RADX Gene Variant May Predispose to Familial Asperger Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Assessment
2.2. Exome Sequencing and Bioinformatics Analysis
2.3. Sanger Sequencing
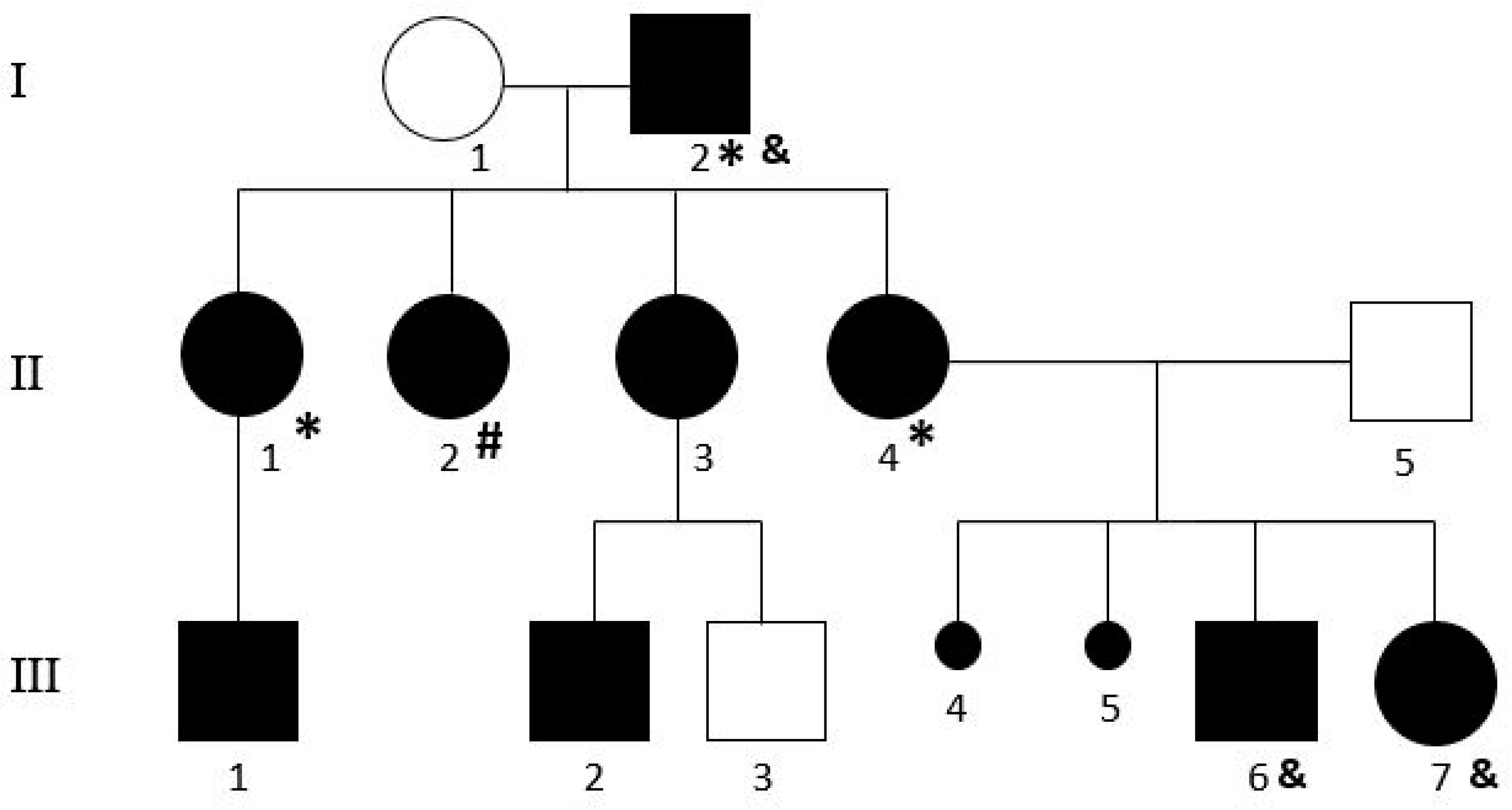
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism Spectrum Disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Kanner, L. Autistic Disturbances of Affective Contact. Nerv. Child 1943, 2, 217–250. [Google Scholar]
- Simonoff, E.; Pickles, A.; Charman, T.; Chandler, S.; Loucas, T.; Baird, G. Psychiatric Disorders in Children with Autism Spectrum Disorders: Prevalence, Comorbidity, and Associated Factors in a Population-Derived Sample. J. Am. Acad. Child Adolesc. Psychiatry 2008, 47, 921–929. [Google Scholar] [CrossRef] [PubMed]
- Vorstman, J.A.S.; Parr, J.R.; Moreno-De-Luca, D.; Anney, R.J.L.; Nurnberger, J.I.; Hallmayer, J.F. Autism Genetics: Opportunities and Challenges for Clinical Translation. Nat. Rev. Genet. 2017, 18, 362–376. [Google Scholar] [CrossRef] [PubMed]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The Contribution of de Novo Coding Mutations to Autism Spectrum Disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Masini, E.; Loi, E.; Vega-Benedetti, A.F.; Carta, M.; Doneddu, G.; Fadda, R.; Zavattari, P. An Overview of the Main Genetic, Epigenetic and Environmental Factors Involved in Autism Spectrum Disorder Focusing on Synaptic Activity. Int. J. Mol. Sci. 2020, 21, 8290. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.R.; Scherer, S.W. Detection and Characterization of Copy Number Variation in Autism Spectrum Disorder. Methods Mol. Biol. 2012, 838, 115–135. [Google Scholar] [CrossRef] [PubMed]
- Devlin, B.; Scherer, S.W. Genetic Architecture in Autism Spectrum Disorder. Curr. Opin. Genet. Dev. 2012, 22, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Gécz, J.; Shoubridge, C.; Corbett, M. The Genetic Landscape of Intellectual Disability Arising from Chromosome X. Trends Genet. 2009, 25, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Willard, H.F. X Chromosome Inactivation and X-Linked Mental Retardation. Am. J. Med. Genet. 1996, 64, 21–26. [Google Scholar] [CrossRef]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; The Galaxy Team. Galaxy: A Comprehensive Approach for Supporting Accessible, Reproducible, and Transparent Computational Research in the Life Sciences. Genome Biol. 2010, 11, R86. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, K. Genomic Variant Annotation and Prioritization with ANNOVAR and WANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Coldren, C.; Karunamurthy, A.; Kip, N.S.; Klee, E.W.; Lincoln, S.E.; Leon, A.; Pullambhatla, M.; Temple-Smolkin, R.L.; Voelkerding, K.V.; et al. Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines: A Joint Recommendation of the Association for Molecular Pathology and the College of American Pathologists. J. Mol. Diagn. 2018, 20, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Plaschkes, I.; Oz-Levi, D.; Alkelai, A.; Olender, T.; Zimmerman, S.; Twik, M.; Belinky, F.; Fishilevich, S.; Nudel, R.; et al. VarElect: The Phenotype-Based Variation Prioritizer of the GeneCards Suite. BMC Genom. 2016, 17, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Schubert, L.; Ho, T.; Hoffmann, S.; Haahr, P.; Guérillon, C.; Mailand, N. RADX Interacts with Single-stranded DNA to Promote Replication Fork Stability. EMBO Rep. 2017, 18, 1991–2003. [Google Scholar] [CrossRef] [PubMed]
- Zeman, M.K.; Cimprich, K.A. Causes and Consequences of Replication Stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Dungrawala, H.; Bhat, K.P.; Le Meur, R.; Chazin, W.J.; Ding, X.; Sharan, S.K.; Wessel, S.R.; Sathe, A.A.; Zhao, R.; Cortez, D. RADX Promotes Genome Stability and Modulates Chemosensitivity by Regulating RAD51 at Replication Forks. Mol. Cell 2017, 67, 374–386.e5. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wei, P.C.; Lim, C.K.; Gallina, I.S.; Marshall, S.; Marchetto, M.C.; Alt, F.W.; Gage, F.H. Increased Neural Progenitor Proliferation in a HiPSC Model of Autism Induces Replication Stress-Associated Genome Instability. Cell Stem Cell 2020, 26, 221–233.e6. [Google Scholar] [CrossRef] [PubMed]
- Depienne, C.; Bouteiller, D.; Méneret, A.; Billot, S.; Groppa, S.; Klebe, S.; Charbonnier-Beaupel, F.; Corvol, J.C.; Saraiva, J.P.; Brueggemann, N.; et al. RAD51 Haploinsufficiency Causes Congenital Mirror Movements in Humans. Am. J. Hum. Genet. 2012, 90, 301–307. [Google Scholar] [CrossRef] [PubMed]
| Individual | I2 | II1 | III1 | II2 | II3 | III2 | II4 | III6 | III7 |
|---|---|---|---|---|---|---|---|---|---|
| Formal diagnosis | AS | AS | AS | AS | |||||
| Clinical diagnosis | Behavioral problems | Major psychiatric disorder | Major psychiatric disorder | Major psychiatric disorder | Major psychiatric disorder | ||||
| AS “features” | Mathematical/ engineering skills | Astrophysicist | Drawing skills | Aerospace engineer | Computer skills |
| Position † | RefSeq Gene and HGVS Nomenclature ‡ | II1 | III1 | II2 | II3 | II4 | III2 |
|---|---|---|---|---|---|---|---|
| chr1:150444527 | RPRD2:NM_015203:exon11:c.3103C>T:p.(P1035S) | √ | √ | X | |||
| chr12:2074822 | DCP1B:NM_152640:exon5:c.424A>G:p.(T142A) | X | X | X | |||
| chr12:95528593 | FGD6:NM_018351:exon8:c. 3004 C>T:p.(R1002C) | √ | X | X | X | ||
| chr16:2263822 | PGP:NM_001042371:exon2:c.873T>G:p.(N291K) | X | X | √ | |||
| chr16:84212975 | TAF1C:NM_005679:exon14:c.2182C>G:p.(R728G) | √ | √ | X | X | ||
| chr19:42819206 | TMEM145:NM_173633:exon6:c.482G>A:p.(R161Q) | X | X | X | √ | ||
| chr19:49129547 | SPHK2:NM_001204160:exon2:c.331C>A:p.(R111S) | X | X | ||||
| chr2:15564564 | NBAS:NM_015909:exon23:c.2452G>C:p.(E818Q) | X | √ | √ | |||
| chr2:241569440 | GPR35:NM_001195382:exon6:c.164A>T:p.(Y55F) | √ | X | √ | √ | √ | √ |
| chr2:24522996 | ITSN2:NM_001348182:exon12:c.1126A>G:p.(M376V) | X | √ | X | |||
| chr2:71209151 | ANKRD53:NM_001115116:exon4:c.703G>A:p.(A235T) | X | √ | √ | |||
| chr2:85828148 | TMEM150A:NM_001031738:exon4:c.196A>G:p.(I66V) | X | |||||
| chr20:62164999 | PTK6:NM_005975:exon4:c.575C>T:p.(T192M) | √ | X | X | |||
| chr4:83839213 | THAP9:NM_024672:exon5:c.1848A>G:p.(L616L) | √ | X | X | |||
| chr5:180660683 | TRIM41:NM_033549:exon5:c.1211C>T:p.(P404L) | √ | √ | X | √ | ||
| chr6:44221229 | HSP90AB1:NM_001271969:exon12:c.2069T>C:p.(I690T) | X | X | ||||
| chr9:1056825 | DMRT2:NM_181872:exon4:c.1238C>T:p.(T413M) | √ | X | √ | √ | √ | X |
| chrX:105921420 | RADX:NM_001184782:exon13:c.2209T>A:p.(C737S) | √ | √ | √ | √ | √ | √ |
| Pathogenicity Scores | |||
| MetaLR | prediction | score | rankscore |
| Damaging | 0.5693 | 0.844 | |
| MetaSVM | prediction | score | rankscore |
| Tolerated | −0.2734 | 0.7572 | |
| MetaRNN | prediction | score | rankscore |
| Damaging | 0.9322 | 0.9255 | |
| REVEL | prediction | score | rankscore |
| Uncertain | 0.578 | 0.8317 | |
| Individual Predictions | |||
| BayesDel addAF | addAF prediction | addAF score | addAF rankscore |
| Damaging | 0.1129 | 0.6565 | |
| BayesDel noAF | noAF prediction | noAF score | noAF rankscore |
| Tolerated | −0.0756 | 0.6523 | |
| FATHMM | prediction | score | converted rankscore |
| Tolerated | −1.09, −1.35, −1.45 | 0.8089 | |
| FATHMM-MKL | coding prediction | coding score | coding rankscore |
| Damaging | 0.8163 | 0.4099 | |
| LIST-S2 | prediction | score | rankscore |
| Tolerated | 0.7136, 0.7082, 0.7238 | 0.3374 | |
| LRT | prediction | score | converted rankscore |
| Deleterious | 0 | 0.8433 | |
| M-CAP | prediction | score | rankscore |
| Damaging | 0.1083 | 0.7848 | |
| MVP | prediction | score | rankscore |
| Benign | 0.3697 | 0.3658 | |
| MutPred | prediction | score | rankscore |
| Pathogenic | 0.811 | 0.9272 | |
| MutationTaster | Prediction | Accuracy | converted rankscore |
| Polymorphism, Disease causing | 0.89, 0.9971 | 0.3593 | |
| PROVEAN | prediction | score | converted rankscore |
| Damaging | −4.95, −5.68, −5.47 | 0.8722 | |
| SIFT | prediction | score | converted rankscore |
| Damaging | 0 | 0.9125 | |
| SIFT4G | prediction | score | converted rankscore |
| Damaging | 0 | 0.9282 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzarà, A.; Rumore, R.; Brugnoletti, F.; Tabolacci, E.; Bottillo, I.; Sangiorgi, E.; Gurrieri, F. RADX Gene Variant May Predispose to Familial Asperger Syndrome. Genes 2023, 14, 301. https://doi.org/10.3390/genes14020301
Azzarà A, Rumore R, Brugnoletti F, Tabolacci E, Bottillo I, Sangiorgi E, Gurrieri F. RADX Gene Variant May Predispose to Familial Asperger Syndrome. Genes. 2023; 14(2):301. https://doi.org/10.3390/genes14020301
Chicago/Turabian StyleAzzarà, Alessia, Roberto Rumore, Fulvia Brugnoletti, Elisabetta Tabolacci, Irene Bottillo, Eugenio Sangiorgi, and Fiorella Gurrieri. 2023. "RADX Gene Variant May Predispose to Familial Asperger Syndrome" Genes 14, no. 2: 301. https://doi.org/10.3390/genes14020301
APA StyleAzzarà, A., Rumore, R., Brugnoletti, F., Tabolacci, E., Bottillo, I., Sangiorgi, E., & Gurrieri, F. (2023). RADX Gene Variant May Predispose to Familial Asperger Syndrome. Genes, 14(2), 301. https://doi.org/10.3390/genes14020301









