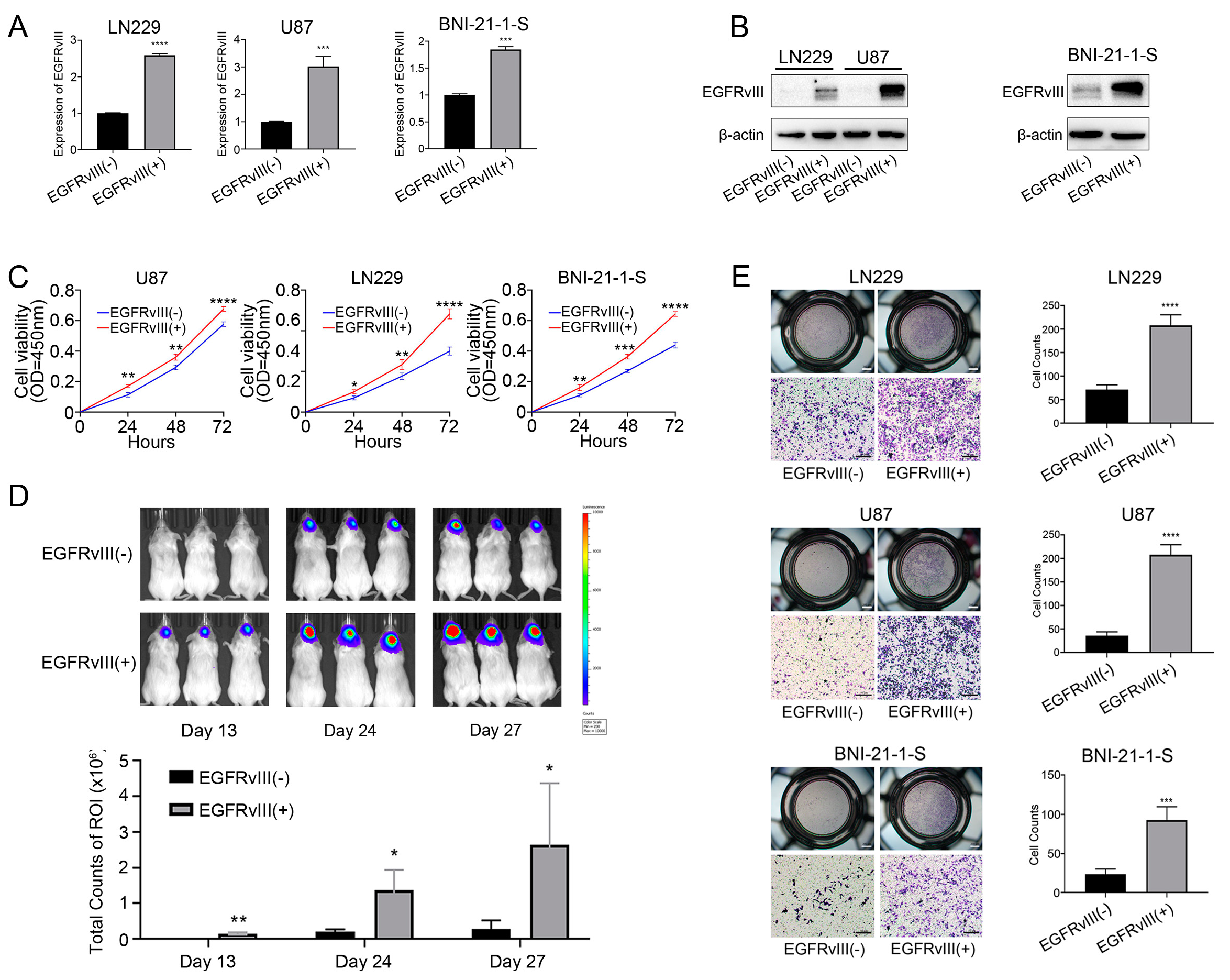
3.2. GBM Cells and Glioma Stem Cells with EGFRvIII (+) Significantly Increased Cell Viability and Cell Migration
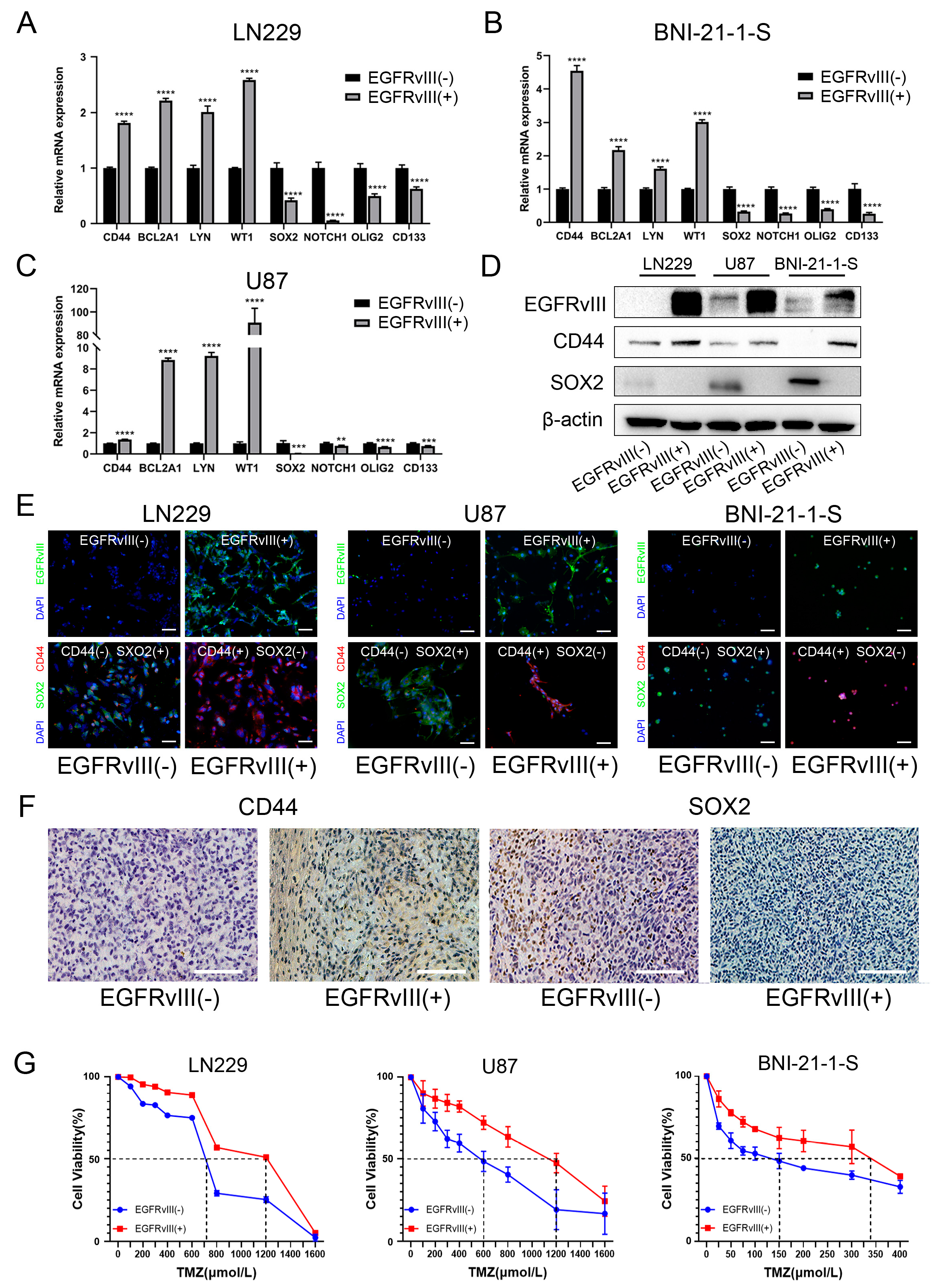
3.3. GBM Cells and Glioma Stem Cells with EGFRvIII (+) Significantly Decreased the Expression of PN GBM Markers, Increased the Expression of MES GBM Markers, and Increased the IC50 Value of TMZ
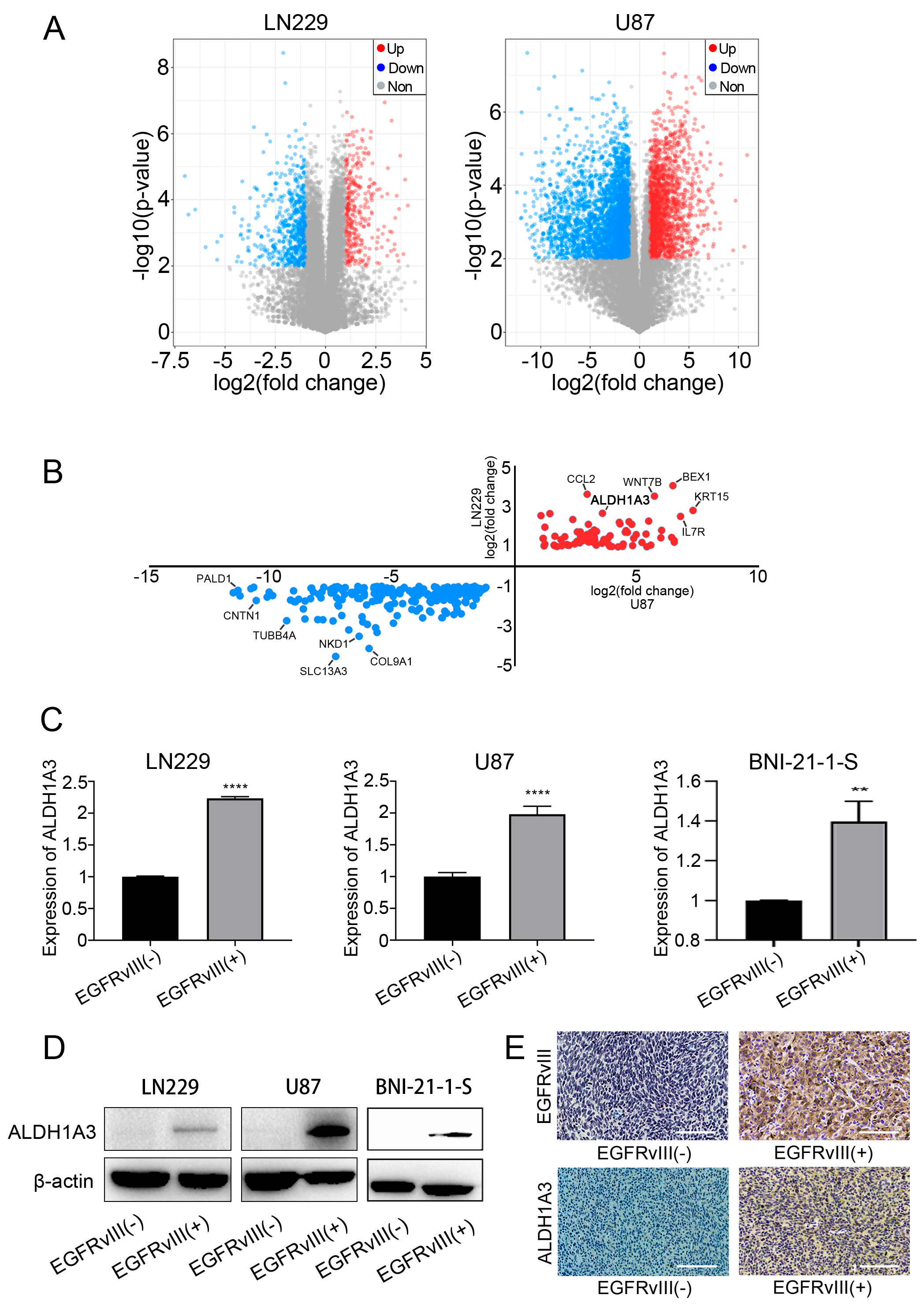
3.4. ALDH1A3 Was Screened and Identified as the Downstream Target Gene of EGFRvIII
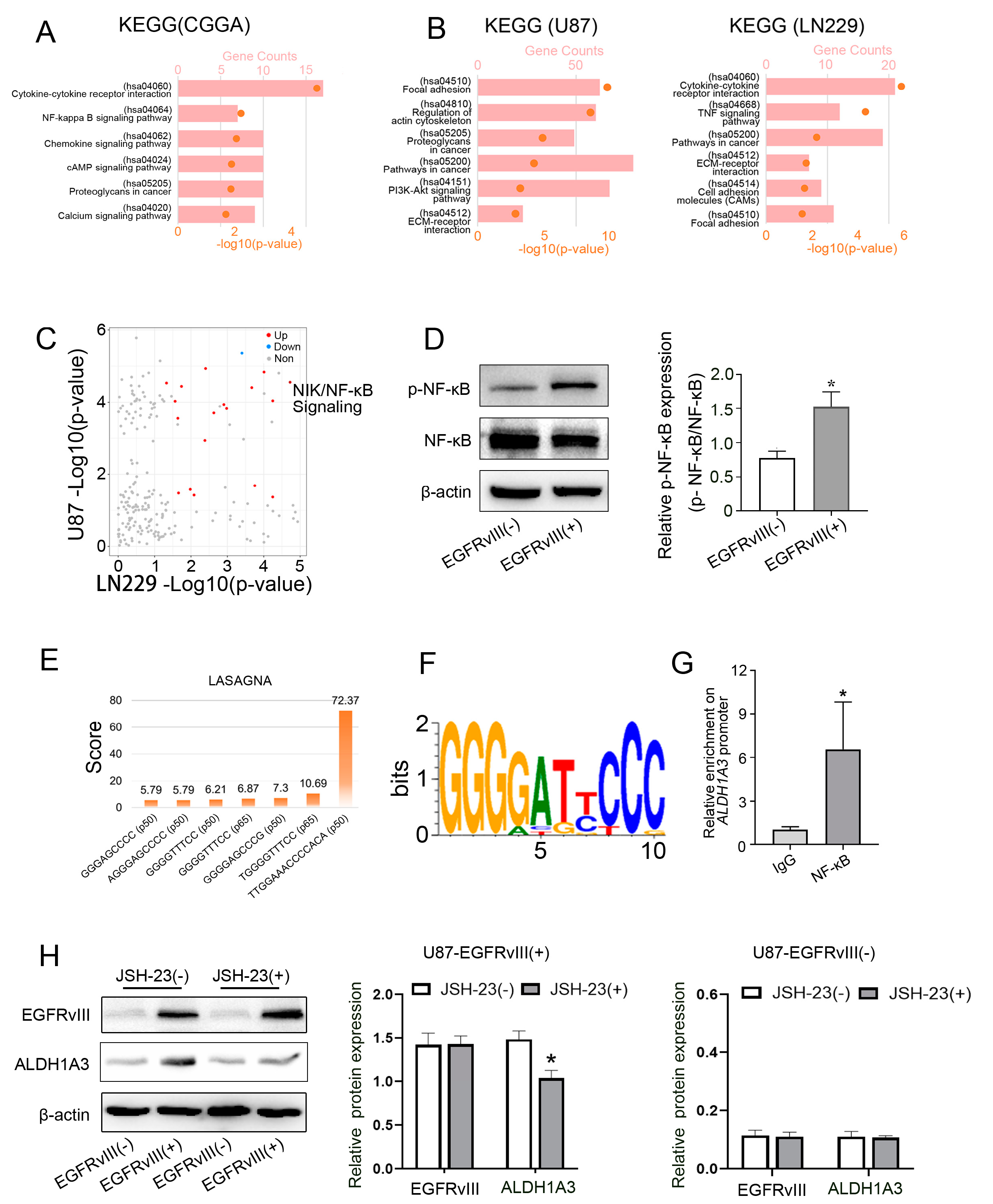
3.5. Screening and Determining That the Downstream Signaling Pathway of EGFRvIII Was Related to the NF-κB Pathway, p- NF-κB in EGFRvIII (+) Cells Was Increased, and NF-κB Bound to the ALDH1A3 Promoter Region
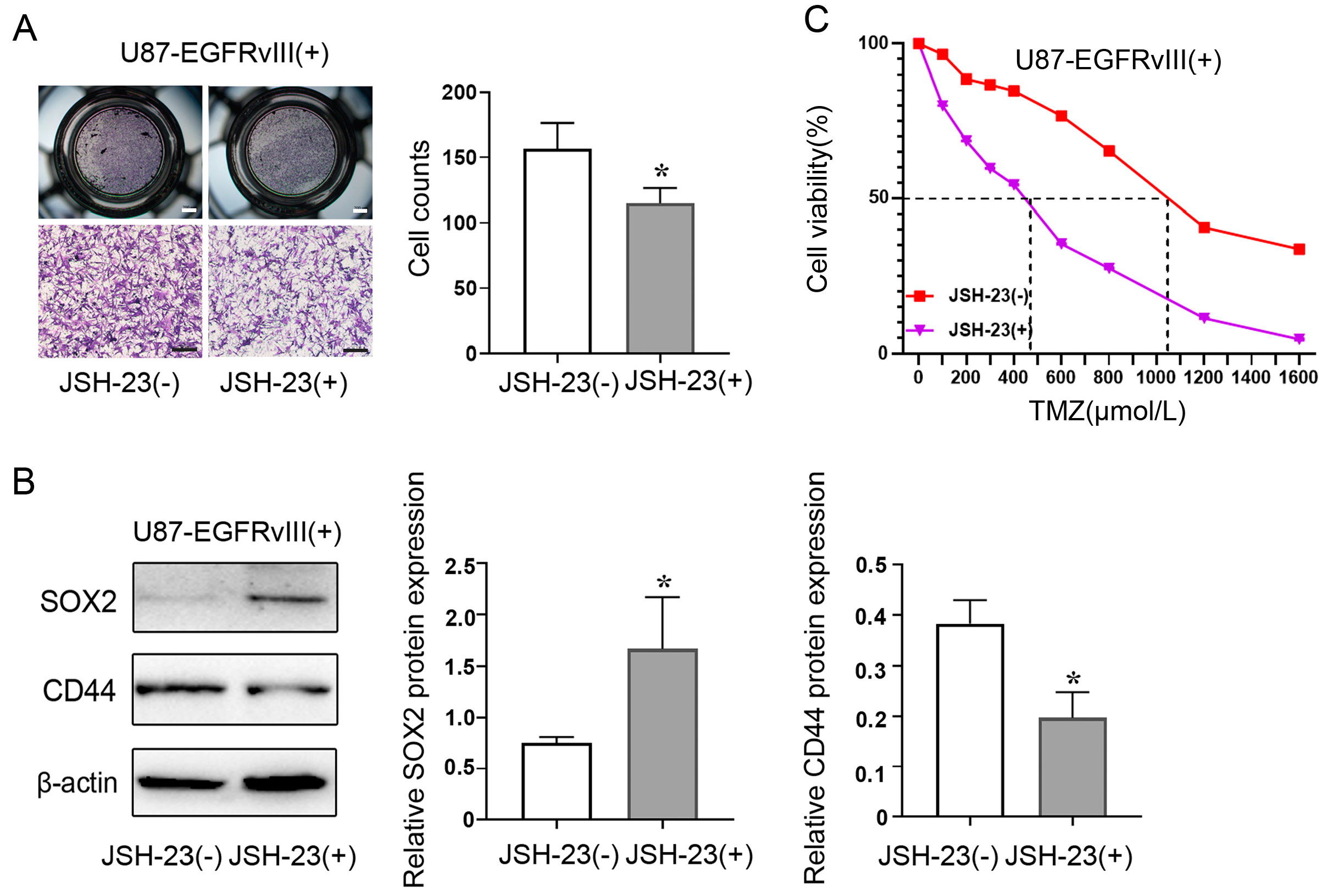
3.6. Inhibition of NF-κB Decreased ALDH1A3 Expression of EGFRvIII (+) Cells, Reversed the Enhanced Cell Migration Ability, Decreased the Expression of PN GBM Marker (SOX2), Increased the Expression of MES GBM Marker (CD44), and Increased the IC50 of TMZ Caused by EGFRvIII (+)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, T.; Nam, D.H.; Ram, Z.; Poon, W.S.; Wang, J.; Boldbaatar, D.; Mao, Y.; Ma, W.; Mao, Q.; You, Y.; et al. Clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2021, 499, 60–72. [Google Scholar] [CrossRef]
- Mooney, J.; Bernstock, J.D.; Ilyas, A.; Ibrahim, A.; Yamashita, D.; Markert, J.M.; Nakano, I. Current Approaches and Challenges in the Molecular Therapeutic Targeting of Glioblastoma. World Neurosurg. 2019, 129, 90–100. [Google Scholar] [CrossRef]
- Chi, A.S.; Cahill, D.P.; Reardon, D.A.; Wen, P.Y.; Mikkelsen, T.; Peereboom, D.M.; Wong, E.T.; Gerstner, E.R.; Dietrich, J.; Plotkin, S.R.; et al. Exploring Predictors of Response to Dacomitinib in EGFR-Amplified Recurrent Glioblastoma. JCO Precis. Oncol. 2020, 4, 593–613. [Google Scholar] [CrossRef]
- Mahinfar, P.; Baradaran, B.; Davoudian, S.; Vahidian, F.; Cho, W.C.; Mansoori, B. Long Non-Coding RNAs in Multidrug Resistance of Glioblastoma. Genes 2021, 12, 455. [Google Scholar] [CrossRef]
- Tomar, M.S.; Kumar, A.; Srivastava, C.; Shrivastava, A. Elucidating the mechanisms of Temozolomide resistance in gliomas and the strategies to overcome the resistance. Biochim. Biophys. Acta. Rev. Cancer 2021, 1876, 188616. [Google Scholar] [CrossRef]
- Vilar, J.B.; Christmann, M.; Tomicic, M.T. Alterations in Molecular Profiles Affecting Glioblastoma Resistance to Radiochemotherapy: Where Does the Good Go? Cancers 2022, 14, 2416. [Google Scholar] [CrossRef]
- Balca-Silva, J.; Matias, D.; Carmo, A.D.; Sarmento-Ribeiro, A.B.; Lopes, M.C.; Moura-Neto, V. Cellular and molecular mechanisms of glioblastoma malignancy: Implications in resistance and therapeutic strategies. Semin. Cancer Biol. 2019, 58, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Tomiyama, A.; Ichimura, K. Signal transduction pathways and resistance to targeted therapies in glioma. Semin. Cancer Biol. 2019, 58, 118–129. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hu, B.; Hu, X.; Kim, H.; Squatrito, M.; Scarpace, L.; deCarvalho, A.C.; Lyu, S.; Li, P.; Li, Y.; et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56.e46. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Cerchia, L.; Pegoraro, S.; Sgarra, R.; Manfioletti, G. Proneural–Mesenchymal Transition: Phenotypic Plasticity to Acquire Multitherapy Resistance in Glioblastoma. Int. J. Mol. Sci. 2019, 20, 2746. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, S.; Li, H.L.; Luo, H.; Wu, X.; Lu, J.; Wang, H.W.; Chen, Y.; Chen, D.; Wu, W.T.; et al. FOSL1 promotes proneural-to-mesenchymal transition of glioblastoma stem cells via UBC9/CYLD/NF-kappaB axis. Mol. Ther. J. Am. Soc. Gene Ther. 2022, 30, 2568–2583. [Google Scholar] [CrossRef]
- Kim, Y.; Varn, F.S.; Park, S.H.; Yoon, B.W.; Park, H.R.; Lee, C.; Verhaak, R.G.W.; Paek, S.H. Perspective of mesenchymal transformation in glioblastoma. Acta Neuropathol. Commun. 2021, 9, 50. [Google Scholar] [CrossRef]
- Kondo, T. Glioblastoma-initiating cell heterogeneity generated by the cell-of-origin, genetic/epigenetic mutation and microenvironment. Semin. Cancer Biol. 2022, 82, 176–183. [Google Scholar] [CrossRef]
- Sharifzad, F.; Ghavami, S.; Verdi, J.; Mardpour, S.; Mollapour Sisakht, M.; Azizi, Z.; Taghikhani, A.; Los, M.J.; Fakharian, E.; Ebrahimi, M.; et al. Glioblastoma cancer stem cell biology: Potential theranostic targets. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer. Chemother. 2019, 42, 35–45. [Google Scholar] [CrossRef]
- Huang, K.; Liu, X.; Li, Y.; Wang, Q.; Zhou, J.; Wang, Y.; Dong, F.; Yang, C.; Sun, Z.; Fang, C.; et al. Genome-Wide CRISPR-Cas9 Screening Identifies NF-kappaB/E2F6 Responsible for EGFRvIII-Associated Temozolomide Resistance in Glioblastoma. Adv. Sci. 2019, 6, 1900782. [Google Scholar] [CrossRef]
- Oprita, A.; Baloi, S.C.; Staicu, G.A.; Alexandru, O.; Tache, D.E.; Danoiu, S.; Micu, E.S.; Sevastre, A.S. Updated Insights on EGFR Signaling Pathways in Glioma. Int. J. Mol. Sci. 2021, 22, 587. [Google Scholar] [CrossRef]
- Sledzinska, P.; Bebyn, M.G.; Furtak, J.; Kowalewski, J.; Lewandowska, M.A. Prognostic and Predictive Biomarkers in Gliomas. Int. J. Mol. Sci. 2021, 22, 373. [Google Scholar] [CrossRef]
- Choi, J.; Bordeaux, Z.A.; McKeel, J.; Nanni, C.; Sutaria, N.; Braun, G.; Davis, C.; Miller, M.N.; Alphonse, M.P.; Kwatra, S.G.; et al. GZ17-6.02 Inhibits the Growth of EGFRvIII+ Glioblastoma. Int. J. Mol. Sci. 2022, 23, 4174. [Google Scholar] [CrossRef]
- Kim, H.M.; Lee, S.H.; Lim, J.; Yoo, J.; Hwang, D.Y. The epidermal growth factor receptor variant type III mutation frequently found in gliomas induces astrogenesis in human cerebral organoids. Cell Prolif. 2021, 54, e12965. [Google Scholar] [CrossRef]
- Hoogstrate, Y.; Vallentgoed, W.; Kros, J.M.; de Heer, I.; de Wit, M.; Eoli, M.; Sepulveda, J.M.; Walenkamp, A.M.E.; Frenel, J.S.; Franceschi, E.; et al. EGFR mutations are associated with response to depatux-m in combination with temozolomide and result in a receptor that is hypersensitive to ligand. Neuro-Oncol. Adv. 2020, 2, vdz051. [Google Scholar] [CrossRef]
- Vaidya, K.S.; Mitten, M.J.; Zelaya-Lazo, A.L.; Oleksijew, A.; Alvey, C.; Falls, H.D.; Mishra, S.; Palma, J.; Ansell, P.; Phillips, A.C.; et al. Synergistic therapeutic benefit by combining the antibody drug conjugate, depatux-m with temozolomide in pre-clinical models of glioblastoma with overexpression of EGFR. J. Neuro-Oncol. 2021, 152, 233–243. [Google Scholar] [CrossRef]
- Wu, W.; Schecker, J.; Wurstle, S.; Schneider, F.; Schonfelder, M.; Schlegel, J. Aldehyde dehydrogenase 1A3 (ALDH1A3) is regulated by autophagy in human glioblastoma cells. Cancer Lett. 2018, 417, 112–123. [Google Scholar] [CrossRef]
- Wang, Z.; Mo, Y.; Tan, Y.; Wen, Z.; Dai, Z.; Zhang, H.; Zhang, X.; Feng, S.; Liang, X.; Song, T.; et al. The ALDH Family Contributes to Immunocyte Infiltration, Proliferation and Epithelial-Mesenchymal Transformation in Glioma. Front. Immunol. 2021, 12, 756606. [Google Scholar] [CrossRef]
- Zhang, W.; Yan, W.; You, G.; Bao, Z.; Wang, Y.; Liu, Y.; You, Y.; Jiang, T. Genome-wide DNA methylation profiling identifies ALDH1A3 promoter methylation as a prognostic predictor in G-CIMP- primary glioblastoma. Cancer Lett. 2013, 328, 120–125. [Google Scholar] [CrossRef]
- Mao, P.; Joshi, K.; Li, J.; Kim, S.H.; Li, P.; Santana-Santos, L.; Luthra, S.; Chandran, U.R.; Benos, P.V.; Smith, L.; et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc. Natl. Acad. Sci. USA 2013, 110, 8644–8649. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Hu, H.; Huang, H.; Bao, Z.; Yang, P.; Wang, Y.; You, G.; Yan, W.; Jiang, T.; et al. ALDH1A3: A Marker of Mesenchymal Phenotype in Gliomas Associated with Cell Invasion. PLoS ONE 2015, 10, e0142856. [Google Scholar] [CrossRef]
- Medeiros, M.; Candido, M.F.; Valera, E.T.; Brassesco, M.S. The multifaceted NF-kB: Are there still prospects of its inhibition for clinical intervention in pediatric central nervous system tumors? Cell. Mol. Life Sci. 2021, 78, 6161–6200. [Google Scholar] [CrossRef]
- Zhang, Q.; Lenardo, M.J.; Baltimore, D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell 2017, 168, 37–57. [Google Scholar] [CrossRef]
- Chai, R.C.; Chang, Y.Z.; Chang, X.; Pang, B.; An, S.Y.; Zhang, K.N.; Chang, Y.H.; Jiang, T.; Wang, Y.Z. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m(6)A modification to activate NF-kappaB and promote the malignant progression of glioma. J. Hematol. Oncol. 2021, 14, 109. [Google Scholar] [CrossRef]
- Chang, Y.Z.; Chai, R.C.; Pang, B.; Chang, X.; An, S.Y.; Zhang, K.N.; Jiang, T.; Wang, Y.Z. METTL3 enhances the stability of MALAT1 with the assistance of HuR via m6A modification and activates NF-kappaB to promote the malignant progression of IDH-wildtype glioma. Cancer Lett. 2021, 511, 36–46. [Google Scholar] [CrossRef]
- Tanaka, K.; Babic, I.; Nathanson, D.; Akhavan, D.; Guo, D.; Gini, B.; Dang, J.; Zhu, S.; Yang, H.; De Jesus, J.; et al. Oncogenic EGFR signaling activates an mTORC2-NF-kappaB pathway that promotes chemotherapy resistance. Cancer Discov. 2011, 1, 524–538. [Google Scholar] [CrossRef]
- Canino, C.; Luo, Y.; Marcato, P.; Blandino, G.; Pass, H.I.; Cioce, M. A STAT3-NFkB/DDIT3/CEBPbeta axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget 2015, 6, 12637–12653. [Google Scholar] [CrossRef]
- Huang, K.; Yang, C.; Wang, Q.X.; Li, Y.S.; Fang, C.; Tan, Y.L.; Wei, J.W.; Wang, Y.F.; Li, X.; Zhou, J.H.; et al. The CRISPR/Cas9 system targeting EGFR exon 17 abrogates NF-kappaB activation via epigenetic modulation of UBXN1 in EGFRwt/vIII glioma cells. Cancer Lett. 2017, 388, 269–280. [Google Scholar] [CrossRef]
- Shi, Z.F.; Fang, Q.; Chen, Y.; Xu, L.X.; Wu, M.; Jia, M.; Lu, Y.; Wang, X.X.; Wang, Y.J.; Yan, X.; et al. Methylene blue ameliorates brain edema in rats with experimental ischemic stroke via inhibiting aquaporin 4 expression. Acta Pharm. Sin. 2021, 42, 382–392. [Google Scholar] [CrossRef]
- Lee, C.; Huang, C.H. LASAGNA-Search 2.0: Integrated transcription factor binding site search and visualization in a browser. Bioinformatics 2014, 30, 1923–1925. [Google Scholar] [CrossRef]
- Zadeh, G.; Bhat, K.P.; Aldape, K. EGFR and EGFRvIII in glioblastoma: Partners in crime. Cancer Cell 2013, 24, 403–404. [Google Scholar] [CrossRef]
- Lauretti, L.; Cenci, T.; Montano, N.; Offi, M.; Giordano, M.; Caccavella, V.M.; Mangraviti, A.; Agostini, L.; Olivi, A.; Gabriele, L.; et al. Molecular Analysis in a Glioblastoma Cohort-Results of a Prospective Analysis. J. Pers. Med. 2022, 12, 685. [Google Scholar] [CrossRef]
- Lindberg, O.R.; McKinney, A.; Engler, J.R.; Koshkakaryan, G.; Gong, H.; Robinson, A.E.; Ewald, A.J.; Huillard, E.; David James, C.; Molinaro, A.M.; et al. GBM heterogeneity as a function of variable epidermal growth factor receptor variant III activity. Oncotarget 2016, 7, 79101–79116. [Google Scholar] [CrossRef]
- Struve, N.; Binder, Z.A.; Stead, L.F.; Brend, T.; Bagley, S.J.; Faulkner, C.; Ott, L.; Muller-Goebel, J.; Weik, A.S.; Hoffer, K.; et al. EGFRvIII upregulates DNA mismatch repair resulting in increased temozolomide sensitivity of MGMT gene methylated glioblastoma. Oncogene 2020, 39, 3041–3055. [Google Scholar] [CrossRef]
- Felsberg, J.; Hentschel, B.; Kaulich, K.; Gramatzki, D.; Zacher, A.; Malzkorn, B.; Kamp, M.; Sabel, M.; Simon, M.; Westphal, M.; et al. Epidermal Growth Factor Receptor Variant III (EGFRvIII) Positivity in EGFR-Amplified Glioblastomas: Prognostic Role and Comparison between Primary and Recurrent Tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 6846–6855. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, J.; Fan, Y.; Qi, Y.; Wang, S.; Zhao, S.; Guo, X.; Xue, H.; Deng, L.; Zhao, R.; et al. PDIA3P1 promotes Temozolomide resistance in glioblastoma by inhibiting C/EBPbeta degradation to facilitate proneural-to-mesenchymal transition. J. Exp. Clin. Cancer Res. CR 2022, 41, 223. [Google Scholar] [CrossRef]
- Li, J.; Garavaglia, S.; Ye, Z.; Moretti, A.; Belyaeva, O.V.; Beiser, A.; Ibrahim, M.; Wilk, A.; McClellan, S.; Klyuyeva, A.V.; et al. A specific inhibitor of ALDH1A3 regulates retinoic acid biosynthesis in glioma stem cells. Commun. Biol. 2021, 4, 1420. [Google Scholar] [CrossRef]
- Poturnajova, M.; Kozovska, Z.; Matuskova, M. Aldehyde dehydrogenase 1A1 and 1A3 isoforms-mechanism of activation and regulation in cancer. Cell. Signal. 2021, 87, 110120. [Google Scholar] [CrossRef]
- Zirjacks, L.; Stransky, N.; Klumpp, L.; Prause, L.; Eckert, F.; Zips, D.; Schleicher, S.; Handgretinger, R.; Huber, S.M.; Ganser, K. Repurposing Disulfiram for Targeting of Glioblastoma Stem Cells: An In Vitro Study. Biomolecules 2021, 11, 1561. [Google Scholar] [CrossRef]
- Bauzone, M.; Souidi, M.; Dessein, A.F.; Wisztorski, M.; Vincent, A.; Gimeno, J.P.; Monte, D.; Van Seuningen, I.; Gespach, C.; Huet, G. Cross-talk between YAP and RAR-RXR Drives Expression of Stemness Genes to Promote 5-FU Resistance and Self-Renewal in Colorectal Cancer Cells. Mol. Cancer Res. MCR 2021, 19, 612–622. [Google Scholar] [CrossRef]
- Li, G.; Li, Y.; Liu, X.; Wang, Z.; Zhang, C.; Wu, F.; Jiang, H.; Zhang, W.; Bao, Z.; Wang, Y.; et al. ALDH1A3 induces mesenchymal differentiation and serves as a predictor for survival in glioblastoma. Cell Death Dis. 2018, 9, 1190. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, J.; Chen, Z.; Wang, H.; Xue, H.; Yang, C.; Guo, Q.; Qi, Y.; Guo, X.; Qian, M.; et al. Transfer of MicroRNA via Macrophage-Derived Extracellular Vesicles Promotes Proneural-to-Mesenchymal Transition in Glioma Stem Cells. Cancer Immunol. Res. 2020, 8, 966–981. [Google Scholar] [CrossRef]
- Keller, M.; Blom, M.; Conze, L.L.; Guo, M.; Hagerstrand, D.; Aspenstrom, P. Altered cytoskeletal status in the transition from proneural to mesenchymal glioblastoma subtypes. Sci. Rep. 2022, 12, 9838. [Google Scholar] [CrossRef]
- Li, M.; Xiao, A.; Floyd, D.; Olmez, I.; Lee, J.; Godlewski, J.; Bronisz, A.; Bhat, K.P.L.; Sulman, E.P.; Nakano, I.; et al. CDK4/6 inhibition is more active against the glioblastoma proneural subtype. Oncotarget 2017, 8, 55319–55331. [Google Scholar] [CrossRef]
- Lee, S.Y. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016, 3, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.L.; Balasubramaniyan, V.; Vaillant, B.; Ezhilarasan, R.; Hummelink, K.; Hollingsworth, F.; Wani, K.; Heathcock, L.; James, J.D.; Goodman, L.D.; et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell 2013, 24, 331–346. [Google Scholar] [CrossRef] [PubMed]
- Bonavia, R.; Inda, M.M.; Vandenberg, S.; Cheng, S.Y.; Nagane, M.; Hadwiger, P.; Tan, P.; Sah, D.W.; Cavenee, W.K.; Furnari, F.B. EGFRvIII promotes glioma angiogenesis and growth through the NF-kappaB, interleukin-8 pathway. Oncogene 2012, 31, 4054–4066. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.H.; Jiao, B.H.; Wang, C.Y.; Wu, J.L. Regulation of temozolomide resistance in glioma cells via the RIP2/NF-kappaB/MGMT pathway. CNS Neurosci. Ther. 2021, 27, 552–563. [Google Scholar] [CrossRef]
- Velpula, K.K.; Guda, M.R.; Sahu, K.; Tuszynski, J.; Asuthkar, S.; Bach, S.E.; Lathia, J.D.; Tsung, A.J. Metabolic targeting of EGFRvIII/PDK1 axis in temozolomide resistant glioblastoma. Oncotarget 2017, 8, 35639–35655. [Google Scholar] [CrossRef]
- Wang, K.; Huang, R.; Wu, C.; Li, G.; Zhao, Z.; Hu, H.; Liu, Y. Receptor tyrosine kinase expression in high-grade gliomas before and after chemoradiotherapy. Oncol. Lett. 2019, 18, 6509–6515. [Google Scholar] [CrossRef]
- Su, I.C.; Su, Y.K.; Chuang, H.Y.; Yadav, V.K.; Setiawan, S.A.; Fong, I.H.; Yeh, C.T.; Huang, H.C.; Lin, C.M. Ubiquitin-Specific Protease 6 n-Terminal-like Protein (USP6NL) and the Epidermal Growth Factor Receptor (EGFR) Signaling Axis Regulates Ubiquitin-Mediated DNA Repair and Temozolomide-Resistance in Glioblastoma. Biomedicines 2022, 10, 1531. [Google Scholar] [CrossRef]
- Xin, S.; Huang, K.; Zhu, X.G. Non-coding RNAs: Regulators of glioma cell epithelial-mesenchymal transformation. Pathol. Res. Pract. 2019, 215, 152539. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, Z.; Zheng, H.; Xu, H.; Wang, Y.; Chen, C.; Liu, J.; Yi, G.; Li, Z.; Wang, X.; et al. Molecular mechanism by which CDCP1 promotes proneural–mesenchymal transformation in primary glioblastoma. Cancer Cell Int. 2022, 22, 151. [Google Scholar] [CrossRef]
- Zhao, K.; Cui, X.; Wang, Q.; Fang, C.; Tan, Y.; Wang, Y.; Yi, K.; Yang, C.; You, H.; Shang, R.; et al. RUNX1 contributes to the mesenchymal subtype of glioblastoma in a TGFbeta pathway-dependent manner. Cell Death Dis. 2019, 10, 877. [Google Scholar] [CrossRef]
- Gan, C.; Pierscianek, D.; El Hindy, N.; Ahmadipour, Y.; Keyvani, K.; Sure, U.; Zhu, Y. The predominant expression of cancer stem cell marker ALDH1A3 in tumor infiltrative area is associated with shorter overall survival of human glioblastoma. BMC Cancer 2020, 20, 672. [Google Scholar] [CrossRef] [PubMed]
- Suwala, A.K.; Koch, K.; Rios, D.H.; Aretz, P.; Uhlmann, C.; Ogorek, I.; Felsberg, J.; Reifenberger, G.; Kohrer, K.; Deenen, R.; et al. Inhibition of Wnt/beta-catenin signaling downregulates expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) to reduce resistance against temozolomide in glioblastoma in vitro. Oncotarget 2018, 9, 22703–22716. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, H.W.; Wang, S.; Fan, L.; Feng, S.; Cai, X.; Peng, C.; Wu, X.; Lu, J.; Chen, D.; et al. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J. Clin. Investig. 2019, 129, 2043–2055. [Google Scholar] [CrossRef] [PubMed]
- Voce, D.J.; Bernal, G.M.; Wu, L.; Crawley, C.D.; Zhang, W.; Mansour, N.M.; Cahill, K.E.; Szymura, S.J.; Uppal, A.; Raleigh, D.R.; et al. Temozolomide Treatment Induces lncRNA MALAT1 in an NF-kappaB and p53 Codependent Manner in Glioblastoma. Cancer Res. 2019, 79, 2536–2548. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Chen, Y.; Wang, S.; Li, P.; Zhou, G.; Yuan, Y. Inhibition of NF-kappaB results in anti-glioma activity and reduces temozolomide-induced chemoresistance by down-regulating MGMT gene expression. Cancer Lett. 2018, 428, 77–89. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | EGFRvIII (−) n = 15 | EGFRvIII (+) n = 51 | Variation (p-Value) |
|---|---|---|---|
| Age at diagnosis | 0.2136 a | ||
| Mean | 44.13 | 48.92 | |
| Standard Deviation | 15.50 | 12.18 | |
| Gender | 0.7654 b | ||
| Male | 10 | 30 | |
| Female | 5 | 21 | |
| IDH1 Mutation | 0.9999 b | ||
| Mutation | 2 | 9 | |
| Wildtype | 13 | 42 | |
| Not Available | |||
| 1p/19q Codeletion Status | 0.9999 b | ||
| Codeletion | 0 | 0 | |
| Non-Codeletion | 15 | 50 | |
| Not Available | 0 | 1 | |
| MGMT Promoter Status | 0.2361 b | ||
| Methylated | 4 | 24 | |
| Un-Methylated | 11 | 27 | |
| Not Available | |||
| Radiotherapy | 0.4200 b | ||
| Yes | 12 | 44 | |
| No | 3 | 6 | |
| Not Available | 0 | 1 | |
| Chemotherapy | 0.2825 b | ||
| Yes | 10 | 41 | |
| No | 5 | 9 | |
| Not Available | 0 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Z.-F.; Li, G.-Z.; Zhai, Y.; Pan, C.-Q.; Wang, D.; Yu, M.-C.; Liu, C.; Zhang, W.; Yu, X.-G. EGFRvIII Promotes the Proneural–Mesenchymal Transition of Glioblastoma Multiforme and Reduces Its Sensitivity to Temozolomide by Regulating the NF-κB/ALDH1A3 Axis. Genes 2023, 14, 651. https://doi.org/10.3390/genes14030651
Shi Z-F, Li G-Z, Zhai Y, Pan C-Q, Wang D, Yu M-C, Liu C, Zhang W, Yu X-G. EGFRvIII Promotes the Proneural–Mesenchymal Transition of Glioblastoma Multiforme and Reduces Its Sensitivity to Temozolomide by Regulating the NF-κB/ALDH1A3 Axis. Genes. 2023; 14(3):651. https://doi.org/10.3390/genes14030651
Chicago/Turabian StyleShi, Zhong-Fang, Guan-Zhang Li, You Zhai, Chang-Qing Pan, Di Wang, Ming-Chen Yu, Chi Liu, Wei Zhang, and Xiao-Guang Yu. 2023. "EGFRvIII Promotes the Proneural–Mesenchymal Transition of Glioblastoma Multiforme and Reduces Its Sensitivity to Temozolomide by Regulating the NF-κB/ALDH1A3 Axis" Genes 14, no. 3: 651. https://doi.org/10.3390/genes14030651
APA StyleShi, Z.-F., Li, G.-Z., Zhai, Y., Pan, C.-Q., Wang, D., Yu, M.-C., Liu, C., Zhang, W., & Yu, X.-G. (2023). EGFRvIII Promotes the Proneural–Mesenchymal Transition of Glioblastoma Multiforme and Reduces Its Sensitivity to Temozolomide by Regulating the NF-κB/ALDH1A3 Axis. Genes, 14(3), 651. https://doi.org/10.3390/genes14030651





