Altered Nucleotide Insertion Mechanisms of Disease-Associated TERT Variants
Abstract
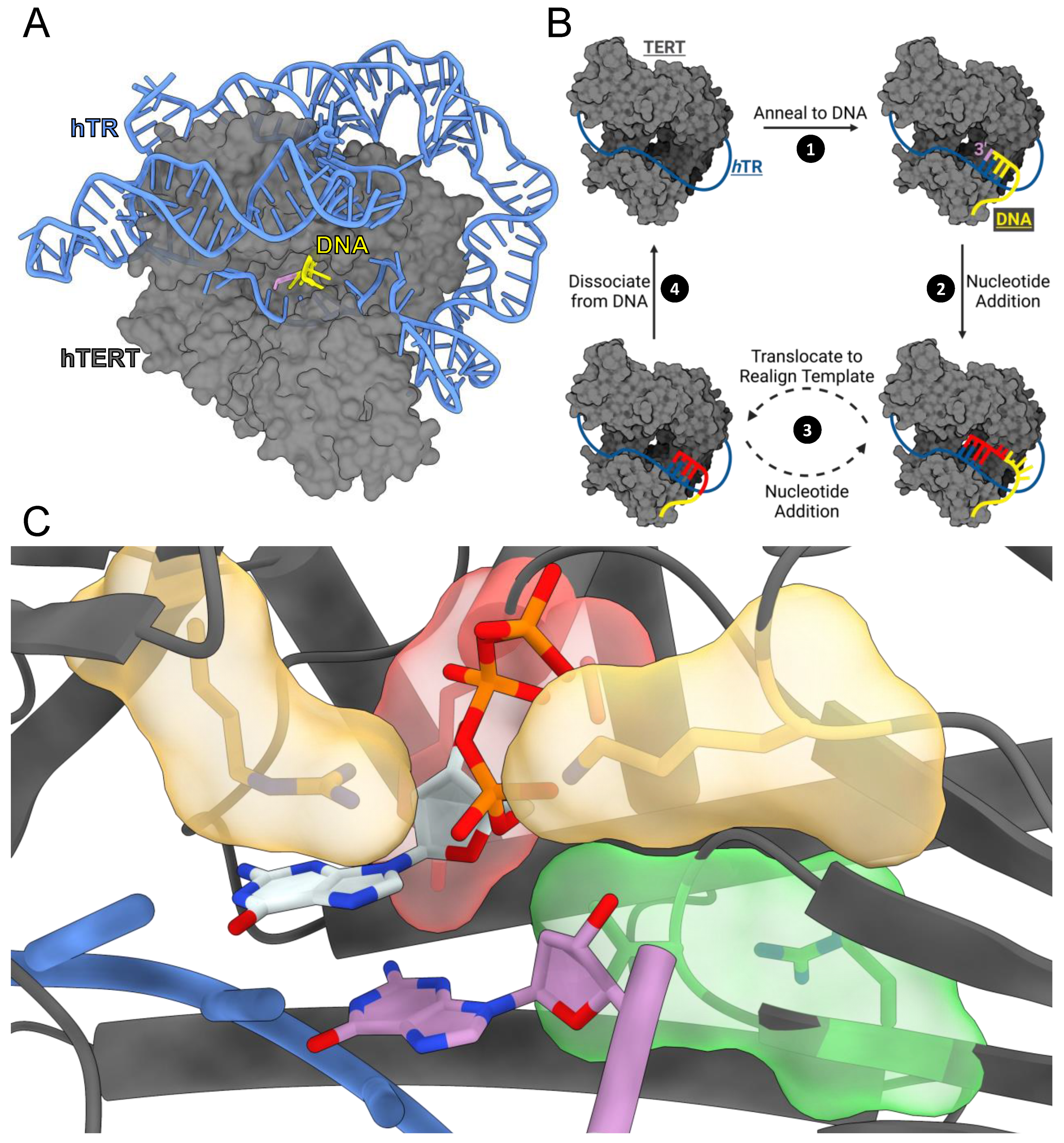
1. Introduction
2. Materials and Methods
2.1. Nucleic Acid Sequences
2.2. Expression and Purification of tcTERT
2.3. tcTERT Single-Turnover Kinetics
2.4. Computer Simulations
3. Results
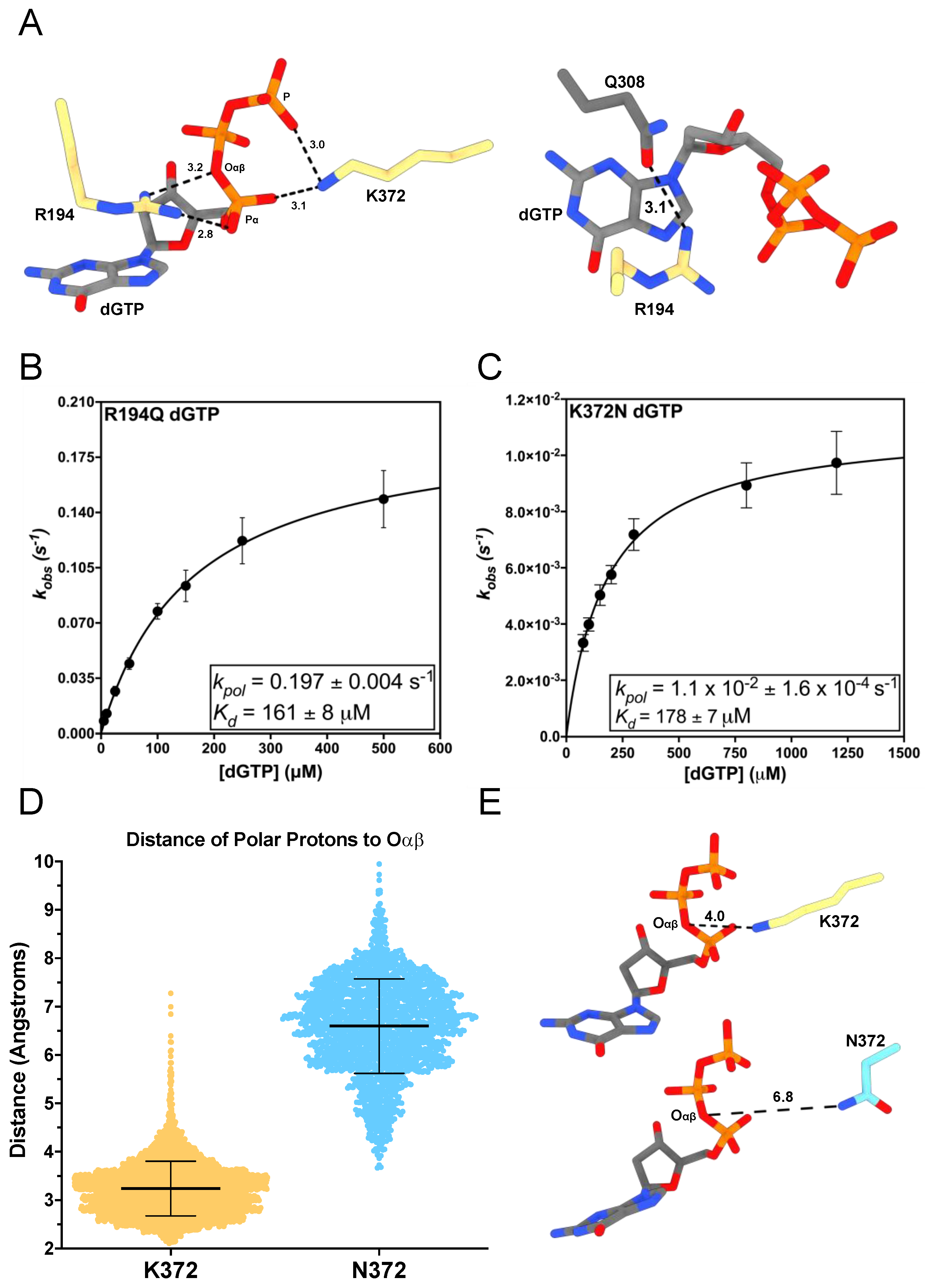
3.1. R194Q and K372N Disrupt the Electrostatic Environment of the dNTP’s Triphosphate
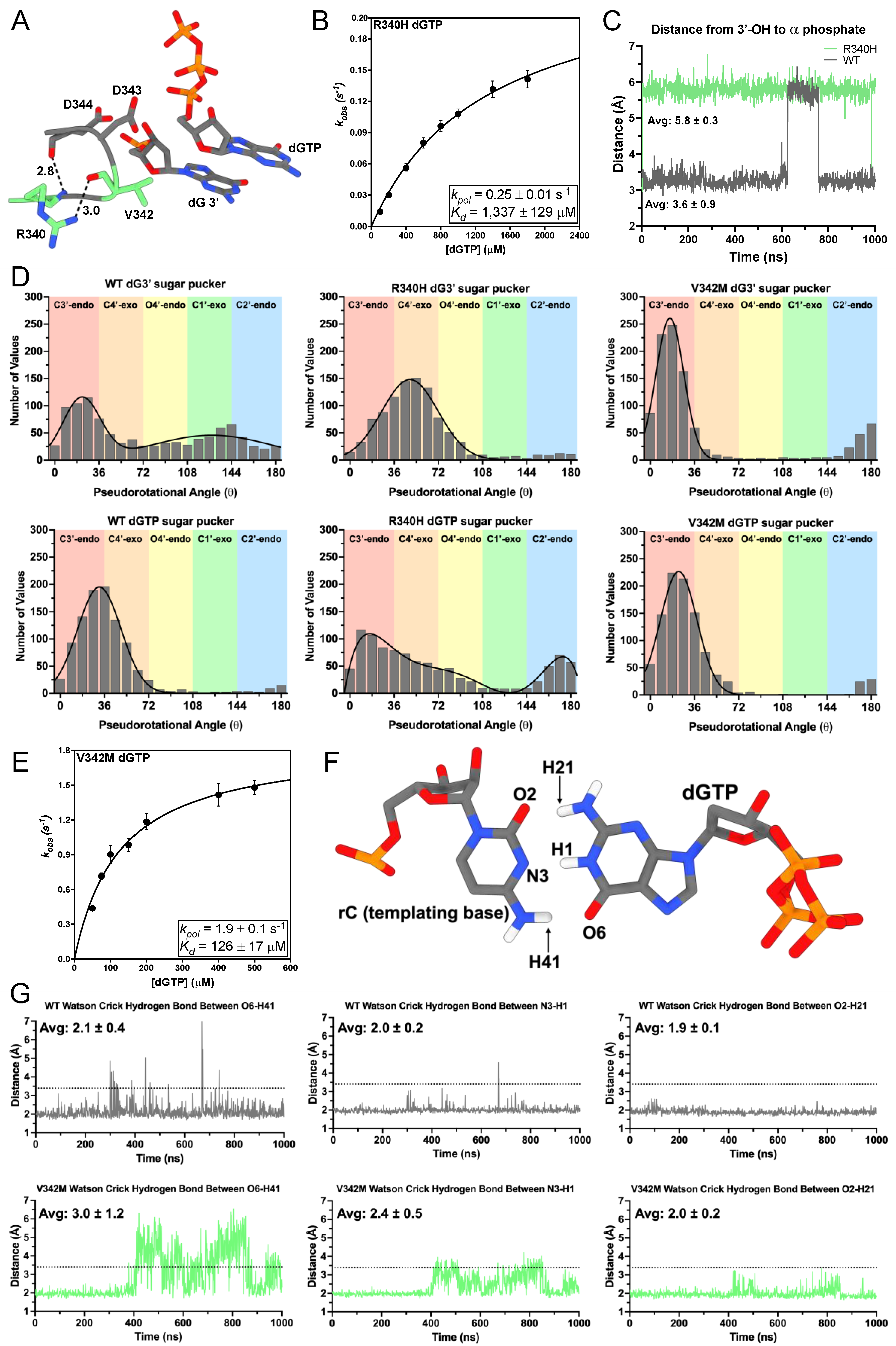
3.2. R340H and V342M Disrupt the DNA 3′-Terminus
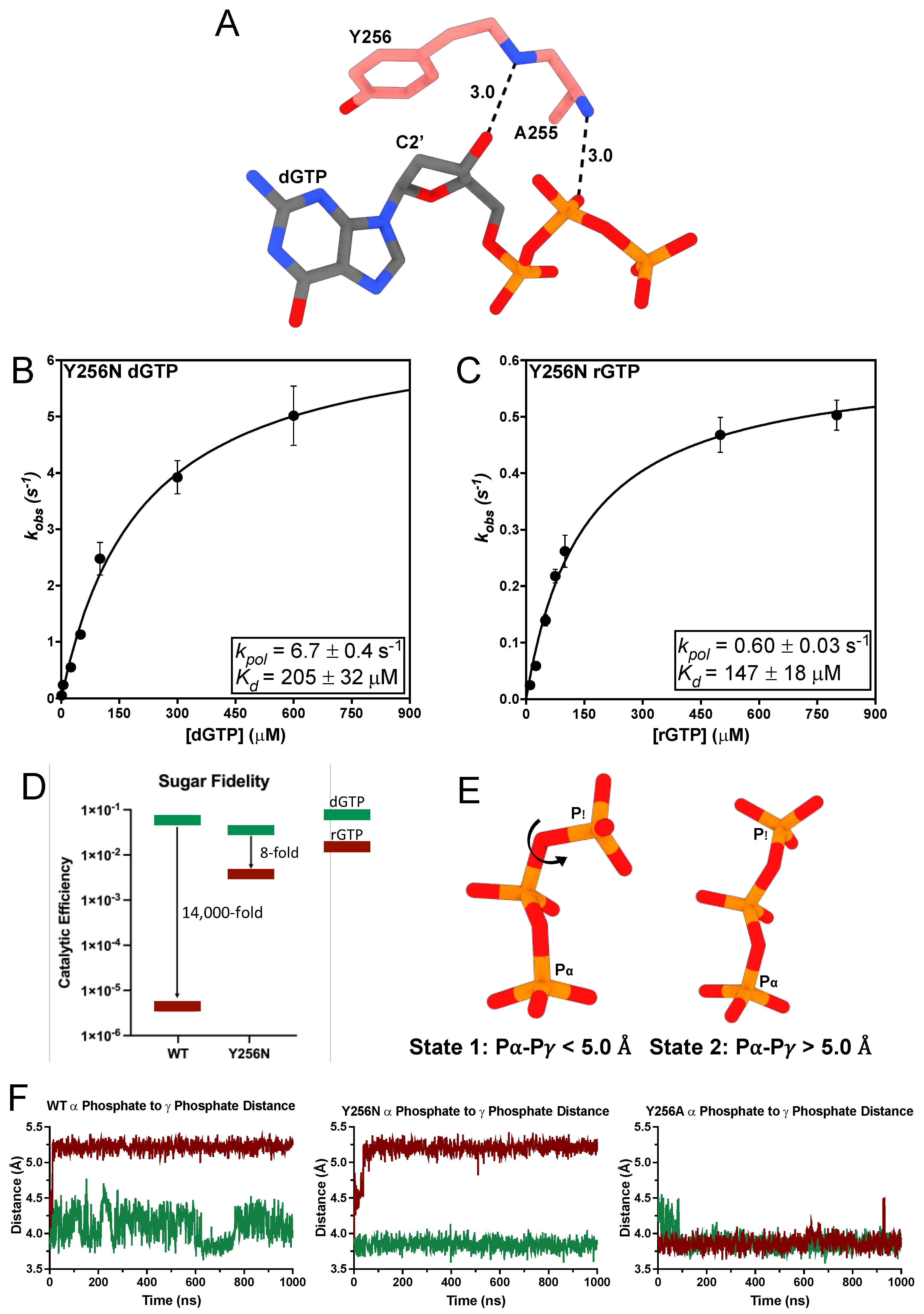
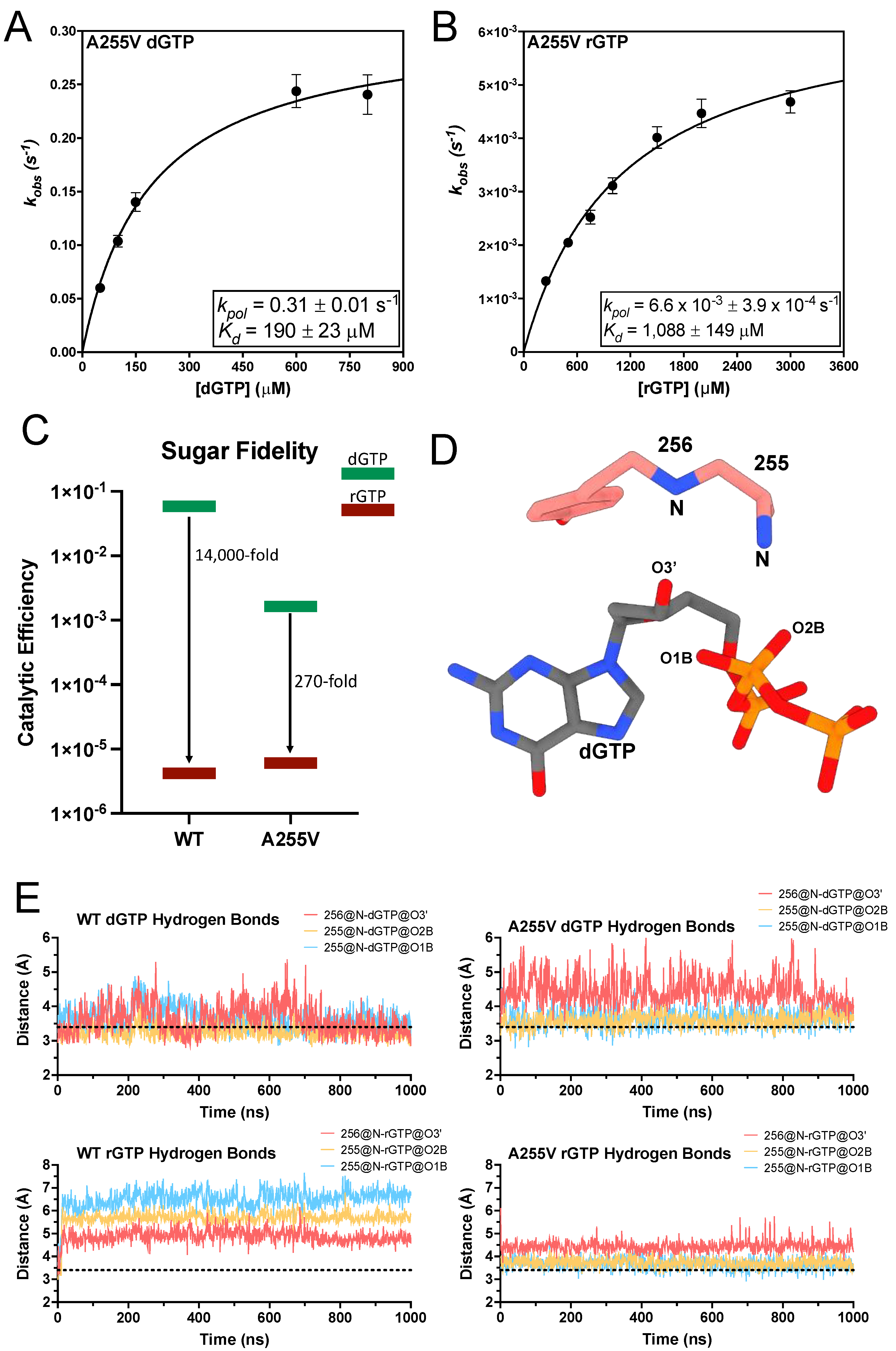
3.3. A255V and Y256N Decrease TERT Sugar Fidelity through Distinct Mechanisms
| tcTERT Variant | Incoming Nucleotide | kpol (s−1) | Kd (μM) | Catalytic Efficiency (μM−1 s−1) |
|---|---|---|---|---|
| R194Q | dGTP | 0.197 ± 0.004 | 161 ± 8 | 1.2 × 10−3 ± 6.7 × 10−5 |
| A255V | dGTP | 0.31 ± 0.01 | 190 ± 23 | 1.6 × 10−3 ± 2.1 × 10−4 |
| A255V | rGTP | 6.6 × 10−3 ± 3.9 × 10−4 | 1088 ± 149 | 6.1 × 10−6 ± 3.7 × 10−6 |
| Y256N | dGTP | 6.7 ± 0.4 | 205 ± 32 | 3.3 × 10−2 ± 5.5 × 10−3 |
| Y256N | rGTP | 0.60 ± 0.03 | 147 ± 18 | 4.1 × 10−3 ± 5.2 × 10−4 |
| R340H | dGTP | 0.25 ± 0.01 | 1337 ± 129 | 1.9 × 10−4 ± 4.0 × 10−5 |
| V342M | dGTP | 1.9 ± 0.1 | 126 ± 17 | 1.5 × 10−2 ± 2.1 × 10−3 |
| K372N | dGTP | 10−2 ± 1.6 × 10−4 | 178 ± 8 | 6.2 × 10−5 ± 2.8 × 10−6 |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, A.M. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 1973, 41, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, L. The Limited In Vitro Lifetime of Human Diploid Cell Strains. Exp. Cell Res. 1965, 37, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Reddel, R.R. The role of senescence and immortalization in carcinogenesis. Carcinogenesis 2000, 21, 477–484. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Barranco, C. Alternative splicing regulates telomerase repression. Nat. Rev. Genet. 2021, 22, 414. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Shay, J.W.; Bacchetti, S. A survey of telomerase activity in human cancer. Eur. J. Cancer 1997, 33, 787–791. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Grill, S.; Nandakumar, J. Molecular mechanisms of telomere biology disorders. J. Biol. Chem. 2021, 296, 100064. [Google Scholar] [CrossRef] [PubMed]
- Savage, S.A.; Bertuch, A.A. The genetics and clinical manifestations of telomere biology disorders. Genet. Med. 2010, 12, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Armanios, M.; Chen, J.L.; Chang, Y.P.; Brodsky, R.A.; Hawkins, A.; Griffin, C.A.; Eshleman, J.R.; Cohen, A.R.; Chakravarti, A.; Hamosh, A.; et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl. Acad. Sci. USA 2005, 102, 15960–15964. [Google Scholar] [CrossRef]
- Armanios, M.Y.; Chen, J.J.; Cogan, J.D.; Alder, J.K.; Ingersoll, R.G.; Markin, C.; Lawson, W.E.; Xie, M.; Vulto, I.; Phillips, J.A., 3rd; et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007, 356, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Bertuch, A.A. The molecular genetics of the telomere biology disorders. RNA Biol. 2016, 13, 696–706. [Google Scholar] [CrossRef]
- Nguyen, T.H.D.; Tam, J.; Wu, R.A.; Greber, B.J.; Toso, D.; Nogales, E.; Collins, K. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 2018, 557, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Ghanim, G.E.; Fountain, A.J.; van Roon, A.M.; Rangan, R.; Das, R.; Collins, K.; Nguyen, T.H.D. Structure of human telomerase holoenzyme with bound telomeric DNA. Nature 2021, 593, 449–453. [Google Scholar] [CrossRef]
- Blackburn, E.H.; Collins, K. Telomerase: An RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2011, 3, a003558. [Google Scholar] [CrossRef]
- Wan, F.; Ding, Y.; Zhang, Y.; Wu, Z.; Li, S.; Yang, L.; Yan, X.; Lan, P.; Li, G.; Wu, J.; et al. Zipper head mechanism of telomere synthesis by human telomerase. Cell Res. 2021, 31, 1275–1290. [Google Scholar] [CrossRef]
- Cohen, S.B.; Graham, M.E.; Lovrecz, G.O.; Bache, N.; Robinson, P.J.; Reddel, R.R. Protein composition of catalytically active human telomerase from immortal cells. Science 2007, 315, 1850–1853. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. The telomere terminal transferase of Tetrahymena is a ribonucleoprotein enzyme with two kinds of primer specificity. Cell 1987, 51, 887–898. [Google Scholar] [CrossRef]
- Schaich, M.A.; Sanford, S.L.; Welfer, G.A.; Johnson, S.A.; Khoang, T.H.; Opresko, P.L.; Freudenthal, B.D. Mechanisms of nucleotide selection by telomerase. eLife 2020, 9, e55438. [Google Scholar] [CrossRef]
- Chen, J.L.; Greider, C.W. Template boundary definition in mammalian telomerase. Genes Dev. 2003, 17, 2747–2752. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.J.; Akiyama, B.M.; Stone, M.D.; Cech, T.R. The RNA accordion model for template positioning by telomerase RNA during telomeric DNA synthesis. Nat. Struct. Mol. Biol. 2011, 18, 1371–1375. [Google Scholar] [CrossRef]
- Wu, R.A.; Upton, H.E.; Vogan, J.M.; Collins, K. Telomerase Mechanism of Telomere Synthesis. Annu. Rev. Biochem. 2017, 86, 439–460. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, C.G.; Sasaki, N.; Jurczyluk, J.; Bryan, T.M.; Cohen, S.B. Quantitative assays for measuring human telomerase activity and DNA binding properties. Methods 2017, 114, 85–95. [Google Scholar] [CrossRef]
- Sun, D.; Lopez-Guajardo, C.C.; Quada, J.; Hurley, L.H.; Von Hoff, D.D. Regulation of catalytic activity and processivity of human telomerase. Biochemistry 1999, 38, 4037–4044. [Google Scholar] [CrossRef] [PubMed]
- Zaug, A.J.; Crary, S.M.; Jesse Fioravanti, M.; Campbell, K.; Cech, T.R. Many disease-associated variants of hTERT retain high telomerase enzymatic activity. Nucleic Acids Res. 2013, 41, 8969–8978. [Google Scholar] [CrossRef]
- Bagshaw, C.R.; Hentschel, J.; Stone, M.D. The Processivity of Telomerase: Insights from Kinetic Simulations and Analyses. Molecules 2021, 26, 7532. [Google Scholar] [CrossRef]
- Palka, C.; Forino, N.M.; Hentschel, J.; Das, R.; Stone, M.D. Folding heterogeneity in the essential human telomerase RNA three-way junction. RNA 2020, 26, 1787–1800. [Google Scholar] [CrossRef]
- Hentschel, J. Multiplexed single-molecule analysis of human telomerase synthesizing DNA. bioRxiv 2021. [Google Scholar] [CrossRef]
- Powers, K.T.; Washington, M.T. Analyzing the Catalytic Activities and Interactions of Eukaryotic Translesion Synthesis Polymerases. Methods Enzymol. 2017, 592, 329–356. [Google Scholar] [CrossRef] [PubMed]
- Gillis, A.J.; Schuller, A.P.; Skordalakes, E. Structure of the Tribolium castaneum telomerase catalytic subunit TERT. Nature 2008, 455, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Wang, Y.; Susac, L.; Chan, H.; Basu, R.; Zhou, Z.H.; Feigon, J. Structure of Telomerase with Telomeric DNA. Cell 2018, 173, 1179–1190.e1113. [Google Scholar] [CrossRef]
- Petrova, O.A.; Mantsyzov, A.B.; Rodina, E.V.; Efimov, S.V.; Hackenberg, C.; Hakanpaa, J.; Klochkov, V.V.; Lebedev, A.A.; Chugunova, A.A.; Malyavko, A.N.; et al. Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase. Nucleic Acids Res. 2018, 46, 1525–1540. [Google Scholar] [CrossRef]
- Choi, W.S.; Weng, P.J.; Yang, W. Flexibility of telomerase in binding the RNA template and DNA telomeric repeat. Proc. Natl. Acad. Sci. USA 2022, 119, e2116159118. [Google Scholar] [CrossRef]
- He, Y.; Wang, Y.; Liu, B.; Helmling, C.; Susac, L.; Cheng, R.; Zhou, Z.H.; Feigon, J. Structures of telomerase at several steps of telomere repeat synthesis. Nature 2021, 593, 454–459. [Google Scholar] [CrossRef]
- Wang, Y.; Gallagher-Jones, M.; Susac, L.; Song, H.; Feigon, J. A structurally conserved human and Tetrahymena telomerase catalytic core. Proc. Natl. Acad. Sci. USA 2020, 117, 31078–31087. [Google Scholar] [CrossRef]
- Liu, B.; He, Y.; Wang, Y.; Song, H.; Zhou, Z.H.; Feigon, J. Structure of active human telomerase with telomere shelterin protein TPP1. Nature 2022, 604, 578–583. [Google Scholar] [CrossRef]
- Sekne, Z.; Ghanim, G.E.; van Roon, A.M.; Nguyen, T.H.D. Structural basis of human telomerase recruitment by TPP1-POT1. Science 2022, 375, 1173–1176. [Google Scholar] [CrossRef]
- Renders, M.; Frere, J.M.; Toye, D.; Herdewijn, P. Full Pre-Steady-State Kinetic Analysis of Single Nucleotide Incorporation by DNA Polymerases. Curr. Protoc. Nucleic Acid Chem. 2019, 78, e98. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Peter Guengerich, F. Pre-Steady-State Kinetic Analysis of Single-Nucleotide Incorporation by DNA Polymerases. Curr. Protoc. Nucleic Acid Chem. 2016, 65, 7.23.1–7.23.10. [Google Scholar] [CrossRef] [PubMed]
- de Leon, A.D.; Cronkhite, J.T.; Katzenstein, A.L.; Godwin, J.D.; Raghu, G.; Glazer, C.S.; Rosenblatt, R.L.; Girod, C.E.; Garrity, E.R.; Xing, C.; et al. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS ONE 2010, 5, e10680. [Google Scholar] [CrossRef] [PubMed]
- Norberg, A.; Rosen, A.; Raaschou-Jensen, K.; Kjeldsen, L.; Moilanen, J.S.; Paulsson-Karlsson, Y.; Baliakas, P.; Lohi, O.; Ahmed, A.; Kittang, A.O.; et al. Novel variants in Nordic patients referred for genetic testing of telomere-related disorders. Eur. J. Hum. Genet. 2018, 26, 858–867. [Google Scholar] [CrossRef] [PubMed]
- Du, H.Y.; Pumbo, E.; Ivanovich, J.; An, P.; Maziarz, R.T.; Reiss, U.M.; Chirnomas, D.; Shimamura, A.; Vlachos, A.; Lipton, J.M.; et al. TERC and TERT gene mutations in patients with bone marrow failure and the significance of telomere length measurements. Blood 2009, 113, 309–316. [Google Scholar] [CrossRef]
- Vulliamy, T.J.; Kirwan, M.J.; Beswick, R.; Hossain, U.; Baqai, C.; Ratcliffe, A.; Marsh, J.; Walne, A.; Dokal, I. Differences in disease severity but similar telomere lengths in genetic subgroups of patients with telomerase and shelterin mutations. PLoS ONE 2011, 6, e24383. [Google Scholar] [CrossRef]
- Seshagiri, S.; Stawiski, E.W.; Durinck, S.; Modrusan, Z.; Storm, E.E.; Conboy, C.B.; Chaudhuri, S.; Guan, Y.; Janakiraman, V.; Jaiswal, B.S.; et al. Recurrent R-spondin fusions in colon cancer. Nature 2012, 488, 660–664. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Tsakiri, K.D.; Cronkhite, J.T.; Kuan, P.J.; Xing, C.; Raghu, G.; Weissler, J.C.; Rosenblatt, R.L.; Shay, J.W.; Garcia, C.K. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc. Natl. Acad. Sci. USA 2007, 104, 7552–7557. [Google Scholar] [CrossRef]
- Beard, W.A.; Shock, D.D.; Batra, V.K.; Prasad, R.; Wilson, S.H. Substrate-induced DNA polymerase β activation. J. Biol. Chem. 2014, 289, 31411–31422. [Google Scholar] [CrossRef]
- Weaver, T.M.; Cortez, L.M.; Khoang, T.H.; Washington, M.T.; Agarwal, P.K.; Freudenthal, B.D. Visualizing Rev1 catalyze protein-template DNA synthesis. Proc. Natl. Acad. Sci. USA 2020, 117, 25494–25504. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Berryman, J.T.; Betz, R.M.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Götz, A.W.; et al. AMBER 15; University of California: San Francisco, CA, USA, 2015. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Savol, A.J.; Agarwal, P.K.; Chennubhotla, C.S. Event detection and sub-state discovery from biomolecular simulations using higher-order statistics: Application to enzyme adenylate kinase. Proteins 2012, 80, 2536–2551. [Google Scholar] [CrossRef] [PubMed]
- Gagne, D.; Narayanan, C.; Nguyen-Thi, N.; Roux, L.D.; Bernard, D.N.; Brunzelle, J.S.; Couture, J.F.; Agarwal, P.K.; Doucet, N. Ligand Binding Enhances Millisecond Conformational Exchange in Xylanase B2 from Streptomyces lividans. Biochemistry 2016, 55, 4184–4196. [Google Scholar] [CrossRef]
- Hoitsma, N.M.; Click, T.H.; Agarwal, P.K.; Freudenthal, B.D. Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker. Comput. Struct. Biotechnol. J. 2021, 19, 3293–3302. [Google Scholar] [CrossRef]
- Hoitsma, N.M.; Whitaker, A.M.; Beckwitt, E.C.; Jang, S.; Agarwal, P.K.; Van Houten, B.; Freudenthal, B.D. AP-endonuclease 1 sculpts DNA through an anchoring tyrosine residue on the DNA intercalating loop. Nucleic Acids Res. 2020, 48, 7345–7355. [Google Scholar] [CrossRef]
- Ramanathan, A.; Agarwal, P.K. Evolutionarily Conserved Linkage between Enzyme Fold, Flexibility, and Catalysis. PLoS Biol. 2011, 9, e1001193. [Google Scholar] [CrossRef]
- Duff, M.R., Jr.; Borreguero, J.M.; Cuneo, M.J.; Ramanathan, A.; He, J.; Kamath, G.; Chennubhotla, S.C.; Meilleur, F.; Howell, E.E.; Herwig, K.W. Modulating enzyme activity by altering protein dynamics with solvent. Biochemistry 2018, 57, 4263–4275. [Google Scholar] [CrossRef]
- Beck, D.A.; Daggett, V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods 2004, 34, 112–120. [Google Scholar] [CrossRef]
- Shukla, S.; Bafna, K.; Gullett, C.; Myles, D.A.; Agarwal, P.K.; Cuneo, M.J. Differential substrate recognition by maltose binding proteins influenced by structure and dynamics. Biochemistry 2018, 57, 5864–5876. [Google Scholar] [CrossRef]
- Nakamura, T.; Zhao, Y.; Yamagata, Y.; Hua, Y.J.; Yang, W. Watching DNA polymerase eta make a phosphodiester bond. Nature 2012, 487, 196–201. [Google Scholar] [CrossRef]
- Freudenthal, B.D.; Beard, W.A.; Shock, D.D.; Wilson, S.H. Observing a DNA polymerase choose right from wrong. Cell 2013, 154, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Delagoutte, E. DNA polymerases: Mechanistic insight from biochemical and biophysical studies. Front. Biosci. 2012, 17, 509–544. [Google Scholar] [CrossRef]
- Castro, C.; Smidansky, E.; Maksimchuk, K.R.; Arnold, J.J.; Korneeva, V.S.; Gotte, M.; Konigsberg, W.; Cameron, C.E. Two proton transfers in the transition state for nucleotidyl transfer catalyzed by RNA- and DNA-dependent RNA and DNA polymerases. Proc. Natl. Acad. Sci. USA 2007, 104, 4267–4272. [Google Scholar] [CrossRef] [PubMed]
- Castro, C.; Smidansky, E.D.; Arnold, J.J.; Maksimchuk, K.R.; Moustafa, I.; Uchida, A.; Gotte, M.; Konigsberg, W.; Cameron, C.E. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009, 16, 212–218. [Google Scholar] [CrossRef]
- Newton, C.A.; Batra, K.; Torrealba, J.; Kozlitina, J.; Glazer, C.S.; Aravena, C.; Meyer, K.; Raghu, G.; Collard, H.R.; Garcia, C.K. Telomere-related lung fibrosis is diagnostically heterogeneous but uniformly progressive. Eur. Respir. J. 2016, 48, 1710–1720. [Google Scholar] [CrossRef]
- Justet, A.; Klay, D.; Porcher, R.; Cottin, V.; Ahmad, K.; Molina Molina, M.; Nunes, H.; Reynaud-Gaubert, M.; Naccache, J.M.; Manali, E.; et al. Safety and efficacy of pirfenidone and nintedanib in patients with idiopathic pulmonary fibrosis and carrying a telomere-related gene mutation. Eur. Respir. J. 2021, 57, 2003198. [Google Scholar] [CrossRef] [PubMed]
- Altona, C.; Sundaralingam, M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. J. Am. Chem. Soc. 1973, 95, 2333–2344. [Google Scholar] [CrossRef] [PubMed]
- Altona, C.; Sundaralingam, M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972, 94, 8205–8212. [Google Scholar] [CrossRef]
- Williams, A.A.; Darwanto, A.; Theruvathu, J.A.; Burdzy, A.; Neidigh, J.W.; Sowers, L.C. Impact of sugar pucker on base pair and mispair stability. Biochemistry 2009, 48, 11994–12004. [Google Scholar] [CrossRef]
- Cavanaugh, N.A.; Beard, W.A.; Wilson, S.H. DNA polymerase β ribonucleotide discrimination: Insertion, misinsertion, extension, and coding. J. Biol. Chem. 2010, 285, 24457–24465. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.; Greider, C.W. Utilization of ribonucleotides and RNA primers by Tetrahymena telomerase. EMBO J. 1995, 14, 5422–5432. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Suo, Z. Unlocking the sugar “steric gate” of DNA polymerases. Biochemistry 2011, 50, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Genna, V.; Gaspari, R.; Dal Peraro, M.; De Vivo, M. Cooperative motion of a key positively charged residue and metal ions for DNA replication catalyzed by human DNA Polymerase-eta. Nucleic Acids Res. 2016, 44, 2827–2836. [Google Scholar] [CrossRef] [PubMed]
- Drosopoulos, W.C.; Prasad, V.R. The active site residue Valine 867 in human telomerase reverse transcriptase influences nucleotide incorporation and fidelity. Nucleic Acids Res. 2007, 35, 1155–1168. [Google Scholar] [CrossRef]
- Johnson, M.K.; Kottur, J.; Nair, D.T. A polar filter in DNA polymerases prevents ribonucleotide incorporation. Nucleic Acids Res. 2019, 47, 10693–10705. [Google Scholar] [CrossRef]
| hTERT Residue | tcTERT Residue | Disease Association | Reference |
|---|---|---|---|
| R631Q | R194Q | IPF | [43,44] |
| A716V | A255V | AA, IPF, Cancer | [45,46,47] |
| Y717N | Y256N | Cancer | [48] |
| R865H | R340H | AA, IPF | [49] |
| V867M | V342M | PF, DC | [43] |
| K902N | K372N | DC | [13] |
| D868 | D343 | ||
| D869 | D344 | ||
| Q833 | Q308 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Welfer, G.A.; Borin, V.A.; Cortez, L.M.; Opresko, P.L.; Agarwal, P.K.; Freudenthal, B.D. Altered Nucleotide Insertion Mechanisms of Disease-Associated TERT Variants. Genes 2023, 14, 281. https://doi.org/10.3390/genes14020281
Welfer GA, Borin VA, Cortez LM, Opresko PL, Agarwal PK, Freudenthal BD. Altered Nucleotide Insertion Mechanisms of Disease-Associated TERT Variants. Genes. 2023; 14(2):281. https://doi.org/10.3390/genes14020281
Chicago/Turabian StyleWelfer, Griffin A., Veniamin A. Borin, Luis M. Cortez, Patricia L. Opresko, Pratul K. Agarwal, and Bret D. Freudenthal. 2023. "Altered Nucleotide Insertion Mechanisms of Disease-Associated TERT Variants" Genes 14, no. 2: 281. https://doi.org/10.3390/genes14020281
APA StyleWelfer, G. A., Borin, V. A., Cortez, L. M., Opresko, P. L., Agarwal, P. K., & Freudenthal, B. D. (2023). Altered Nucleotide Insertion Mechanisms of Disease-Associated TERT Variants. Genes, 14(2), 281. https://doi.org/10.3390/genes14020281





