A Network-Based Approach for Improving Annotation of Transcription Factor Functions and Binding Sites in Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
2.1. Method Overview
2.2. Data Sources
- Gene Expression: Large-scale gene expression data [29] for Arabidopsis thaliana. The dataset contains genome-wide expressions from various treated/control samples collected at different time intervals from various tissue source sites, spanning approximately 7000 Affymetrix ATH1 profiles. Out of the 6057 treatment conditions, we used 3317, spanning four broad categories: abiotic stress, biotic stress, development, and hormone response.
- TFs and associated motifs: List of Arabidopsis thaliana transcription factors with a curated list of binding motifs from PlantTFDB [30]. Corresponding expression data were available for 1384 out of 1717 TFs.
- TF functions: Currently known biological functions associated with TFs from gene ontology [31].
- Gene promoter sequences: 25,516 promoter sequences associated with genes [32].
- Gene loci: Gene loci, lengths, and attributes acquired from TAIR [33].
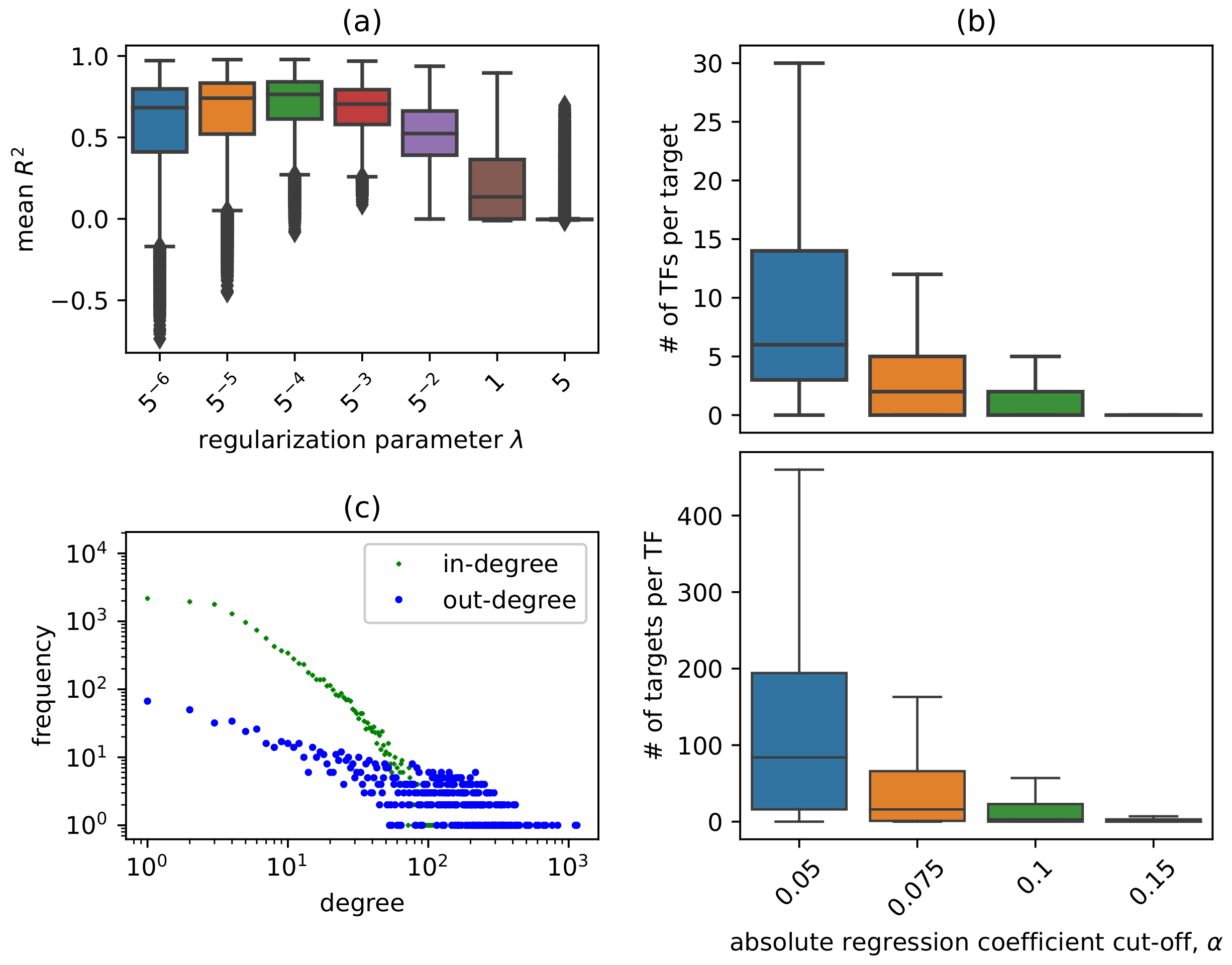
2.3. Network Construction Using Lasso Regression
2.4. Network Analysis
2.5. Biological Function Enrichment Analysis
2.5.1. Evaluating the Significance of the Number of Annotations
2.5.2. Comparison of Functional Annotations to Known Annotations
2.5.3. Predicting Novel Functional Annotations for Transcription Factors
2.6. Motif Discovery and Alignment with Reference PWMs
3. Results and Discussion
3.1. Network Construction Process and Topological Properties
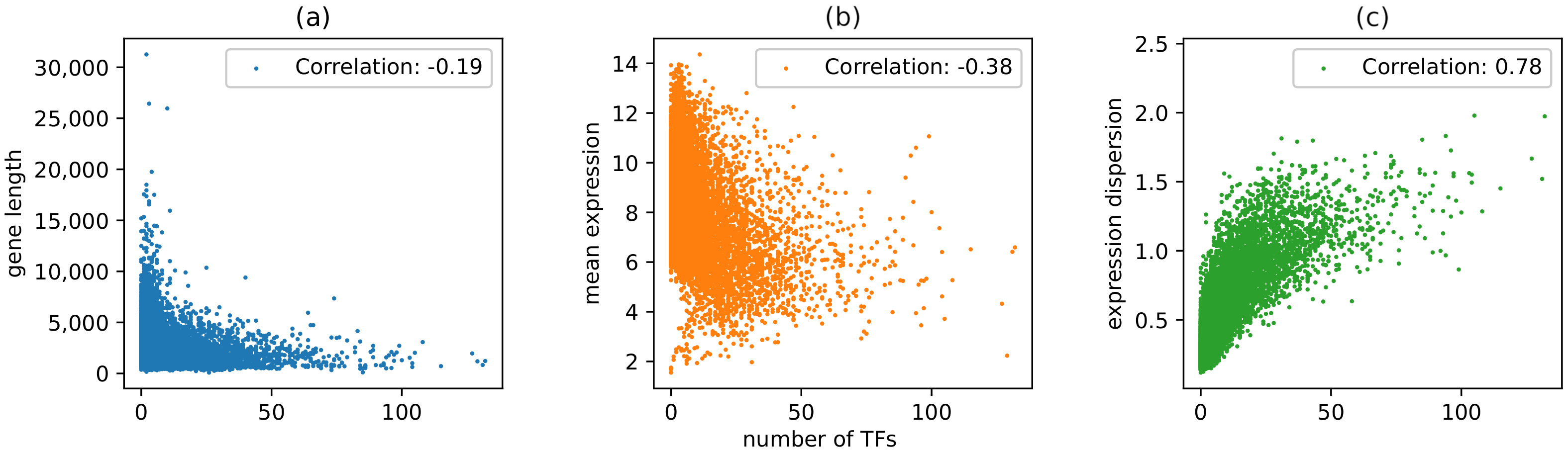
3.2. Network Analysis Results Reveal Interesting Relationships among Gene Length, Gene Expression Patterns, and Gene Regulatory Complexity
3.3. Biological Function Enrichment Analysis Results
3.3.1. Target Pools of TFs Are Enriched in a Significant Number of Functional Terms
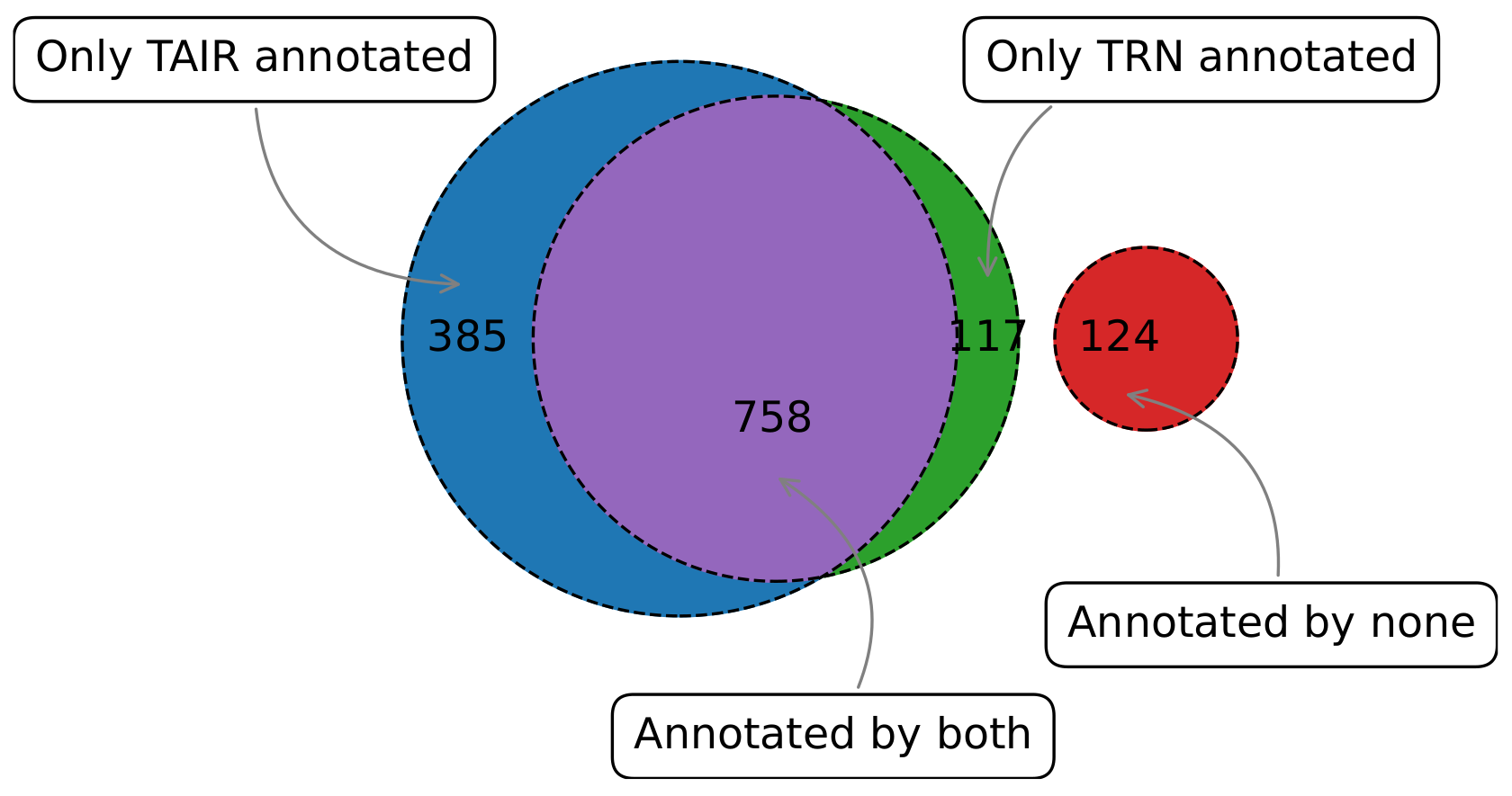
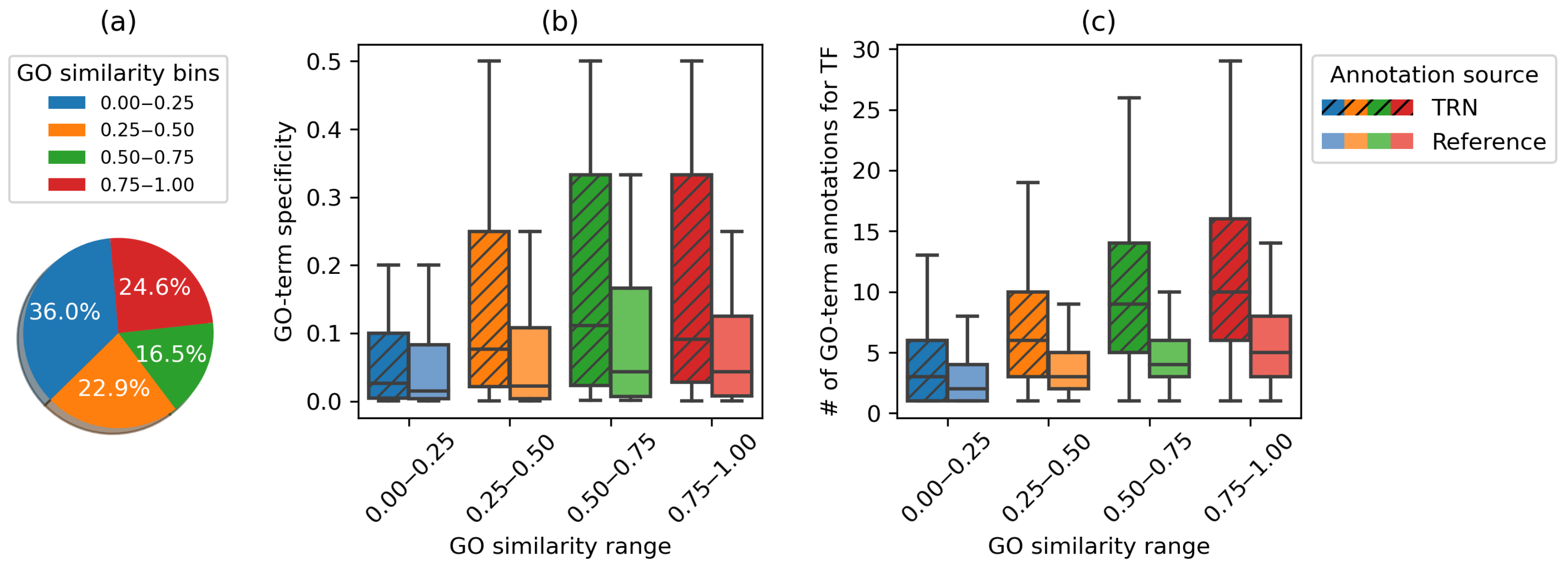
3.3.2. Predicted Functional Annotations Reflect and Improve Reference Annotations
3.3.3. TRN Predicts Reliable Novel Functional Annotations
3.3.4. TFs Exhibit Unique Condition-Specific Functional Roles
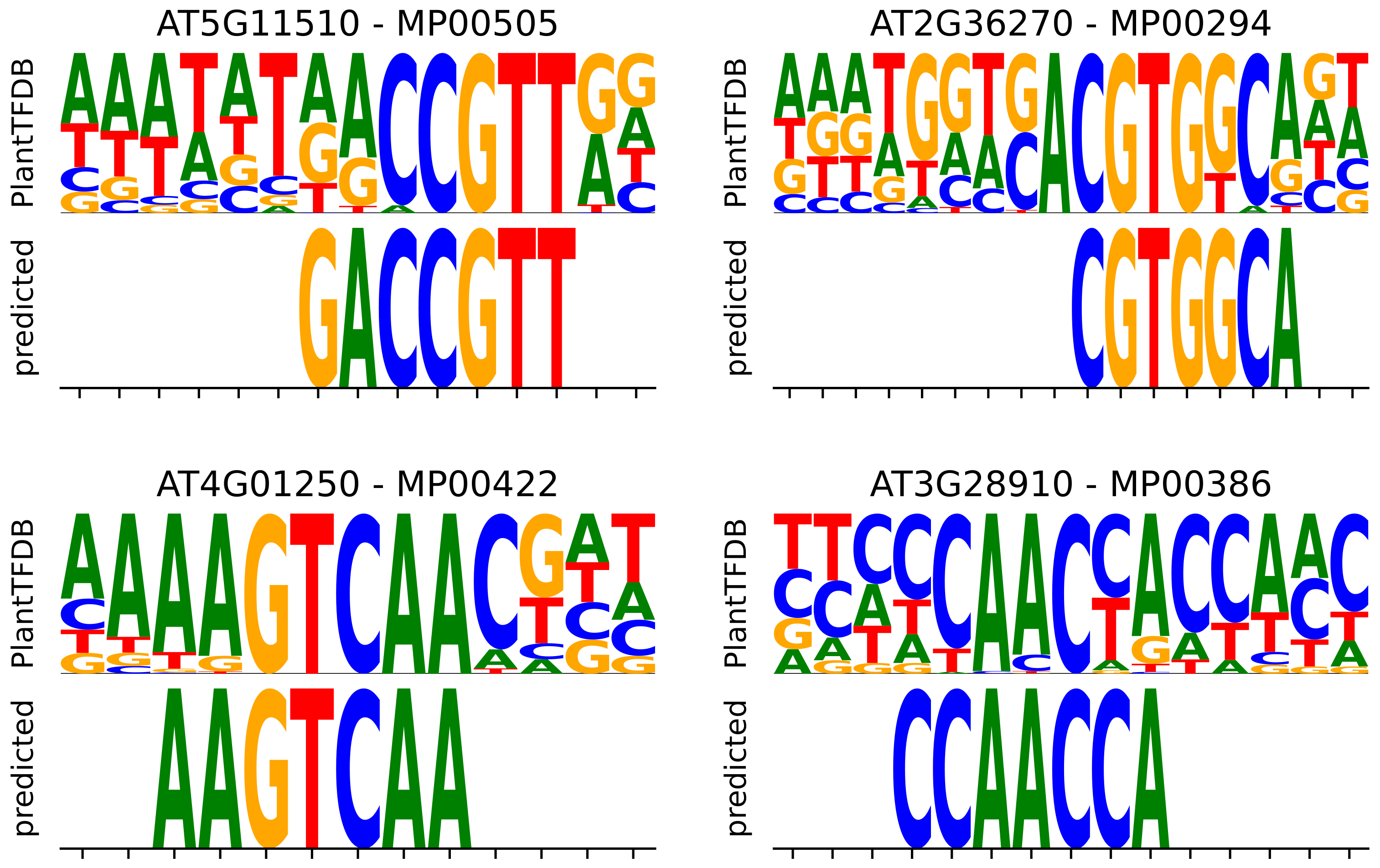
3.4. Motif Enrichment Recovers Reference PWMs and Identifies Candidate Binding Motifs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FDR | False discovery rate |
| GO | Gene ontology |
| TRN | Transcriptional regulatory network |
| PWM | Position-weight matrix |
| TF | Transcription factor |
References
- Thijs, G.; Lescot, M.; Marchal, K.; Rombauts, S.; De Moor, B.; Rouze, P.; Moreau, Y. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics 2001, 17, 1113–1122. [Google Scholar] [CrossRef]
- Hashim, F.A.; Mabrouk, M.S.; Al-Atabany, W. Review of different sequence motif finding algorithms. Avicenna J. Med. Biotechnol. 2019, 11, 130. [Google Scholar]
- Bailey, T.L. DREME: Motif discovery in transcription factor ChIP-seq data. Bioinformatics 2011, 27, 1653–1659. [Google Scholar] [CrossRef]
- Ma, S.; Bachan, S.; Porto, M.; Bohnert, H.J.; Snyder, M.; Dinesh-Kumar, S.P. Discovery of Stress Responsive DNA Regulatory Motifs in Arabidopsis. PLoS ONE 2012, 7, e43198. [Google Scholar] [CrossRef]
- Davey, N.E.; Cowan, J.L.; Shields, D.C.; Gibson, T.J.; Coldwell, M.J.; Edwards, R.J. SLiMPrints: Conservation-based discovery of functional motif fingerprints in intrinsically disordered protein regions. Nucleic Acids Res. 2012, 40, 10628–10641. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef]
- Bar-Joseph, Z.; Gerber, G.K.; Lee, T.I.; Rinaldi, N.J.; Yoo, J.Y.; Robert, F.; Gordon, D.B.; Fraenkel, E.; Jaakkola, T.S.; Young, R.A.; et al. Computational discovery of gene modules and regulatory networks. Nat. Biotechnol. 2003, 21, 1337–1342. [Google Scholar] [CrossRef] [PubMed]
- Segal, E.; Shapira, M.; Regev, A.; Pe’er, D.; Botstein, D.; Koller, D.; Friedman, N. Module networks: Identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 2003, 34, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Janky, R.; Verfaillie, A.; Imrichová, H.; Van de Sande, B.; Standaert, L.; Christiaens, V.; Hulselmans, G.; Herten, K.; Naval Sanchez, M.; Potier, D.; et al. iRegulon: From a gene list to a gene regulatory network using large motif and track collections. PLoS Comput. Biol. 2014, 10, e1003731. [Google Scholar] [CrossRef]
- Pe’er, D.; Regev, A.; Elidan, G.; Friedman, N. Inferring subnetworks from perturbed expression profiles. Bioinformatics 2001, 17, S215–S224. [Google Scholar] [CrossRef]
- Huynh-Thu, V.A.; Irrthum, A.; Wehenkel, L.; Geurts, P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE 2010, 5, e12776. [Google Scholar] [CrossRef] [PubMed]
- Haury, A.C.; Mordelet, F.; Vera-Licona, P.; Vert, J.P. TIGRESS: Trustful inference of gene regulation using stability selection. BMC Syst. Biol. 2012, 6, 145. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Hao, J.; Gautam, Y.; Mersha, T.B.; Kang, M. DiffGRN: Differential gene regulatory network analysis. Int. J. Data Min. Bioinform. 2018, 20, 362–379. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, S.R.; Vaneechoutte, D.; Van de Velde, J.; Vandepoele, K. TF2Network: Predicting transcription factor regulators and gene regulatory networks in Arabidopsis using publicly available binding site information. Nucleic Acids Res. 2018, 46, e31. [Google Scholar] [CrossRef]
- Marbach, D.; Prill, R.J.; Schaffter, T.; Mattiussi, C.; Floreano, D.; Stolovitzky, G. Revealing strengths and weaknesses of methods for gene network inference. Proc. Natl. Acad. Sci. USA 2010, 107, 6286–6291. [Google Scholar] [CrossRef]
- Berri, S.; Abbruscato, P.; Faivre-Rampant, O.; Brasileiro, A.; Fumasoni, I.; Satoh, K.; Kikuchi, S.; Mizzi, L.; Morandini, P.; Pè, M.E.; et al. Characterization of WRKYco-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009, 9, 120. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed]
- Sazegari, S.; Niazi, A.; Ahmadi, F.S. A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 2015, 11, 101. [Google Scholar] [CrossRef]
- Van den Broeck, L.; Dubois, M.; Vermeersch, M.; Storme, V.; Matsui, M.; Inzé, D. From network to phenotype: The dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol. Syst. Biol. 2017, 13, 961. [Google Scholar] [CrossRef]
- Taylor-Teeples, M.; Lin, L.; De Lucas, M.; Turco, G.; Toal, T.; Gaudinier, A.; Young, N.; Trabucco, G.; Veling, M.; Lamothe, R.; et al. An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 2015, 517, 571–575. [Google Scholar] [CrossRef]
- Brady, S.M.; Zhang, L.; Megraw, M.; Martinez, N.J.; Jiang, E.; Yi, C.S.; Liu, W.; Zeng, A.; Taylor-Teeples, M.; Kim, D.; et al. A stele-enriched gene regulatory network in the Arabidopsis root. Mol. Syst. Biol. 2011, 7, 459. [Google Scholar] [CrossRef]
- González-Morales, S.I.; Chávez-Montes, R.A.; Hayano-Kanashiro, C.; Alejo-Jacuinde, G.; Rico-Cambron, T.Y.; de Folter, S.; Herrera-Estrella, L. Regulatory network analysis reveals novel regulators of seed desiccation tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, E5232–E5241. [Google Scholar] [CrossRef] [PubMed]
- Keurentjes, J.J.; Fu, J.; Terpstra, I.R.; Garcia, J.M.; van den Ackerveken, G.; Snoek, L.B.; Peeters, A.J.; Vreugdenhil, D.; Koornneef, M.; Jansen, R.C. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc. Natl. Acad. Sci. USA 2007, 104, 1708–1713. [Google Scholar] [CrossRef]
- Yu, C.P.; Lin, J.J.; Li, W.H. Positional distribution of transcription factor binding sites in Arabidopsis thaliana. Sci. Rep. 2016, 6, 25164. [Google Scholar] [CrossRef]
- De Jong, H. Modeling and simulation of genetic regulatory systems: A literature review. J. Comput. Biol. 2002, 9, 67–103. [Google Scholar] [CrossRef] [PubMed]
- Van Someren, E.P.; Vaes, B.L.; Steegenga, W.T.; Sijbers, A.M.; Dechering, K.J.; Reinders, M.J. Least absolute regression network analysis of the murine osteoblast differentiation network. Bioinformatics 2006, 22, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Marbach, D.; Costello, J.C.; Küffner, R.; Vega, N.M.; Prill, R.J.; Camacho, D.M.; Allison, K.R.; Kellis, M.; Collins, J.J.; Stolovitzky, G. Wisdom of crowds for robust gene network inference. Nat. Methods 2012, 9, 796–804. [Google Scholar] [CrossRef]
- Wu, H.; Lu, T.; Xue, H.; Liang, H. Sparse additive ordinary differential equations for dynamic gene regulatory network modeling. J. Am. Stat. Assoc. 2014, 109, 700–716. [Google Scholar] [CrossRef]
- He, F.; Yoo, S.; Wang, D.; Kumari, S.; Gerstein, M.; Ware, D.; Maslov, S. Large-scale atlas of microarray data reveals the distinct expression landscape of different tissues in Arabidopsis. Plant J. 2016, 86, 472–480. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.C.; Meng, Y.Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2007, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Davuluri, R.V.; Sun, H.; Palaniswamy, S.K.; Matthews, N.; Molina, C.; Kurtz, M.; Grotewold, E. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinform. 2003, 4, 25. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Kaiser, M. Mean clustering coefficients: The role of isolated nodes and leafs on clustering measures for small-world networks. New J. Phys. 2008, 10, 083042. [Google Scholar] [CrossRef]
- Newman, M.E.J. Mixing patterns in networks. Phys. Rev. E 2003, 67, 026126. [Google Scholar] [CrossRef] [PubMed]
- Hagberg, A.A.; Schult, D.A.; Swart, P.J. Exploring Network Structure, Dynamics, and Function using NetworkX. In Proceedings of the 7th Python in Science Conference, Pasadena, CA, USA, 19–24 August 2008; Varoquaux, G., Vaught, T., Millman, J., Eds.; pp. 11–15. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44. [Google Scholar]
- EASE Score, a Modified Fisher Exact p-value. Available online: https://david.ncifcrf.gov/helps/functional_annotation.html#fisher (accessed on 10 January 2023).
- Ge, W.; Fazal, Z.; Jakobsson, E. Using optimal f-measure and random resampling in gene ontology enrichment calculations. Front. Appl. Math. Stat. 2019, 5, 20. [Google Scholar] [CrossRef]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO:: TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed]
- Holmans, P.; Green, E.K.; Pahwa, J.S.; Ferreira, M.A.; Purcell, S.M.; Sklar, P.; Owen, M.J.; O’Donovan, M.C.; Craddock, N.; Consortium, W.T.C.C.; et al. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am. J. Hum. Genet. 2009, 85, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z. Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 2003, 6, 430–440. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Vasques Filho, D.; O’Neale, D.R. Degree distributions of bipartite networks and their projections. Phys. Rev. E 2018, 98, 022307. [Google Scholar] [CrossRef]
- Eyre-Walker, A. Synonymous codon bias is related to gene length in Escherichia coli: Selection for translational accuracy? Mol. Biol. Evol. 1996, 13, 864–872. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, E.N.; Powell, J.R. Gene length and codon usage bias in Drosophila melanogaster, Saccharomyces cerevisiae and Escherichia coli. Nucleic Acids Res. 1998, 26, 3188–3193. [Google Scholar] [CrossRef] [PubMed]
- Colinas, J.; Schmidler, S.C.; Bohrer, G.; Iordanov, B.; Benfey, P.N. Intergenic and genic sequence lengths have opposite relationships with respect to gene expression. PLoS ONE 2008, 3, e3670. [Google Scholar] [CrossRef] [PubMed]
- Seoighe, C.; Gehring, C.; Hurst, L.D. Gametophytic selection in Arabidopsis thaliana supports the selective model of intron length reduction. PLoS Genet. 2005, 1, e13. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Davis, C.I.; Mekhedov, S.L.; Hartl, D.L.; Koonin, E.V.; Kondrashov, F.A. Selection for short introns in highly expressed genes. Nat. Genet. 2002, 31, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.Y.; Vorst, O.; Fiers, M.W.; Stiekema, W.J.; Nap, J.P. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef]
- Wang, J.Z.; Du, Z.; Payattakool, R.; Yu, P.S.; Chen, C.F. A new method to measure the semantic similarity of GO terms. Bioinformatics 2007, 23, 1274–1281. [Google Scholar] [CrossRef]
- Valentini, G. Hierarchical ensemble methods for protein function prediction. Int. Sch. Res. Not. 2014, 2014, 901419. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Higo, K.; Takano, M. Circadian clock-and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ. 2009, 32, 592–603. [Google Scholar] [CrossRef]
- Prochetto, S.; Reinheimer, R. Step by step evolution of Indeterminate Domain (IDD) transcriptional regulators: From algae to angiosperms. Ann. Bot. 2020, 126, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Liu, J.; Zhong, G.; Wang, B. Genome-Wide Identification and Expression Patterns of the C2H2-Zinc Finger Gene Family Related to Stress Responses and Catechins Accumulation in Camellia sinensis [L.] O. Kuntze. Int. J. Mol. Sci. 2021, 22, 4197. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.A.; Mukhtar, M.S.; Blanco, F.; Boatwright, J.L.; Moreno, I.; Jordan, M.R.; Chen, Y.; Brandizzi, F.; Dong, X.; Orellana, A.; et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 2012, 7, e31944. [Google Scholar] [CrossRef] [PubMed]




| Network Attribute | Value |
|---|---|
| Number of nodes | 14,043 |
| Number of edges | 117,587 |
| Mean degree | |
| Largest degree | 1156 |
| Number of TFs | 1213 |
| Number of targets | 13,813 |
| Mean (median) out-degree of TFs | (54) |
| Mean (median) in-degree of targets | (4) |
| Diameter 1 | 6 |
| Clustering coefficient | |
| Degree coefficient |
| Gene ID | Common Name | GO Term Description | p-Value | Benjamini | Fold Enrichment |
|---|---|---|---|---|---|
| translation | |||||
| AT2G37120 | S1FA2 | ribosome biogenesis | |||
| cytoplasmic translation | |||||
| plant-type cell wall organization | |||||
| AT2G28510 | DOF2.1 | hydrogen peroxide catabolic process | |||
| response to oxidative stress | |||||
| photosynthesis | |||||
| AT1G68520 | BBX14 | protein–chromophore linkage | |||
| photosynthetic electron transport in photosystem I | |||||
| flavonoid biosynthetic process | |||||
| AT3G47500 | CDF3 | cellular response to hypoxia | |||
| response to karrikin | |||||
| AT1G76580 | F14G6.18 | removal of superoxide radicals | |||
| hydrogen peroxide catabolic process | |||||
| AT3G21270 | ADOF2 | thalianol metabolic process | |||
| response to toxic substance | |||||
| oxidation-reduction process | |||||
| AT1G21450 | F24J8.8 | response to oxidative stress | |||
| embryo development | |||||
| embryo development ending in seed dormancy | |||||
| AT5G10120 | EIL4 | lipid storage | |||
| seed oilbody biogenesis | |||||
| AT1G28050 | BBX13 | circadian rhythm | |||
| response to hydrogen peroxide | |||||
| AT4G29190 | AtC3H49 | response to toxic substance | |||
| AT3G18400 | NAC058 | suberin biosynthetic process | |||
| lipid catabolic process | |||||
| AT2G23290 | AtMYB70 | plant-type cell wall organization | |||
| removal of superoxide radicals | |||||
| AT2G37650 | F13M22.15 | cellular response to high light intensity | |||
| response to copper ion | |||||
| AT2G40740 | ATWRKY55 | defense response | |||
| embryo development ending in seed dormancy | |||||
| AT5G52010 | AT5G52010 | protein refolding | |||
| response to heat | |||||
| AT5G05790 | AT5G05790 | hydrogen peroxide catabolic process | |||
| response to oxidative stress | |||||
| photosynthesis | |||||
| AT3G24490 | AT3G24490 | photosynthesis, light harvesting in photosystem I | |||
| reductive pentose-phosphate cycle | |||||
| AT1G32360 | F27G20.10 | response to bacterium | |||
| amino acid transmembrane export | |||||
| AT1G64620 | DOF1.8 | response to salicylic acid | |||
| seed development | |||||
| AT1G60240 | AT1G60240 | cell wall modification | |||
| pectin catabolic process |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Najnin, T.; Saimon, S.H.; Sunter, G.; Ruan, J. A Network-Based Approach for Improving Annotation of Transcription Factor Functions and Binding Sites in Arabidopsis thaliana. Genes 2023, 14, 282. https://doi.org/10.3390/genes14020282
Najnin T, Saimon SH, Sunter G, Ruan J. A Network-Based Approach for Improving Annotation of Transcription Factor Functions and Binding Sites in Arabidopsis thaliana. Genes. 2023; 14(2):282. https://doi.org/10.3390/genes14020282
Chicago/Turabian StyleNajnin, Tanzira, Sakhawat Hossain Saimon, Garry Sunter, and Jianhua Ruan. 2023. "A Network-Based Approach for Improving Annotation of Transcription Factor Functions and Binding Sites in Arabidopsis thaliana" Genes 14, no. 2: 282. https://doi.org/10.3390/genes14020282
APA StyleNajnin, T., Saimon, S. H., Sunter, G., & Ruan, J. (2023). A Network-Based Approach for Improving Annotation of Transcription Factor Functions and Binding Sites in Arabidopsis thaliana. Genes, 14(2), 282. https://doi.org/10.3390/genes14020282






