Investigation of UTR Variants by Computational Approaches Reveal Their Functional Significance in PRKCI Gene Regulation
Abstract
1. Introduction
2. Methods
2.1. Retrieval of UTR SNPs and Annotation Using RegulomeDB
2.2. Assessment of Effect of 3′ UTR SNPs on miRNA Binding Sites
2.3. Effect of UTR SNPs on Tissue Expression Using eQTL Analysis
2.4. Determination of Effect of SNPs on Secondary Structure of mRNA
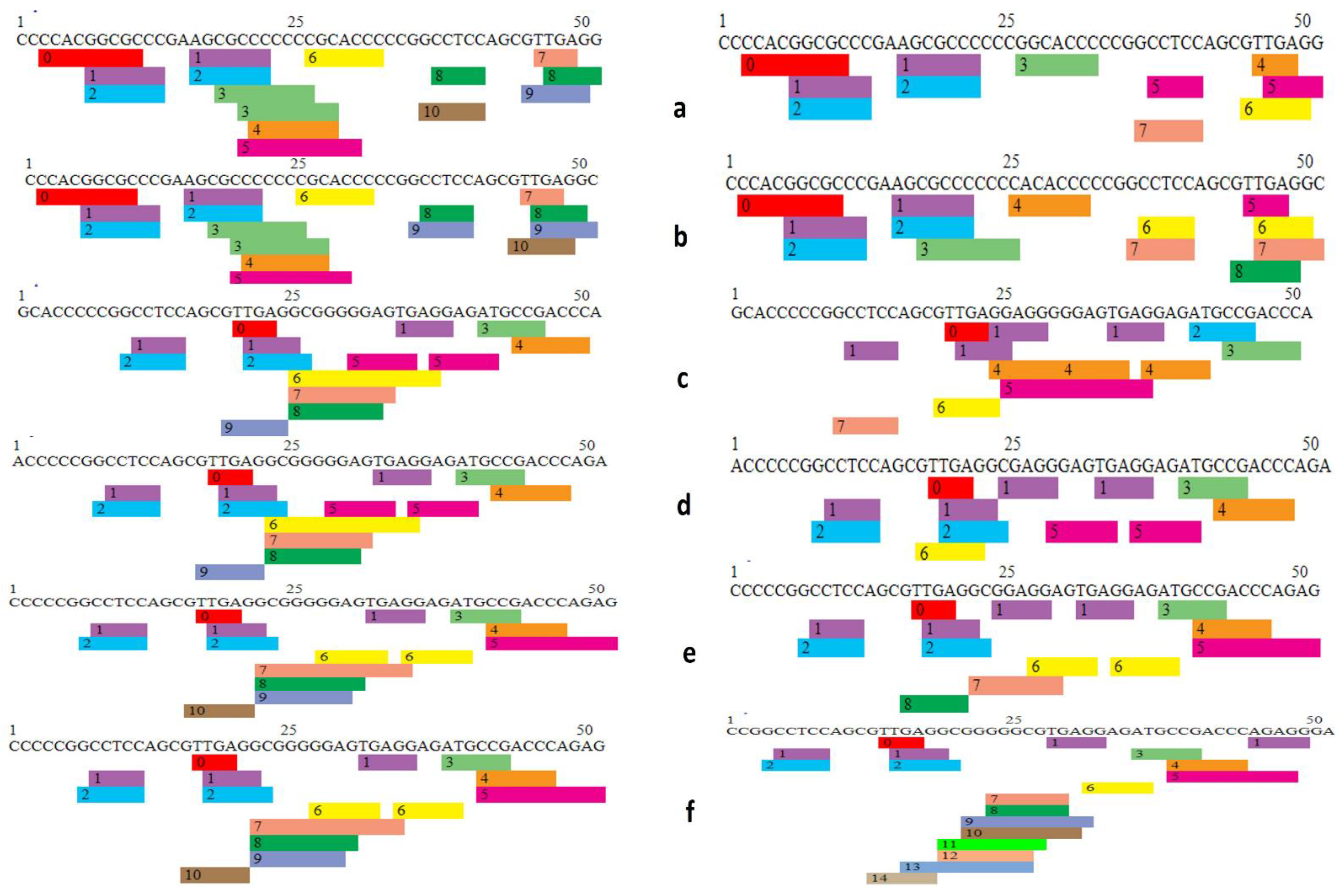
2.5. Analysis of 5′ UTR SNPs on Transcription Factor Binding Sites (TFBS)
2.6. Variation Tolerance in PRKCI
3. Results
3.1. Retrieval of UTR SNPs of PRKCI from Ensembl and Scoring on RegulomeDB
3.2. Association of UTR SNPs with miRNAs
3.3. Determination of UTR SNPs eQTLs
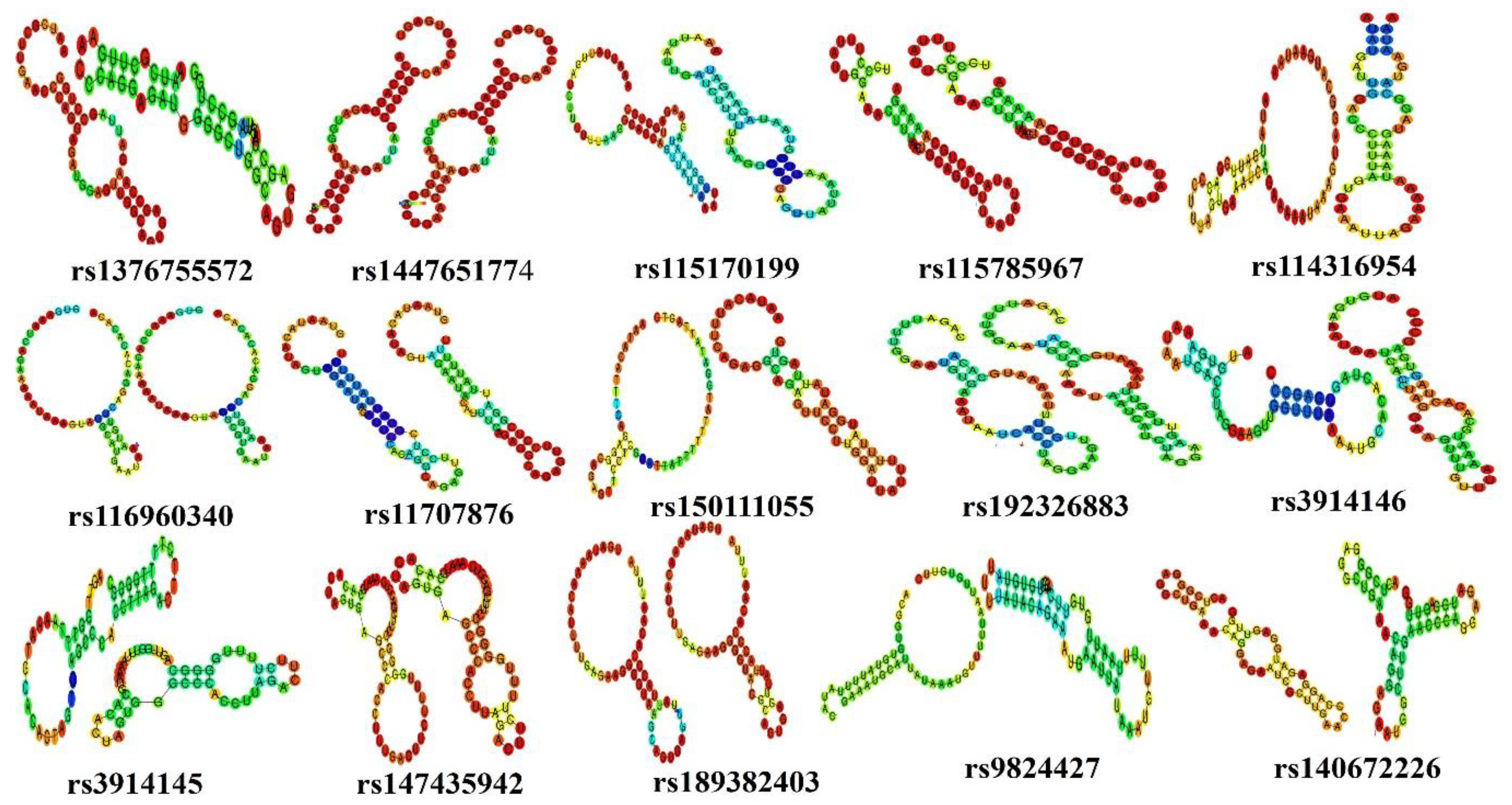
3.4. Impact of UTR SNPs on Secondary Structure of mRNA
3.5. Effect of 5′ UTR SNPs on Transcription Binding Factors
3.6. Variation Tolerance in PRKCI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parker, P.J.; Justilien, V.; Riou, P.; Linch, M.; Fields, A.P. Atypical protein kinase Cι as a human oncogene and therapeutic target. Biochem. Pharmacol. 2014, 88, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; FuJII, T.; Ogita, K.; Kikkawa, U.; Igarashi, K.; Nishizuka, Y. Protein kinase C zeta subspecies from rat brain: Its structure, expression, and properties. Proc. Natl. Acad. Sci. USA 1989, 86, 3099–3103. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, K.; Mizuno, K.; Osada, S.-I.; Hirai, S.-I.; Tanuma, S.-I.; Suzuki, K.; Ohno, S. A new member of the third class in the protein kinase C family, PKC lambda, expressed dominantly in an undifferentiated mouse embryonal carcinoma cell line and also in many tissues and cells. J. Biol. Chem. 1994, 269, 12677–12683. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.H.; Ridden, J.; Parker, P.J. Cloning and expression patterns of two members of a novel protein-kinase-C-related kinase family. Eur. J. Biochem. 1995, 227, 344–351. [Google Scholar] [CrossRef]
- Fields, A.P.; Regala, R.P. Protein kinase Cι: Human oncogene, prognostic marker and therapeutic target. Pharmacol. Res. 2007, 55, 487–497. [Google Scholar] [CrossRef]
- Jamieson, L.; Carpenter, L.; Biden, T.J.; Fields, A.P. Protein kinase Cι activity is necessary for Bcr-Abl-mediated resistance to drug-induced apoptosis. J. Biol. Chem. 1999, 274, 3927–3930. [Google Scholar] [CrossRef]
- Murray, N.R.; Jamieson, L.; Yu, W.; Zhang, J.; Gökmen-Polar, Y.; Sier, D.; Anastasiadis, P.; Gatalica, Z.; Thompson, E.A.; Fields, A.P. Protein kinase Cι is required for Ras transformation and colon carcinogenesis in vivo. J. Cell Biol. 2004, 164, 797–802. [Google Scholar] [CrossRef]
- Bjørkøy, G.; Perander, M.; Øvervatn, A.; Johansen, T. Reversion of Ras-and phosphatidylcholine-hydrolyzing phospholipase C-mediated transformation of NIH 3T3 cells by a dominant interfering mutant of protein kinase C λ is accompanied by the loss of constitutive nuclear mitogen-activated protein kinase/extracellular signal-regulated kinase activity. J. Biol. Chem. 1997, 272, 11557–11565. [Google Scholar]
- Regala, R.P.; Weems, C.; Jamieson, L.; Khoor, A.; Edell, E.S.; Lohse, C.M.; Fields, A.P. Atypical protein kinase Cι is an oncogene in human non–small cell lung cancer. Cancer Res. 2005, 65, 8905–8911. [Google Scholar] [CrossRef]
- Yang, Y.L.; Chu, J.Y.; Luo, M.L.; Wu, Y.P.; Zhang, Y.; Feng, Y.B.; Shi, Z.Z.; Xu, X.; Han, Y.L.; Cai, Y.; et al. Amplification of PRKCI, located in 3q26, is associated with lymph node metastasis in esophageal squamous cell carcinoma. Genes Chromosomes Cancer 2008, 47, 127–136. [Google Scholar] [CrossRef]
- Rehmani, H.; Li, Y.; Li, T.; Padia, R.; Calbay, O.; Jin, L.; Chen, H.; Huang, S. Addiction to protein kinase Cɩ due to PRKCI gene amplification can be exploited for an aptamer-based targeted therapy in ovarian cancer. Signal Transduct. Target. Ther. 2020, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Brookes, A.J. The essence of SNPs. Gene 1999, 234, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Sekar, R.B.; Hansen, N.F.; Fox-Talbot, K.; Morley, M.; Pihur, V.; Chatterjee, S.; Brandimarto, J.; Moravec, C.S.; Pulit, S.L. An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am. J. Hum. Genet. 2014, 94, 854–869. [Google Scholar] [CrossRef]
- Spieler, D.; Kaffe, M.; Knauf, F.; Bessa, J.; Tena, J.J.; Giesert, F.; Schormair, B.; Tilch, E.; Lee, H.; Horsch, M. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res. 2014, 24, 592–603. [Google Scholar] [CrossRef]
- Bauer, D.E.; Kamran, S.C.; Lessard, S.; Xu, J.; Fujiwara, Y.; Lin, C.; Shao, Z.; Canver, M.C.; Smith, E.C.; Pinello, L. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 2013, 342, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Meulendijks, D.; Jacobs, B.A.; Aliev, A.; Pluim, D.; Van Werkhoven, E.; Deenen, M.J.; Beijnen, J.H.; Cats, A.; Schellens, J.H. Increased risk of severe fluoropyrimidine-associated toxicity in patients carrying a G to C substitution in the first 28-bp tandem repeat of the thymidylate synthase 2 R allele. Int. J. Cancer 2016, 138, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Lurje, G.; Manegold, P.C.; Ning, Y.; Pohl, A.; Zhang, W.; Lenz, H.-J. Thymidylate synthase gene variations: Predictive and prognostic markersTS Gene Variations: Predictive and Prognostic Markers. Mol. Cancer Ther. 2009, 8, 1000–1007. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; He, Y.; Ding, J.; Wu, K.; Hu, B.; Liu, Y.; Wu, Y.; Guo, B.; Shen, Y.; Landi, D. An insertion/deletion polymorphism at miRNA-122-binding site in the interleukin-1α 3′ untranslated region confers risk for hepatocellular carcinoma. Carcinogenesis 2009, 30, 2064–2069. [Google Scholar] [CrossRef]
- Christensen, B.C.; Moyer, B.J.; Avissar, M.; Ouellet, L.G.; Plaza, S.L.; McClean, M.D.; Marsit, C.J.; Kelsey, K.T. A let-7 microRNA-binding site polymorphism in the KRAS 3′ UTR is associated with reduced survival in oral cancers. Carcinogenesis 2009, 30, 1003–1007. [Google Scholar] [CrossRef]
- Hashemi, M.; Shahkar, G.; Simforoosh, N.; Basiri, A.; Ziaee, S.; Narouie, B.; Taheri, M.J.C.; Biology, M. Association of polymorphisms in PRKCI gene and risk of prostate cancer in a sample of Iranian Population. Cell. Mol. Biol. 2015, 61, 16–21. [Google Scholar]
- Shah, H.; Khan, K.; Khan, N.; Badshah, Y.; Ashraf, N.M.; Shabbir, M. Impact of deleterious missense PRKCI variants on structural and functional dynamics of protein. Sci. Rep. 2022, 12, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Ziebarth, J.D.; Cui, Y. PolymiRTS Database 3.0: Linking polymorphisms in microRNAs and their target sites with human diseases and biological pathways. Nucleic Acids Res. 2014, 42, D86–D91. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Fu, X.; Xia, M.; Zhang, Q.; Gu, Z.; Guo, A.-Y. miRNASNP-v3: A comprehensive database for SNPs and disease-related variations in miRNAs and miRNA targets. Nucleic Acids Res. 2021, 49, D1276–D1281. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Keen, J.C.; Moore, H.M. The genotype-tissue expression (GTEx) project: Linking clinical data with molecular analysis to advance personalized medicine. J. Pers. Med. 2015, 5, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef] [PubMed]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Firth, H.V.; Richards, S.M.; Bevan, A.P.; Clayton, S.; Corpas, M.; Rajan, D.; Van Vooren, S.; Moreau, Y.; Pettett, R.M.; Carter, N.P. DECIPHER: Database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 2009, 84, 524–533. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv 2019, 531210. [Google Scholar] [CrossRef]
- Khan, K.; Shah, H.; Rehman, A.; Badshah, Y.; Ashraf, N.M.; Shabbir, M. Influence of PRKCE non-synonymous variants on protein dynamics and functionality. Hum. Mol. Genet. 2022, 31, 2236–2261. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, M.; Badshah, Y.; Khan, K.; Trembley, J.H.; Rizwan, A.; Faraz, F.; Shah, S.A.; Farooqi, M.; Ashraf, N.M.; Afsar, T. Association of CTLA-4 and IL-4 polymorphisms in viral induced liver cancer. BMC Cancer 2022, 22, 518. [Google Scholar] [CrossRef] [PubMed]
- Tak, Y.G.; Farnham, P.J. Making sense of GWAS: Using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 2015, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. Non-Coding RNA 2021, 7, 47. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, X.; Jin, Y. ADAR-mediated RNA editing in non-coding RNA sequences. Sci. China Life Sci. 2013, 56, 944–952. [Google Scholar] [CrossRef]
- Khan, K.; Zafar, S.; Hafeez, A.; Badshah, Y.; Shahid, K.; Mahmood Ashraf, N.; Shabbir, M. PRKCE non-coding variants influence on transcription as well as translation of its gene. RNA Biol. 2022, 19, 1115–1129. [Google Scholar] [CrossRef]
- Wilkie, G.S.; Dickson, K.S.; Gray, N.K. Regulation of mRNA translation by 5′-and 3′-UTR-binding factors. Trends Biochem. Sci. 2003, 28, 182–188. [Google Scholar] [CrossRef]
- Bugaut, A.; Balasubramanian, S. 5′-UTR RNA G-quadruplexes: Translation regulation and targeting. Nucleic Acids Res. 2012, 40, 4727–4741. [Google Scholar] [CrossRef]
- Yoo, S.S.; Jin, C.; Jung, D.K.; Choi, Y.Y.; Choi, J.E.; Lee, W.K.; Lee, S.Y.; Lee, J.; Cha, S.I.; Kim, C.H. Putative functional variants of XRCC1 identified by RegulomeDB were not associated with lung cancer risk in a Korean population. Cancer Genet. 2015, 208, 19–24. [Google Scholar] [CrossRef]
- Cheema, A.N.; Rosenthal, S.L.; Ilyas Kamboh, M. Proficiency of data interpretation: Identification of signaling SNPs/specific loci for coronary artery disease. Database 2017, 2017, bax078. [Google Scholar] [CrossRef]
- Abdi, A.; Zafarpiran, M.; Farsani, Z.S. The computational analysis conducted on miRNA target sites in association with SNPs at 3′UTR of ADHD-implicated genes. Cent. Nerv. Syst. Agents Med. Chem. 2020, 20, 58–75. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.; Awan, F.R.; Mirza, M.R.; Nishat, S.; Rajput, S.A.; Malik, I.R. Mining the SNPs of Human Low Density Lipoprotein (LDL) related Gene APOB through in silico Approaches. Pak. J. Zool 2021, 54, 2315–2327. [Google Scholar] [CrossRef]
- Nimir, M.; Abdelrahim, M.; Abdelrahim, M.; Abdalla, M.; eldin Ahmed, W.; Abdullah, M.; Hamid, M.M.A. In silico analysis of single nucleotide polymorphisms (SNPs) in human FOXC2 gene. F1000Research 2017, 6, 243. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Tian, L.; Cao, H.; Baranova, A.; Zhang, F. Involvement of the long intergenic non-coding RNA LINC00461 in schizophrenia. BMC Psychiatry 2022, 22, 59. [Google Scholar] [CrossRef]
- De Goede, O.M.; Nachun, D.C.; Ferraro, N.M.; Gloudemans, M.J.; Rao, A.S.; Smail, C.; Eulalio, T.Y.; Aguet, F.; Ng, B.; Xu, J. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 2021, 184, 2633–2648.e2619. [Google Scholar] [CrossRef]
- Dodson, R.E.; Shapiro, D.J. Regulation of Pathways of mRNA Destabilization and Stabilization; Elsevier: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Moudi, M.; Sargazi, S.; Heidari Nia, M.; Saravani, R.; Shirvaliloo, M.; Shakiba, M. Polymorphism in the 3′-UTR of LIF but not in the ATF6B gene associates with schizophrenia susceptibility: A case-control study and in silico analyses. J. Mol. Neurosci. 2020, 70, 2093–2101. [Google Scholar] [CrossRef]
- Karthi, S.; Rajeshwari, M.; Francis, A.; Saravanan, M.; Varalakshmi, P.; Houlden, H.; Thangaraj, K.; Ashokkumar, B. 3′-UTR SNP rs2229611 in G6PC1 affects mRNA stability, expression and Glycogen Storage Disease type-Ia risk. Clin. Chim. Acta 2017, 471, 46–54. [Google Scholar] [CrossRef]
- Briones-Orta, M.A.; Avendaño-Vázquez, S.E.; Aparicio-Bautista, D.I.; Coombes, J.D.; Weber, G.F.; Syn, W.-K. Prediction of transcription factor bindings sites affected by SNPs located at the osteopontin promoter. Data Brief 2017, 14, 538–542. [Google Scholar] [CrossRef]
- Itan, Y.; Shang, L.; Boisson, B.; Patin, E.; Bolze, A.; Moncada-Vélez, M.; Scott, E.; Ciancanelli, M.J.; Lafaille, F.G.; Markle, J.G. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA 2015, 112, 13615–13620. [Google Scholar] [CrossRef]





| SNPs | Location | MFE Wild (kcal/mol) | MFE Mutated (kcal/mol) |
|---|---|---|---|
| rs1482898617(C/T) | 3:170222490 | −24.90 | −23.90 |
| rs1237925954 (C/T) | 3:170222491 | −22.60 | −24.80 |
| rs1024270582 (C/T) | 3:170222468 | −26.20 | −27.40 |
| rs999438596 (G/A) | 3:170222469 | −21.60 | −21.20 |
| rs1031689697 (C/T) | 3:170222470 | −16.20 | −19.90 |
| rs370911690 (C/G) | 3:170222471 | −21.60 | −22.00 |
| rs1299372677(C/T) | 3:170222621 | −14.30 | −12.40 |
| rs199584503 (C/T) | 3:170222623 | −14.50 | −11.80 |
| rs1434910430 (C/G) | 3:170222624 | −10.60 | −10.40 |
| rs556899946 (C/G) | 3:170222626 | −10.60 | −12.30 |
| rs763083643 (C/T) | 3:170222629 | −13.40 | −16.40 |
| rs759715886 (C/A) | 3:170222634 | −23.70 | −23.40 |
| rs1448713189 (C/T) | 3:170222635 | −26.90 | −26.40 |
| rs1015429064 (C/T) | 3:170222636 | −24.20 | −26.60 |
| rs1419292188 (G/T) | 3:170222653 | −25.60 | −19.00 |
| rs778557075(C/A) | 3:170222655 | −22.70 | −18.20 |
| rs968409340(G/A) | 3:170222657 | −19.50 | −15.20 |
| rs750297755 (G/A) | 3:170222658 | −19.20 | −14.10 |
| rs542458816 (A/C) | 3:170222661 | −15.70 | −21.50 |
| rs1323399846 (G/C) | 3:170222662 | −15.00 | −16.00 |
| Variant ID | Region | SNP ID | p-Value | NES | Tissue eQTL |
|---|---|---|---|---|---|
| chr3_170303911_C_G_b38 | 3′ UTR | rs140672226 | 0.000064 | 0.82 | Stomach |
| 0.000099 | 0.49 | Esophagus—muscularis | |||
| chr3_170304707_A_G_b38 | 3′ UTR | rs2650220 | 7.9 × 10−7 | 0.15 | Esophagus—mucosa |
| Gene | GDI Value | Residual Variation Tolerance Score | Haploinsufficiency Score | Loss of Function (o:e) Ratio | pLI |
|---|---|---|---|---|---|
| PRKCI | 0.99 | 17.6% | 19.34 | 0.26 (0.16–0.44) | 0.07 |
| SNPs | Location | MFE Wild (kcal/mol) | MFE Mutated (kcal/mol) |
|---|---|---|---|
| rs1376755572(T/C) | 3:170303934 | −13.60 | −11.20 |
| rs1447651774(G/A) | 3:170303943 | −17.70 | −13.60 |
| rs115170199 (C/G) | 3:170303662 | −8.30 | −3.10 |
| rs115785967 (A/G) | 3:170304156 | −13.90 | −13.20 |
| rs114316954 (C/T) | 3:170304522 | −3.10 | −1.20 |
| rs116960340 (A/G) | 3:170304540 | −2.50 | −2.10 |
| rs11707876 (G/C) | 3:170304653 | −5.20 | −1.40 |
| rs150111055 (C/T) | 3:170304667 | −4.50 | −5.00 |
| rs192326883 (C/T) | 3:170305221 | −4.50 | −4.60 |
| rs3914146 (G/T) | 3:170305232 | −5.10 | −5.50 |
| rs3914145 (A/G) | 3:170305253 | −7.40 | −7.30 |
| rs147435942 (A/T) | 3:170305263 | −8.20 | −6.80 |
| rs189382403 (A/C) | 3:170305381 | −7.00 | −6.10 |
| rs9824427 (C/A) | 3:170305608 | −6.00 | −3.20 |
| rs140672226 (C/G) | 3:170303911 | −9.90 | −8.40 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shah, H.; Khan, K.; Badshah, Y.; Mahmood Ashraf, N.; Shabbir, M.; Trembley, J.H.; Afsar, T.; Abusharha, A.; Razak, S. Investigation of UTR Variants by Computational Approaches Reveal Their Functional Significance in PRKCI Gene Regulation. Genes 2023, 14, 247. https://doi.org/10.3390/genes14020247
Shah H, Khan K, Badshah Y, Mahmood Ashraf N, Shabbir M, Trembley JH, Afsar T, Abusharha A, Razak S. Investigation of UTR Variants by Computational Approaches Reveal Their Functional Significance in PRKCI Gene Regulation. Genes. 2023; 14(2):247. https://doi.org/10.3390/genes14020247
Chicago/Turabian StyleShah, Hania, Khushbukhat Khan, Yasmin Badshah, Naeem Mahmood Ashraf, Maria Shabbir, Janeen H. Trembley, Tayyaba Afsar, Ali Abusharha, and Suhail Razak. 2023. "Investigation of UTR Variants by Computational Approaches Reveal Their Functional Significance in PRKCI Gene Regulation" Genes 14, no. 2: 247. https://doi.org/10.3390/genes14020247
APA StyleShah, H., Khan, K., Badshah, Y., Mahmood Ashraf, N., Shabbir, M., Trembley, J. H., Afsar, T., Abusharha, A., & Razak, S. (2023). Investigation of UTR Variants by Computational Approaches Reveal Their Functional Significance in PRKCI Gene Regulation. Genes, 14(2), 247. https://doi.org/10.3390/genes14020247







