Case Report: Rare IKZF1 Gene Fusions Identified in Neonate with Congenital KMT2A-Rearranged Acute Lymphoblastic Leukemia
Abstract
1. Introduction
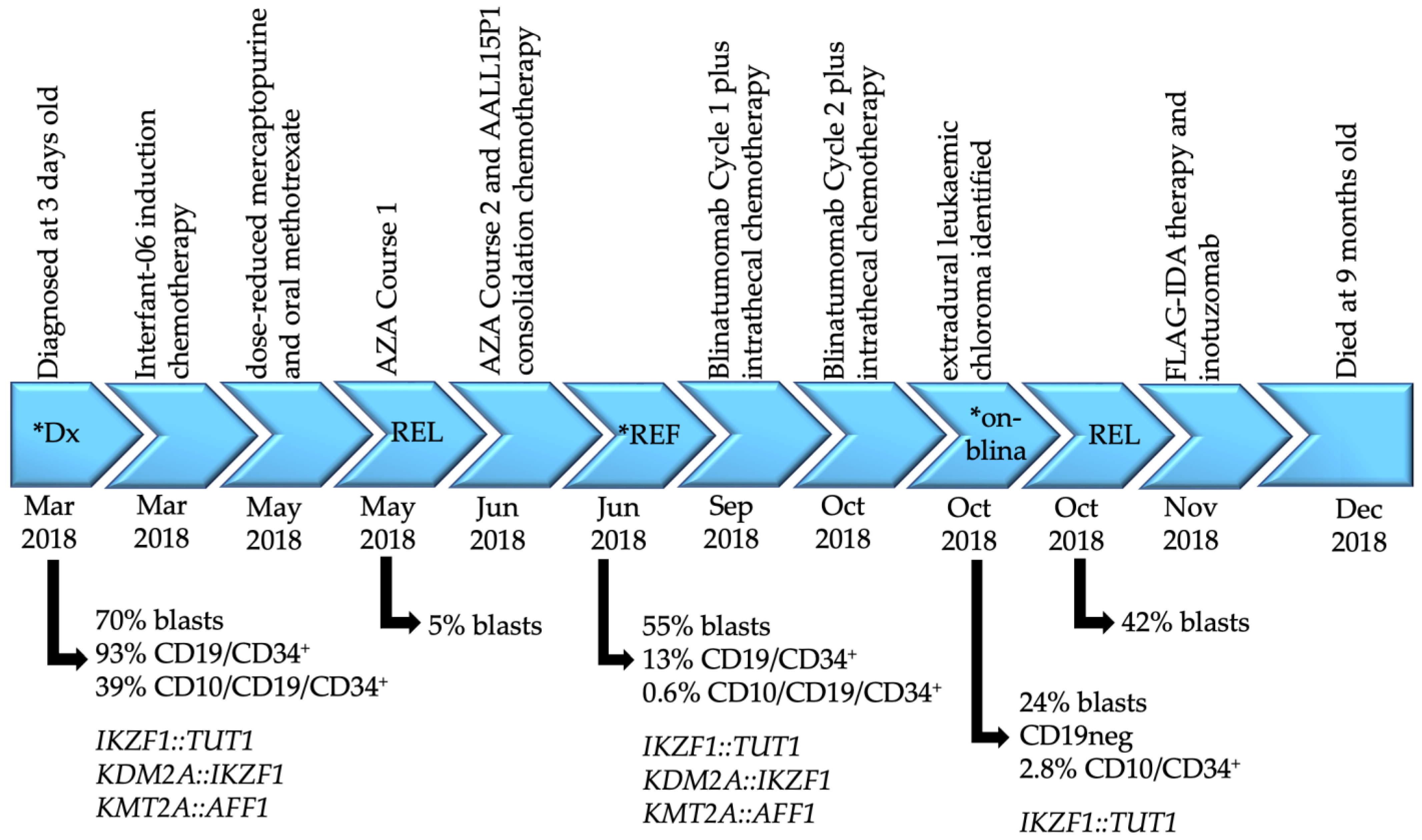
2. Case Report
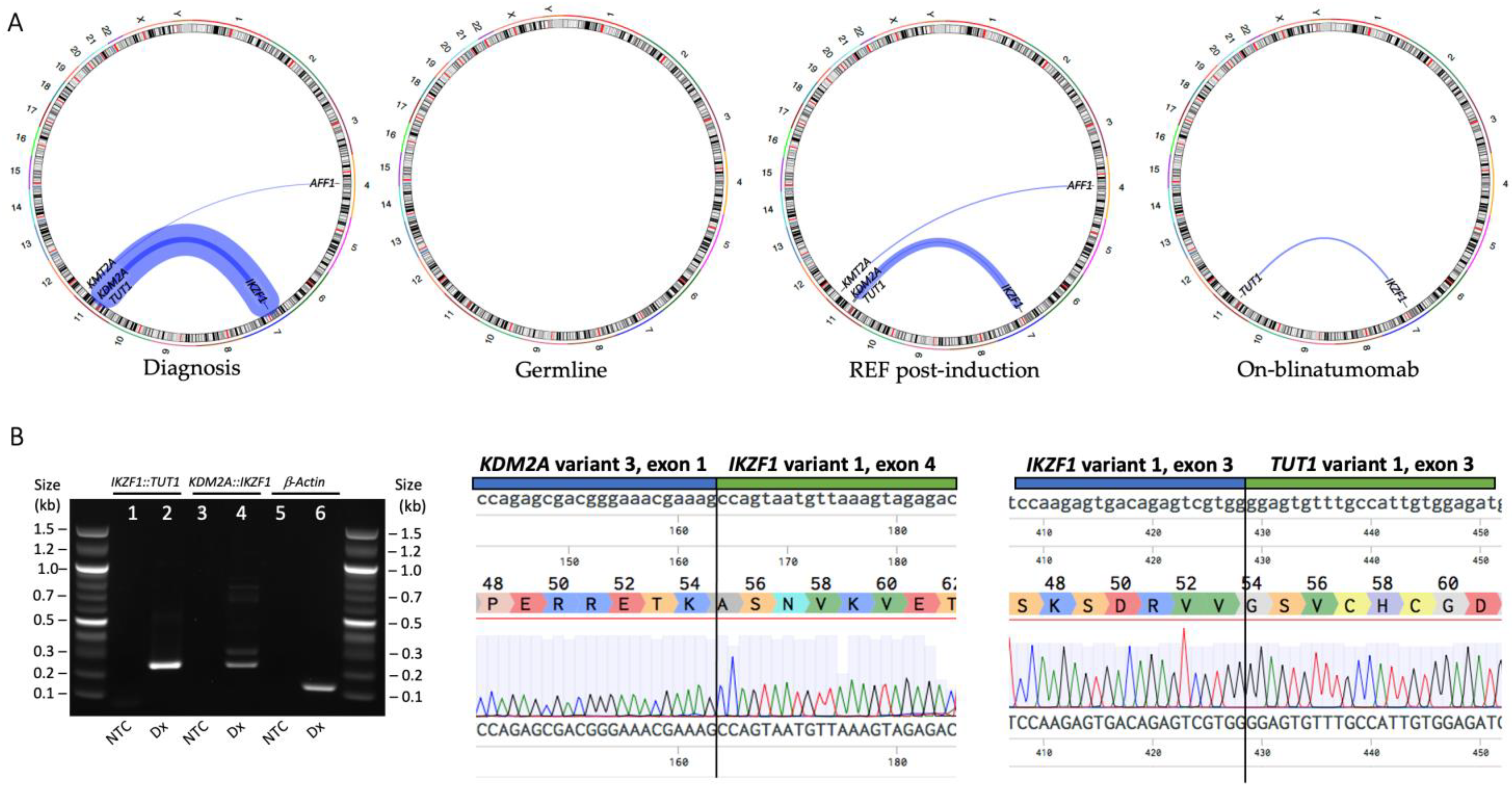
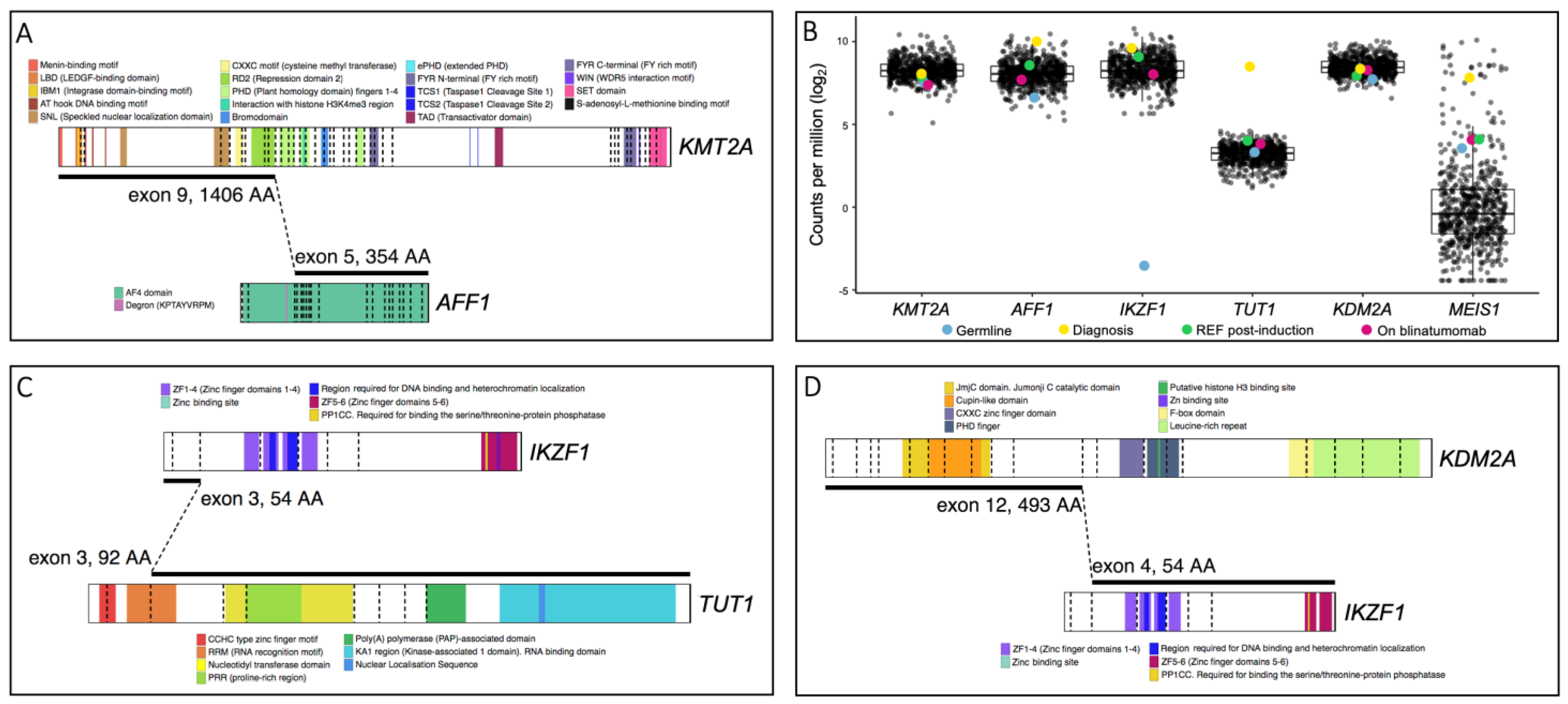
3. Results
| Fusion | Chromosome Abnormality | Disease | Number of Cases | Description | IKZF1 Exon Retention | Predicted Function | Ref |
|---|---|---|---|---|---|---|---|
| DNAH14::IKZF1 | t(1;7)(q42;p12) | B-ALL | 1 | in-frame; exon 36 to exon 4 ZNF2-4 domains (DNA-binding function) and ZNF5-6 domains containing dimerization sites are truncated | exons 1–4 | Similar to the IK6 isoform, loss-of-function allele | [24] |
| ETV6::IKZF1 | t(7;12)(p12;p13) | B-ALL | 1 | out-of-frame; intron 2 to intron 3 No functional IKZF1 domains are present | exons 1–3 | Likely abolishes the function of Ikaros protein | [18] |
| FIGNL1::IKZF1 | t(7;7)(p12;p12) | B-ALL | 1 | out-of-frame; exon 4 to exon 4* Majority of functional IKZF1 domains are retained; ZNF1 domain (DNA-binding function) is truncated | exons 5–8 | Altered transcriptional regulation | [25] |
| IKZF1::CDK2 | t(7;12)(p12;q13) | B-ALL | 1 | out-of-frame; exon 3, no CDK2 breakpoint details provided No functional Ikaros domains are present | exons 1–3 | Likely abolishes the function of Ikaros protein | [26] |
| IKZF1::ETV6 | t(7;12)(p12;p13) | B-ALL | 1 | out-of-frame; intron 3 to intron 2 No functional IKZF1 domains are present | exons 1–3 | Likely abolishes the function of Ikaros protein | [18] |
| IKZF1::FIGNL1 | t(7;7)(p12;p12) | B-ALL | 2 | out-of-frame; intron 3 to 5′ UTR exon 2 and exon 3 to 13691 bp downstream No functional IKZF1 domains are present | exons 1–3 | Likely abolish the function of Ikaros protein | [18,27] |
| IKZF1::NUTM1 | t(7;15)(p12;q14) | B-ALL | 1 | in-frame; exon 7 to exon 2 Some functional IKZF1 domains are retained; ZNF5-6 domains containing dimerization sites are truncated | exons 1–7 | Altered transcriptional regulation | [26] |
| IKZF1::SETD5 | t(3;7)(p25;p12) | B-ALL | 1 # | in-frame; exon 3, no SETD5 breakpoint details provided No functional IKZF1 domains are present | exons 1–3 | Likely abolishes the function of Ikaros protein | [26] |
| IKZF1::TRPV2 | t(7;17)(p12;p11) | B-ALL | 1 | out-of-frame; no exon/intron details provided No functional IKZF1 domains are present | breakpoint not specified | Likely abolishes the function of Ikaros protein | [26] |
| IKZF1::ZEB2 | t(2;7)(q22;p12) | B-ALL | 1 | in-frame; exon 3 to exon 5 No functional IKZF1 domains are present | exons 1–3 | Likely abolishes the function of Ikaros protein | [18] |
| SETD5::IKZF1 | t(3;7)(p25;p12) | B-ALL | 1 # | in-frame; no SETD5 breakpoint details provided, exon 4 All functional IKZF1 domains are retained | exons 4–8 | Altered transcriptional regulation | [26] |
| STIM2::IKZF1 | t(4;7)(p15;p12) | B-ALL | 1 | No fusions details provided | breakpoint not specified | No fusion details provided | [28] |
| IKZF1::ABCA13 | t(7;7)(p12;p12) | T-ALL | 1 | out-of-frame; exon to intron Some functional IKZF1 domains are retained; ZNF4 domain (DNA-binding function) and ZNF5-6 domains containing dimerization sites are truncated | exons 1–5 | Reduced expression of IKZF1; likely abolishes the function of Ikaros protein | [29] |
| IKZF1::NOTCH1 | t(7;9)(p12;q34) | T-ALL | 1 | in-frame; exon to exon Some functional IKZF1 domains are retained; ZNF4 domain (DNA-binding function) and ZNF5-6 domains containing dimerization sites are truncated | exons 1–5 | Altered transcriptional regulation | [30] |
| IKZF1::NOTCH1 | t(7;9)(p12;q34) | T-ALL | 1 $ | out-of-frame; intron to intron Some functional IKZF1 domains are retained; ZNF4 domain (DNA-binding function) and ZNF5-6 domains containing dimerization sites are truncated | exons 1–5 | Altered transcriptional regulation | [27] |
| NOTCH1::IKZF1 | t(7;9)(p12;q34) | T-ALL | 1 $ | out-of-frame; exon to intron Some functional IKZF1 domains are retained; ZNF1-4 domains containing dimerization sites are truncated | exons 6–8 | Altered transcriptional regulation | [27] |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Forgione, M.O.; McClure, B.J.; Eadie, L.N.; Yeung, D.T.; White, D.L. KMT2A rearranged acute lymphoblastic leukaemia: Unravelling the genomic complexity and heterogeneity of this high-risk disease. Cancer Lett. 2020, 469, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Winters, A.C.; Bernt, K.M. MLL-Rearranged Leukemias—An Update on Science and Clinical Approaches. Front. Pediatr. 2017, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Tauchi, H.; Tomizawa, D.; Eguchi, M.; Eguchi-Ishimae, M.; Koh, K.; Hirayama, M.; Miyamura, N.; Kinukawa, N.; Hayashi, Y.; Horibe, K.; et al. Clinical features and outcome of MLL gene rearranged acute lymphoblastic leukemia in infants with additional chromosomal abnormalities other than 11q23 translocation. Leuk. Res. 2008, 32, 1523–1529. [Google Scholar] [CrossRef]
- Sun, L.; Heerema, N.; Crotty, L.; Wu, X.; Navara, C.; Vassilev, A.; Sensel, M.; Reaman, G.H.; Uckun, F.M. Expression of dominant-negative and mutant isoforms of the antileukemic transcription factor Ikaros in infant acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 1999, 96, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.K.; Ma, J.; Wang, J.; Chen, X.; Gedman, A.L.; Dang, J.; Nakitandwe, J.; Holmfeldt, L.; Parker, M.; Easton, J.; et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat. Genet. 2015, 47, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Pieters, R.; Lorenzo, P.D.; Ancliffe, P.; Aversa, L.A.; Brethon, B.; Biondi, A.; Campbell, M.; Escherich, G.; Ferster, A.; Gardner, R.A.; et al. Outcome of Infants Younger Than 1 Year With Acute Lymphoblastic Leukemia Treated with the Interfant-06 Protocol: Results From an International Phase III Randomized Study. J. Clin. Oncol. 2019, 37, 2246–2256. [Google Scholar] [CrossRef] [PubMed]
- Guest, E.; Kairalla, J.; Devidas, M.; Hibbitts, E.; Carroll, A.J.; Heerema, N.A.; Kubaney, H.; August, A.; Pauly, M.; Wechsler, D.; et al. A Pilot Study of Azacitidine As Epigenetic Priming for Chemotherapy in Infants Less Than 1 Year of Age with KMT2A-Rearranged Acute Lymphoblastic Leukemia (ALL); Results from the Children’s Oncology Group (COG) Trial AALL15P1. Blood 2022, 140, 3256–3257. [Google Scholar] [CrossRef]
- Meyer, C.; Burmeister, T.; Gröger, D.; Tsaur, G.; Fechina, L.; Renneville, A.; Sutton, R.; Venn, N.C.; Emerenciano, M.; Pombo-de-Oliveira, M.S.; et al. The MLL recombinome of acute leukemias in 2017. Leukemia 2017, 32, 273. [Google Scholar] [CrossRef]
- Robinson, M.A. Linkage Disequilibrium. In Encyclopedia of Immunology, 2nd ed.; Delves, P.J., Ed.; Elsevier: Oxford, UK, 1998; pp. 1586–1588. [Google Scholar]
- Murai, M.J.; Pollock, J.; He, S.; Miao, H.; Purohit, T.; Yokom, A.; Hess, J.L.; Muntean, A.G.; Grembecka, J.; Cierpicki, T. The same site on the integrase-binding domain of lens epithelium-derived growth factor is a therapeutic target for MLL leukemia and HIV. Blood 2014, 124, 3730–3737. [Google Scholar] [CrossRef]
- Yokoyama, A.; Cleary, M.L. Menin Critically Links MLL Proteins with LEDGF on Cancer-Associated Target Genes. Cancer Cell 2008, 14, 36–46. [Google Scholar] [CrossRef]
- Ferrando, A.A.; Armstrong, S.A.; Neuberg, D.S.; Sallan, S.E.; Silverman, L.B.; Korsmeyer, S.J.; Look, A.T. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: Dominance of HOX dysregulation. Blood 2003, 102, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Montes, R.; Ayllón, V.; Gutierrez-Aranda, I.; Prat, I.; Hernández-Lamas, M.C.; Ponce, L.; Bresolin, S.; te Kronnie, G.; Greaves, M.; Bueno, C.; et al. Enforced expression of MLL-AF4 fusion in cord blood CD34+ cells enhances the hematopoietic repopulating cell function and clonogenic potential but is not sufficient to initiate leukemia. Blood 2011, 117, 4746–4758. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Edmonson, M.N.; Wilkinson, M.R.; Patel, A.; Wu, G.; Liu, Y.; Li, Y.; Zhang, Z.; Rusch, M.C.; Parker, M.; et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 2016, 48, 4–6. [Google Scholar] [CrossRef] [PubMed]
- López-Nieva, P.; Fernández-Navarro, P.; Graña-Castro, O.; Andrés-León, E.; Santos, J.; Villa-Morales, M.; Cobos-Fernández, M.Á.; González-Sánchez, L.; Malumbres, M.; Salazar-Roa, M.; et al. Detection of novel fusion-transcripts by RNA-Seq in T-cell lymphoblastic lymphoma. Sci. Rep. 2019, 9, 5179. [Google Scholar] [CrossRef]
- Georgopoulos, K.; Bigby, M.; Wang, J.-H.; Molnar, A.; Wu, P.; Winandy, S.; Sharpe, A. The ikaros gene is required for the development of all lymphoid lineages. Cell 1994, 79, 143–156. [Google Scholar] [CrossRef]
- Marke, R.; van Leeuwen, F.N.; Scheijen, B. The many faces of IKZF1 in B-cell precursor acute lymphoblastic leukemia. Haematologica 2018, 103, 565–574. [Google Scholar] [CrossRef]
- Ma, X.; Liu, Y.; Liu, Y.; Alexandrov, L.B.; Edmonson, M.N.; Gawad, C.; Zhou, X.; Li, Y.; Rusch, M.C.; Easton, J.; et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 2018, 555, 371. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Su, X.; Zhang, J.; Radtke, I.; Phillips, L.A.A.; Miller, C.B.; Ma, J.; Liu, W.; Cheng, C.; Schulman, B.A.; et al. Deletion of IKZF1 and Prognosis in Acute Lymphoblastic Leukemia. New Engl. J. Med. 2009, 360, 470–480. [Google Scholar] [CrossRef]
- Kuiper, R.P.; Waanders, E.; van der Velden, V.H.J.; van Reijmersdal, S.V.; Venkatachalam, R.; Scheijen, B.; Sonneveld, E.; van Dongen, J.J.M.; Veerman, A.J.P.; van Leeuwen, F.N.; et al. IKZF1 deletions predict relapse in uniformly treated pediatric precursor B-ALL. Leukemia 2010, 24, 1258–1264. [Google Scholar] [CrossRef]
- Waanders, E.; Van Der Velden, V.H.J.; Van Der Schoot, C.E.; Van Leeuwen, F.N.; Van Reijmersdal, S.V.; De Haas, V.; Veerman, A.J.; Van Kessel, A.G.; Hoogerbrugge, P.M.; Kuiper, R.P.; et al. Integrated use of minimal residual disease classification and IKZF1 alteration status accurately predicts 79% of relapses in pediatric acute lymphoblastic leukemia. Leukemia 2011, 25, 254–258. [Google Scholar] [CrossRef]
- Chiaretti, S.; Zini, G.; Bassan, R. Diagnosis and subclassification of acute lymphoblastic leukemia. Mediterr. J. Hematol. Infect. Dis. 2014, 6, e2014073. [Google Scholar] [CrossRef] [PubMed]
- Patton, D.T.; Plumb, A.W.; Abraham, N. The Survival and Differentiation of Pro-B and Pre-B Cells in the Bone Marrow Is Dependent on IL-7Rα Tyr449. J. Immunol. 2014, 193, 3446–3455. [Google Scholar] [CrossRef] [PubMed]
- Mata-Rocha, M.; Rangel-López, A.; Jiménez-Hernández, E.; Morales-Castillo, B.A.; González-Torres, C.; Gaytan-Cervantes, J.; Álvarez-Olmos, E.; Núñez-Enríquez, J.C.; Fajardo-Gutiérrez, A.; Martín-Trejo, J.A.; et al. Identification and Characterization of Novel Fusion Genes with Potential Clinical Applications in Mexican Children with Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2019, 20, 2394. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Zhang, H.; Kham, S.K.; Liu, S.; Jiang, C.; Zhao, X.; Lu, Y.; Goodings, C.; Lin, T.N.; Zhang, R.; et al. Whole-transcriptome sequencing identifies a distinct subtype of acute lymphoblastic leukemia with predominant genomic abnormalities of EP300 and CREBBP. Genome Res. 2017, 27, 185–195. [Google Scholar] [CrossRef]
- Lilljebjörn, H.; Henningsson, R.; Hyrenius-Wittsten, A.; Olsson, L.; Orsmark-Pietras, C.; von Palffy, S.; Askmyr, M.; Rissler, M.; Schrappe, M.; Cario, G.; et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat. Commun. 2016, 7, 11790. [Google Scholar] [CrossRef]
- Li, B.; Brady, S.W.; Ma, X.; Shen, S.; Zhang, Y.; Li, Y.; Szlachta, K.; Dong, L.; Liu, Y.; Yang, F.; et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 2020, 135, 41–55. [Google Scholar] [CrossRef]
- Gu, Z.; Churchman, M.; Roberts, K.; Li, Y.; Liu, Y.; Harvey, R.C.; McCastlain, K.; Reshmi, S.C.; Payne-Turner, D.; Iacobucci, I.; et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat. Commun. 2016, 7, 13331. [Google Scholar] [CrossRef]
- Atak, Z.K.; Gianfelici, V.; Hulselmans, G.; De Keersmaecker, K.; Devasia, A.G.; Geerdens, E.; Mentens, N.; Chiaretti, S.; Durinck, K.; Uyttebroeck, A.; et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet. 2013, 9, e1003997. [Google Scholar] [CrossRef]
- Chen, B.; Jiang, L.; Zhong, M.-L.; Li, J.-F.; Li, B.-S.; Peng, L.-J.; Dai, Y.-T.; Cui, B.-W.; Yan, T.-Q.; Zhang, W.-N.; et al. Identification of fusion genes and characterization of transcriptome features in T-cell acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 2018, 115, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Agraz-Doblas, A.; Bueno, C.; Bashford-Rogers, R.; Roy, A.; Schneider, P.; Bardini, M.; Ballerini, P.; Cazzaniga, G.; Moreno, T.; Revilla, C.; et al. Unraveling the cellular origin and clinical prognostic markers of infant B-cell acute lymphoblastic leukemia using genome-wide analysis. Haematologica 2019, 104, 1176–1188. [Google Scholar] [CrossRef]
- Trentin, L.; Bresolin, S.; Giarin, E.; Bardini, M.; Serafin, V.; Accordi, B.; Fais, F.; Tenca, C.; De Lorenzo, P.; Valsecchi, M.G.; et al. Deciphering KRAS and NRAS mutated clone dynamics in MLL-AF4 paediatric leukaemia by ultra deep sequencing analysis. Sci. Rep. 2016, 6, 34449. [Google Scholar] [CrossRef] [PubMed]
- Ferreirós-Vidal, I.; Carroll, T.; Taylor, B.; Terry, A.; Liang, Z.; Bruno, L.; Dharmalingam, G.; Khadayate, S.; Cobb, B.S.; Smale, S.T.; et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B–cell differentiation. Blood 2013, 121, 1769–1782. [Google Scholar] [CrossRef]
- Chillón, M.C.; Gómez-Casares, M.T.; López-Jorge, C.E.; Rodriguez-Medina, C.; Molines, A.; Sarasquete, M.E.; Alcoceba, M.; Miguel, J.D.G.S.; Bueno, C.; Montes, R.; et al. Prognostic significance of FLT3 mutational status and expression levels in MLL-AF4+ and MLL-germline acute lymphoblastic leukemia. Leukemia 2012, 26, 2360–2366. [Google Scholar] [CrossRef] [PubMed]
- Uckun, F.M.; Qazi, S. Tyrosine kinases in KMT2A/MLL-rearranged acute leukemias as potential therapeutic targets to overcome cancer drug resistance. Cancer Drug Resist. 2022, 5, 902–916. [Google Scholar] [CrossRef]
- Armstrong, S.A.; Kung, A.L.; Mabon, M.E.; Silverman, L.B.; Stam, R.W.; Den Boer, M.L.; Pieters, R.; Kersey, J.H.; Sallan, S.E.; Fletcher, J.A.; et al. Inhibition of FLT3 in MLL. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell 2003, 3, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.; Levis, M.; McIntyre, E.; Griesemer, M.; Small, D. Combinations of the FLT3 inhibitor CEP-701 and chemotherapy synergistically kill infant and childhood MLL-rearranged ALL cells in a sequence-dependent manner. Leukemia 2006, 20, 1368–1376. [Google Scholar] [CrossRef]
- Stam, R.W.; den Boer, M.L.; Schneider, P.; Nollau, P.; Horstmann, M.; Beverloo, H.B.; van der Voort, E.; Valsecchi, M.G.; de Lorenzo, P.; Sallan, S.E.; et al. Targeting FLT3 in primary MLL-gene-rearranged infant acute lymphoblastic leukemia. Blood 2005, 106, 2484–2490. [Google Scholar] [CrossRef]
- Brown, P.A.; Kairalla, J.A.; Hilden, J.M.; Dreyer, Z.E.; Carroll, A.J.; Heerema, N.A.; Wang, C.; Devidas, M.; Gore, L.; Salzer, W.L.; et al. FLT3 inhibitor lestaurtinib plus chemotherapy for newly diagnosed KMT2A-rearranged infant acute lymphoblastic leukemia: Children’s Oncology Group trial AALL0631. Leukemia 2021, 35, 1279–1290. [Google Scholar] [CrossRef]
- Loftus, J.P.; Yahiaoui, A.; Brown, P.A.; Niswander, L.M.; Bagashev, A.; Wang, M.; Schauf, A.; Tannheimer, S.; Tasian, S.K. Combinatorial efficacy of entospletinib and chemotherapy in patient-derived xenograft models of infant acute lymphoblastic leukemia. Haematologica 2020, 106, 1067–1078. [Google Scholar] [CrossRef]
- Clesham, K.; Rao, V.; Bartram, J.; Ancliff, P.; Ghorashian, S.; O’Connor, D.; Pavasovic, V.; Rao, A.; Samarasinghe, S.; Cummins, M.; et al. Blinatumomab for infant acute lymphoblastic leukemia. Blood 2020, 135, 1501–1504. [Google Scholar] [CrossRef]
- Cheung, L.C.; Aya-Bonilla, C.; Cruickshank, M.N.; Chiu, S.K.; Kuek, V.; Anderson, D.; Chua, G.A.; Singh, S.; Oommen, J.; Ferrari, E.; et al. Preclinical efficacy of azacitidine and venetoclax for infant KMT2A-rearranged acute lymphoblastic leukemia reveals a new therapeutic strategy. Leukemia 2022, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kirchhoff, H.; Karsli, U.; Schoenherr, C.; Battmer, K.; Erschow, S.; Talbot, S.R.; Steinemann, D.; Heuser, M.; Heidenreich, O.; Hilfiker-Kleiner, D.; et al. Venetoclax and dexamethasone synergize with inotuzumab ozogamicin–induced DNA damage signaling in B-lineage ALL. Blood 2021, 137, 2657–2661. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eadie, L.N.; Rehn, J.A.; Breen, J.; Osborn, M.P.; Jessop, S.; Downes, C.E.J.; Heatley, S.L.; McClure, B.J.; Yeung, D.T.; Revesz, T.; et al. Case Report: Rare IKZF1 Gene Fusions Identified in Neonate with Congenital KMT2A-Rearranged Acute Lymphoblastic Leukemia. Genes 2023, 14, 264. https://doi.org/10.3390/genes14020264
Eadie LN, Rehn JA, Breen J, Osborn MP, Jessop S, Downes CEJ, Heatley SL, McClure BJ, Yeung DT, Revesz T, et al. Case Report: Rare IKZF1 Gene Fusions Identified in Neonate with Congenital KMT2A-Rearranged Acute Lymphoblastic Leukemia. Genes. 2023; 14(2):264. https://doi.org/10.3390/genes14020264
Chicago/Turabian StyleEadie, Laura N., Jacqueline A. Rehn, James Breen, Michael P. Osborn, Sophie Jessop, Charlotte E. J. Downes, Susan L. Heatley, Barbara J. McClure, David T. Yeung, Tamas Revesz, and et al. 2023. "Case Report: Rare IKZF1 Gene Fusions Identified in Neonate with Congenital KMT2A-Rearranged Acute Lymphoblastic Leukemia" Genes 14, no. 2: 264. https://doi.org/10.3390/genes14020264
APA StyleEadie, L. N., Rehn, J. A., Breen, J., Osborn, M. P., Jessop, S., Downes, C. E. J., Heatley, S. L., McClure, B. J., Yeung, D. T., Revesz, T., Saxon, B., & White, D. L. (2023). Case Report: Rare IKZF1 Gene Fusions Identified in Neonate with Congenital KMT2A-Rearranged Acute Lymphoblastic Leukemia. Genes, 14(2), 264. https://doi.org/10.3390/genes14020264










