Development of Human Rhinovirus RNA Reference Material Using Digital PCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Preparation of RNA
2.2. Reverse Transcription Droplet Digital PCR (RT-ddPCR)
2.3. RT-qPCR Analysis
2.4. Homogeneity and Stability Tests
2.5. Unvertainty and Statistical Analyses
3. Results
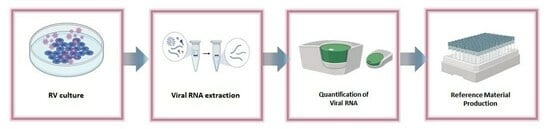
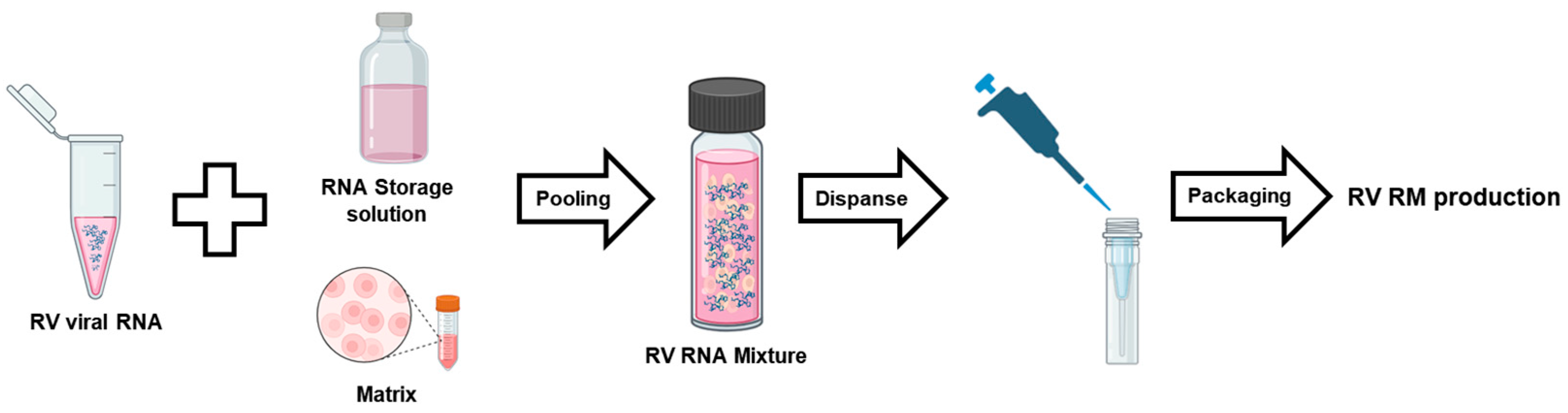
3.1. RV Reference Material Design and Preparation Processes
3.2. RV RMs Serial Dilution
3.3. RV RMs Homogeneity Test
3.4. Short- and Long-Term Stability of RV RMs
3.5. RV RM Freeze–Thaw Repeated Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dee, K.; Goldfarb, D.M.; Haney, J.; Amat, J.A.R.; Herder, V.; Stewart, M.; Szemiel, A.M.; Baguelin, M.; Murcia, P.R. Human Rhinovirus Infection Blocks Severe Acute Respiratory Syndrome Coronavirus 2 Replication Within the Respiratory Epithelium: Implications for COVID-19 Epidemiology. J. Infect. Dis. 2021, 224, 31–38. [Google Scholar] [CrossRef]
- Kutter, J.S.; Spronken, M.I.; Fraaij, P.L.; Fouchier, R.A.; Herfst, S. Transmission routes of respiratory viruses among humans. Curr. Opin. Virol. 2018, 28, 142–151. [Google Scholar] [CrossRef]
- Nickbakhsh, S.; Thorburn, F.; VON Wissmann, B.; McMENAMIN, J.; Gunson, R.N.; Murcia, P.R. Extensive multiplex PCR diagnostics reveal new insights into the epidemiology of viral respiratory infections. Epidemiol. Infect. 2016, 144, 2064–2076. [Google Scholar] [CrossRef]
- Kim, S.R. Viral Infection and Airway Epithelial Immunity in Asthma. Int. J. Mol. Sci. 2022, 23, 9914. [Google Scholar] [CrossRef]
- Novak, N.; Cabanillas, B. Viruses and asthma: The role of common respiratory viruses in asthma and its potential meaning for SARS-CoV-2. Immunology 2020, 161, 83–93. [Google Scholar] [CrossRef]
- Faizi, N.; Kazmi, S. Universal health coverage—There is more to it than meets the eye. J. Fam. Med. Prim. Care 2017, 6, 169–170. [Google Scholar] [CrossRef]
- Busse, W.W.; Lemanske, R.F.; Gern, J.E. Role of viral respiratory infections in asthma and asthma exacerbations. Lancet 2010, 376, 826–834. [Google Scholar] [CrossRef]
- Numata, M.; Sajuthi, S.; Bochkov, Y.A.; Loeffler, J.; Everman, J.; Vladar, E.K.; Cooney, R.A.; Reinhardt, R.L.; Liu, A.H.; Seibold, M.A.; et al. Anionic Pulmonary Surfactant Lipid Treatment Inhibits Rhinovirus A Infection of the Human Airway Epithelium. Viruses 2023, 15, 747. [Google Scholar] [CrossRef]
- Wark, P.A.B. Asthma and the Dysregulated Immune Response to Rhinovirus. Am. J. Respir. Crit. Care Med. 2020, 202, 157–159. [Google Scholar] [CrossRef]
- Touabi, L.; Aflatouni, F.; McLean, G.R. Mechanisms of rhinovirus neutralisation by antibodies. Viruses 2021, 13, 360. [Google Scholar] [CrossRef]
- Friedlander, S.L.; Busse, W.W. The role of rhinovirus in asthma exacerbations. J. Allergy Clin. Immunol. 2005, 116, 267–273. [Google Scholar] [CrossRef]
- Basnet, S.; Palmenberg, A.C.; Gern, J.E. Rhinoviruses and Their Receptors. Chest 2019, 155, 1018–1025. [Google Scholar] [CrossRef]
- Lee, W.-M.; Kiesner, C.; Pappas, T.; Lee, I.; Grindle, K.; Jartti, T.; Jakiela, B.; Lemanske, R.F., Jr.; Shult, P.A.; Gern, J.E. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE 2007, 2, e966. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.N.; Trinh, Q.D.; Pham, N.T.K.; Pham, T.M.H.; Ha, M.T.; Nguyen, T.Q.N.; Okitsu, S.; Shimizu, H.; Hayakawa, S.; Mizuguchi, M.; et al. Human rhinovirus infections in hospitalized children: Clinical, epidemiological and virological features. Epidemiol. Infect. 2015, 144, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, S.E.; Lamson, D.M.; St George, K.; Walsh, T.J. Human rhinoviruses. Clin. Microbiol. Rev. 2013, 26, 135–162. [Google Scholar] [CrossRef]
- Garbino, J.; Soccal, P.M.; Aubert, J.-D.; Rochat, T.; Meylan, P.; Thomas, Y.; Tapparel, C.; Bridevaux, P.-O.; Kaiser, L. Respiratory viruses in bronchoalveolar lavage: A hospital-based cohort study in adults. Thorax 2009, 64, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Garbino, J.; Gerbase, M.W.; Wunderli, W.; Deffernez, C.; Thomas, Y.; Rochat, T.; Ninet, B.; Schrenzel, J.; Yerly, S.; Perrin, L.; et al. Lower respiratory viral illnesses: Improved diagnosis by molecular methods and clinical impact. Am. J. Respir. Crit. Care Med. 2004, 170, 1197–1203. [Google Scholar] [CrossRef]
- Renwick, N.; Schweiger, B.; Kapoor, V.; Liu, Z.; Villari, J.; Bullmann, R.; Miething, R.; Briese, T.; Lipkin, W.I. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J. Infect. Dis. 2007, 196, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Kotaniemi-Syrjänen, A.; Vainionpää, R.; Reijonen, T.M.; Waris, M.; Korhonen, K.; Korppi, M. Rhinovirus-induced wheezing in infancy—The first sign of childhood asthma? J. Allergy Clin. Immunol. 2003, 111, 66–71. [Google Scholar] [CrossRef]
- Honkinen, M.; Lahti, E.; Österback, R.; Ruuskanen, O.; Waris, M. Viruses and bacteria in sputum samples of children with community-acquired pneumonia. Clin. Microbiol. Infect. 2012, 18, 300–307. [Google Scholar] [CrossRef]
- Peltola, V.; Jartti, T.; Putto-Laurila, A.; Mertsola, J.; Vainionpää, R.; Waris, M.; Hyypiä, T.; Ruuskanen, O. Rhinovirus infections in children: A retrospective and prospective hospital-based study. J. Med. Virol. 2009, 81, 1831–1838. [Google Scholar] [CrossRef]
- Stefanska, I.; Romanowska, M.; Donevski, S.; Gawryluk, D.; Brydak, L.B. Co-infections with influenza and other respiratory viruses. Adv. Exp. Med. Biol. 2013, 756, 291–301. [Google Scholar] [CrossRef]
- Wu, A.; Mihaylova, V.T.; Landry, M.L.; Foxman, E.F. Interference between rhinovirus and influenza A virus: A clinical data analysis and experimental infection study. Lancet Microbe 2020, 1, e254–e262. [Google Scholar] [CrossRef] [PubMed]
- Royston, L.; Tapparel, C. Rhinoviruses and Respiratory Enteroviruses: Not as Simple as ABC. Viruses 2016, 8, 16. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Nickbakhsh, S.; Mair, C.; Matthews, L.; Reeve, R.; Johnson, P.C.D.; Thorburn, F.; von Wissmann, B.; Reynolds, A.; McMenamin, J.; Gunson, R.N.; et al. Virus-virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. USA 2019, 116, 27142–27150. [Google Scholar] [CrossRef] [PubMed]
- Al-Dulaimi, A.; Alsayed, A.R.; Maqbali, M.A.; Zihlif, M. Investigating the human rhinovirus co-infection in patients with asthma exacerbations and COVID-19. Pharm. Pract. 2022, 20, 2665. [Google Scholar] [CrossRef]
- Mackenzie, J.S.; Smith, D.W. COVID-19: A novel zoonotic disease caused by a coronavirus from China: What we know and what we don’t. Microbiol. Aust. 2020, 41, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Pujadas, E.; Ibeh, N.; Hernandez, M.M.; Waluszko, A.; Sidorenko, T.; Flores, V.; Shiffrin, B.; Chiu, N.; Young-Francois, A.; Nowak, M.D.; et al. Comparison of SARS-CoV-2 detection from nasopharyngeal swab samples by the Roche cobas 6800 SARS-CoV-2 test and a laboratory-developed real-time RT-PCR test. J. Med. Virol. 2020, 92, 1695–1698. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Unger, S.D.; Zhang, N.; Taka, S.; Michel, S.; Akdağ, N.; Lan, F.; Helfer, M.; Hudemann, C.; Eickmann, M.; et al. Development and characterization of DNAzyme candidates demonstrating significant efficiency against human rhinoviruses. J. Allergy Clin. Immunol. 2019, 143, 1403–1415. [Google Scholar] [CrossRef]
- McIntyre, C.L.; Knowles, N.J.; Simmonds, P. Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types. J. Gen. Virol. 2013, 94, 1791–1806. [Google Scholar] [CrossRef]
- Palmenberg, A.C.; Rathe, J.A.; Liggett, S.B. Analysis of the complete genome sequences of human rhinovirus. J. Allergy Clin. Immunol. 2010, 125, 1190–1199. [Google Scholar] [CrossRef] [PubMed]
- Palmenberg, A.C.; Spiro, D.; Kuzmickas, R.; Wang, S.; Djikeng, A.; Rathe, J.A.; Fraser-liggett, C.M.; Liggett, S.B. Sequencing and Analyses of All Reveal Structure and Evolution. Science 2009, 324, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.; Gül, D.C.; Grunert, H.P.; Zeichhardt, H.; Erdmann, V.A.; Kurreck, J. RNA cleaving “10-23” DNAzymes with enhanced stability and activity. Nucleic Acids Res. 2003, 31, 5982–5992. [Google Scholar] [CrossRef] [PubMed]
- Homburg, U.; Renz, H.; Timmer, W.; Hohlfeld, J.M.; Seitz, F.; Lüer, K.; Mayer, A.; Wacker, A.; Schmidt, O.; Kuhlmann, J.; et al. Safety and tolerability of a novel inhaled GATA3 mRNA targeting DNAzyme in patients with TH2-driven asthma. J. Allergy Clin. Immunol. 2015, 136, 797–800. [Google Scholar] [CrossRef]
- Potaczek, D.P.; Garn, H.; Unger, S.D.; Renz, H. Antisense molecules: A new class of drugs. J. Allergy Clin. Immunol. 2016, 137, 1334–1346. [Google Scholar] [CrossRef] [PubMed]
- Tuschl, T.; Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar]
- Phipps, K.M.; Martinez, A.; Lu, J.; Heinz, B.A.; Zhao, G. Small interfering RNA molecules as potential anti-human rhinovirus agents: In vitro potency, specificity, and mechanism. Antivir. Res. 2004, 61, 49–55. [Google Scholar] [CrossRef]
- Burns, J.L.; Emerson, J.; Kuypers, J.; Campbell, A.P.; Gibson, R.L.; McNamara, S.; Worrell, K.; Englund, J.A. Respiratory viruses in children with cystic fibrosis: Viral detection and clinical findings. Influenza Other Respir. Viruses 2012, 6, 218–223. [Google Scholar] [CrossRef]
- Seemungal, T.A.; Harper-Owen, R.; Bhowmik, A.; Jeffries, D.J.; Wedzicha, J.A. Detection of rhinovirus in induced sputum at exacerbation of chronic obstructive pulmonary disease. Eur. Respir. J. 2000, 16, 677–683. [Google Scholar] [CrossRef][Green Version]
- Sedlak, R.H.; Nguyen, T.; Palileo, I.; Jerome, K.R.; Kuypers, J. Superiority of Digital Reverse Transcription-PCR (RT-PCR) over Real-Time RT-PCR for Quantitation of Highly Divergent Human Rhinoviruses. J. Clin. Microbiol. 2017, 55, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G. Rhinovirus and the lower respiratory tract. Rev. Med. Virol. 2004, 14, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, N.G.; Psarras, S. Rhinoviruses in the pathogenesis of asthma. Curr. Allergy Asthma Rep. 2003, 3, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Do, D.H.; Laus, S.; Leber, A.; Marcon, M.J.; Jordan, J.A.; Martin, J.M.; Wadowsky, R.M. A one-step, real-time PCR assay for rapid detection of rhinovirus. J. Mol. Diagn. 2010, 12, 102–108. [Google Scholar] [CrossRef]
- Tapparel, C.; Cordey, S.; Van Belle, S.; Turin, L.; Lee, W.-M.; Regamey, N.; Meylan, P.; Mühlemann, K.; Gobbini, F.; Kaiser, L. New molecular detection tools adapted to emerging rhinoviruses and enteroviruses. J. Clin. Microbiol. 2009, 47, 1742–1749. [Google Scholar] [CrossRef] [PubMed]
- Schibler, M.; Yerly, S.; Vieille, G.; Docquier, M.; Turin, L.; Kaiser, L.; Tapparel, C. Critical analysis of rhinovirus RNA load quantification by real-time reverse transcription-PCR. J. Clin. Microbiol. 2012, 50, 2868–2872. [Google Scholar] [CrossRef] [PubMed]
- Scheltinga, S.A.; Templeton, K.E.; Beersma, M.F.C.; Claas, E.C.J. Diagnosis of human metapneumovirus and rhinovirus in patients with respiratory tract infections by an internally controlled multiplex real-time RNA PCR. J. Clin. Virol. 2005, 33, 306–311. [Google Scholar] [CrossRef]
- Stults, J.R.; Snoeyenbos-West, O.; Methe, B.; Lovley, D.R.; Chandler, D.P. Application of the 5′ Fluorogenic Exonuclease Assay (TaqMan) for Quantitative Ribosomal DNA and rRNA Analysis in Sediments. Appl. Environ. Microbiol. 2001, 67, 2781–2789. [Google Scholar] [CrossRef]
- Smith, C.J.; Osborn, A.M. Advantages and limitations of quantitative PCR (Q-PCR)-based approaches in microbial ecology. FEMS Microbiol. Ecol. 2009, 67, 6–20. [Google Scholar] [CrossRef]
- Mehta, N. RT-qPCR Made Simple: A Comprehensive Guide on the Methods, Advantages, Disadvantages, and Everything in Between. Undergrad. Res. Nat. Clin. Sci. Technol. J. 2022, 6, 1–6. [Google Scholar] [CrossRef]
- Bustin, S.A.; Nolan, T. Pitfalls of quantitative real- time reverse-transcription polymerase chain reaction. J. Biomol. Tech. JBT 2004, 15, 155–166. [Google Scholar]
- Paul, N.; Shum, J.; Le, T. Hot Start PCR; Springer: Berlin/Heidelberg, Germany, 2010; Volume 630, ISBN 9781607616283. [Google Scholar]
- Ling, C.L.; McHugh, T.D. Rapid Detection of Atypical Respiratory Bacterial Pathogens by Real-Time PCR; Springer: Berlin/Heidelberg, Germany, 2013; Volume 943, ISBN 9781603273527. [Google Scholar]
- Kalle, E.; Kubista, M.; Rensing, C. Multi-template polymerase chain reaction. Biomol. Detect. Quantif. 2014, 2, 11–29. [Google Scholar] [CrossRef]
- Uchiyama, A.; Naritomi, Y.; Hashimoto, Y.; Hanada, T.; Watanabe, K.; Kitta, K.; Suzuki, G.; Komatsuno, T.; Nakamura, T. Understanding quantitative polymerase chain reaction bioanalysis issues before validation planning: Japan Bioanalysis Forum discussion group. Bioanalysis 2022, 14, 1391–1405. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Alm, E.; Karlsson, M.; Enkirch, T.; Norder, H.; Eriksson, R.; Simonsson, M.; Ellström, P. A new assay for quantitative detection of hepatitis A virus. J. Virol. Methods 2021, 288, 114010. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Karlsson, M.; Borsch-Reniers, H.; Ellström, P.; Eriksson, R.; Simonsson, M. Missing the Match Might Not Cost You the Game: Primer-Template Mismatches Studied in Different Hepatitis A Virus Variants. Food Environ. Virol. 2019, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Coudray-Meunier, C.; Fraisse, A.; Martin-Latil, S.; Guillier, L.; Delannoy, S.; Fach, P.; Perelle, S. A comparative study of digital RT-PCR and RT-qPCR for quantification of Hepatitis A virus and Norovirus in lettuce and water samples. Int. J. Food Microbiol. 2015, 201, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Persson, S.; Eriksson, R.; Lowther, J.; Ellström, P.; Simonsson, M. Comparison between RT droplet digital PCR and RT real-time PCR for quantification of noroviruses in oysters. Int. J. Food Microbiol. 2018, 284, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Hayden, R.T.; Gu, Z.; Ingersoll, J.; Abdul-Ali, D.; Shi, L.; Pounds, S.; Caliendo, A.M. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 2013, 51, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Dong, L.; Meng, Y.; Sui, Z.; Wang, J.; Wu, L.; Fu, B. Comparison of four digital PCR platforms for accurate quantification of DNA copy number of a certified plasmid DNA reference material. Sci. Rep. 2015, 5, 13174. [Google Scholar] [CrossRef]
- Huggett, J.F.; Cowen, S.; Foy, C.A. Considerations for Digital PCR as an Accurate Molecular Diagnostic Tool. Clin. Chem. 2015, 61, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Vynck, M.; Trypsteen, W.; Thas, O.; Vandekerckhove, L.; De Spiegelaere, W. The Future of Digital Polymerase Chain Reaction in Virology. Mol. Diagn. Ther. 2016, 20, 437–447. [Google Scholar] [CrossRef]
- Hall Sedlak, R.; Jerome, K.R. The potential advantages of digital PCR for clinical virology diagnostics. Expert Rev. Mol. Diagn. 2014, 14, 501–507. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R.; Mason, D.J.; Foy, C.A.; Huggett, J.F. Evaluation of digital PCR for absolute RNA quantification. PLoS ONE 2013, 8, e75296. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Kim, S.; Yoo, H.M.; Lee, D.H.; Bae, Y.K. Development of SARS-CoV-2 packaged RNA reference material for nucleic acid testing. Anal. Bioanal. Chem. 2022, 414, 1773–1785. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Bustin, S.A.; Huggett, J.F. Reproducibility of biomedical research—The importance of editorial vigilance. Biomol. Detect. Quantif. 2017, 11, 1–3. [Google Scholar] [CrossRef]
- Huggett, J.F.; Foy, C.A.; Benes, V.; Emslie, K.; Garson, J.A.; Haynes, R.; Hellemans, J.; Kubista, M.; Mueller, R.D.; Nolan, T.; et al. The digital MIQE guidelines: Minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013, 59, 892–902. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Lindner, L.; Cayrou, P.; Jacquot, S.; Birling, M.C.; Herault, Y.; Pavlovic, G. Reliable and robust droplet digital PCR (ddPCR) and RT-ddPCR protocols for mouse studies. Methods 2021, 191, 95–106. [Google Scholar] [CrossRef]
- Dybkaer, R. Metrology in laboratory medicine—Reference measurement systems. Accredit. Qual. Assur. 2001, 6, 16–19. [Google Scholar] [CrossRef]
- Thienpont, L.M.; Van Uytfanghe, K.; De Leenheer, A.P. Reference measurement systems in clinical chemistry. Clin. Chim. Acta 2002, 323, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.G.; Myers, G.L.; Ashwood, E.R.; Killeen, A.A.; Wang, E.; Thienpont, L.M.; Siekmann, L. Creatinine measurement: State of the art in accuracy and interlaboratory harmonization. Arch. Pathol. Lab. Med. 2005, 129, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Gullett, J.C.; Nolte, F.S. Quantitative nucleic acid amplification methods for viral infections. Clin. Chem. 2015, 61, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lin, G.; Li, J. Quantitative nucleic acid amplification by digital PCR for clinical viral diagnostics. Clin. Chem. Lab. Med. 2016, 54, 1427–1433. [Google Scholar] [CrossRef] [PubMed]
- Vesper, H.W.; Miller, W.G.; Myers, G.L. Reference materials and commutability. Clin. Biochem. Rev. 2007, 28, 139–147. [Google Scholar]
- Vim, I.S.O. International vocabulary of basic and general terms in metrology (VIM). Int. Organ. 2004, 2004, 9–14. [Google Scholar]
- Rutkowska, M.; Namieśnik, J.; Konieczka, P. Production of certified reference materials—Homogeneity and stability study based on the determination of total mercury and methylmercury. Microchem. J. 2020, 153, 104338. [Google Scholar] [CrossRef]
- Niu, C.; Dong, L.; Zhang, J.; Wang, D.; Gao, Y. Reference material development for detection of human respiratory syncytial virus using digital PCR. Anal. Bioanal. Chem. 2023, 415, 3131–3135. [Google Scholar] [CrossRef]
- Schiel, J.E.; Turner, A. The NISTmAb Reference Material 8671 lifecycle management and quality plan. Anal. Bioanal. Chem. 2018, 410, 2067–2078. [Google Scholar] [CrossRef]
- Schiel, J.E.; Turner, A.; Mouchahoir, T.; Yandrofski, K.; Telikepalli, S.; King, J.; DeRose, P.; Ripple, D.; Phinney, K. The NISTmAb Reference Material 8671 value assignment, homogeneity, and stability. Anal. Bioanal. Chem. 2018, 410, 2127–2139. [Google Scholar] [CrossRef]
- Trapmann, S.; Botha, A.; Linsinger, T.P.J.; Mac Curtain, S.; Emons, H. The new International Standard ISO 17034: General requirements for the competence of reference material producers. Accredit. Qual. Assur. 2017, 22, 381–387. [Google Scholar] [CrossRef]
- Park, C.; Lee, J.; Hassan, Z.U.; Ku, K.B.; Kim, S.J.; Kim, H.G.; Park, E.C.; Park, G.S.; Park, D.; Baek, S.H.; et al. Comparison of Digital PCR and Quantitative PCR with Various SARS-CoV-2 Primer-Probe Sets. J. Microbiol. Biotechnol. 2021, 31, 358–367. [Google Scholar] [CrossRef]
- Emslie, K.R.; McLaughlin, J.L.H.; Griffiths, K.; Forbes-Smith, M.; Pinheiro, L.B.; Burke, D.G. Droplet volume variability and impact on digital pcr copy number concentration measurements. Anal. Chem. 2019, 91, 4124–4131. [Google Scholar] [CrossRef] [PubMed]
- Corbisier, P.; Pinheiro, L.; Mazoua, S.; Kortekaas, A.M.; Chung, P.Y.J.; Gerganova, T.; Roebben, G.; Emons, H.; Emslie, K. DNA copy number concentration measured by digital and droplet digital quantitative PCR using certified reference materials. Anal. Bioanal. Chem. 2015, 407, 1831–1840. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, S.A.; Deveson, I.W.; Mercer, T.R. Reference standards for next-generation sequencing. Nat. Rev. Genet. 2017, 18, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, W.; Ma, S.; Li, Z.; Yao, Y.; Fei, T. A chemical-enhanced system for CRISPR-Based nucleic acid detection. Biosens. Bioelectron. 2021, 192, 113493. [Google Scholar] [CrossRef]
- Mei, H.; Zha, Z.; Wang, W.; Xie, Y.; Huang, Y.; Li, W.; Wei, D.; Zhang, X.; Qu, J.; Liu, J. Surfaceome CRISPR screen identifies OLFML3 as a rhinovirus-inducible IFN antagonist. Genome Biol. 2021, 22, 297. [Google Scholar] [CrossRef]
- Sathiamoorthy, S.; Malott, R.J.; Gisonni-Lex, L.; Ng, S.H.S. Selection and evaluation of an efficient method for the recovery of viral nucleic acid extraction from complex biologicals. Npj Vaccines 2018, 3, 31. [Google Scholar] [CrossRef]
- Ambrosi, C.; Prezioso, C.; Checconi, P.; Scribano, D.; Sarshar, M.; Capannari, M.; Tomino, C.; Fini, M.; Garaci, E.; Palamara, A.T.; et al. SARS-CoV-2: Comparative analysis of different RNA extraction methods. J. Virol. Methods 2021, 287, 114008. [Google Scholar] [CrossRef]
- Iwane, M.K.; Prill, M.M.; Lu, X.; Miller, E.K.; Edwards, K.M.; Hall, C.B.; Griffin, M.R.; Staat, M.A.; Anderson, L.J.; Williams, J.V.; et al. Human Rhinovirus Species Associated With Hospitalizations for Acute Respiratory Illness in Young US Children. J. Infect. Dis. 2011, 204, 1702–1710. [Google Scholar] [CrossRef]
- Tapparel, C.; Cordey, S.; Junier, T.; Farinelli, L.; Van Belle, S.; Soccal, P.M.; Aubert, J.-D.; Zdobnov, E.; Kaiser, L. Rhinovirus genome variation during chronic upper and lower respiratory tract infections. PLoS ONE 2011, 6, e21163. [Google Scholar] [CrossRef] [PubMed]
- Kiang, D.; Yagi, S.; Kantardjieff, K.A.; Kim, E.J.; Louie, J.K.; Schnurr, D.P. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J. Clin. Virol. 2007, 38, 227–237. [Google Scholar] [CrossRef]
- Jin, Y.; Yuan, X.H.; Xie, Z.P.; Gao, H.C.; Song, J.R.; Zhang, R.F.; Xu, Z.Q.; Zheng, L.S.; De Hou, Y.; Duan, Z.J. Prevalence and clinical characterization of a newly identified human rhinovirus C species in children with acute respiratory tract infection. J. Clin. Microbiol. 2009, 47, 2895–2900. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Yip, C.C.Y.; Tsoi, H.W.; Lee, R.A.; So, L.Y.; Lau, Y.L.; Chan, K.H.; Woo, P.C.Y.; Yuen, K.Y. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J. Clin. Microbiol. 2007, 45, 3655–3664. [Google Scholar] [CrossRef] [PubMed]
- Winther, B. Rhinoviruses. In International Encyclopedia of Public Health; Academic Press: Cambridge, MA, USA, 2008; pp. 577–581. [Google Scholar]
- Peltola, V.; Waris, M.; Österback, R.; Susi, P.; Hyypiä, T.; Ruuskanen, O. Clinical effects of rhinovirus infections. J. Clin. Virol. 2008, 43, 411–414. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, S.; Shafiei-Jandaghi, N.Z.; Shadab, A.; Hassani, S.A.; Foroushani, A.R.; Hosseinkhan, N.; Aghamir, F.; Mokhtari-Azad, T.; Yavarian, J. Phylogenetic characterization of rhinovirus and adenovirus in hospitalized children aged ≤ 18 years with severe acute respiratory infection in Iran. Iran. J. Microbiol. 2023, 15, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Spencer, J.A.; Shutt, D.P.; Moser, S.K.; Clegg, H.; Wearing, H.J.; Mukundan, H.; Manore, C.A. Epidemiological parameter review and comparative dynamics of influenza, respiratory syncytial virus, rhinovirus, human coronavirus, and adenovirus. medRxiv 2020. [Google Scholar] [CrossRef]
- Vitetta, L.; Du, S. The common cold. J. Altern. Complement. Med. 2008, 7, 56–57. [Google Scholar] [CrossRef]
- Florkowski, C.; Don-Wauchope, A.; Gimenez, N.; Rodriguez-Capote, K.; Wils, J.; Zemlin, A. Point-of-care testing (POCT) and evidence-based laboratory medicine (EBLM)—Does it leverage any advantage in clinical decision making? Crit. Rev. Clin. Lab. Sci. 2017, 54, 471–494. [Google Scholar] [CrossRef]
- Luppa, P.B.; Müller, C.; Schlichtiger, A.; Schlebusch, H. Point-of-care testing (POCT): Current techniques and future perspectives. TrAC Trends Anal. Chem. 2011, 30, 887–898. [Google Scholar] [CrossRef]
- Miller, W.G.; Myers, G.; Cobbaert, C.M.; Young, I.S.; Theodorsson, E.; Wielgosz, R.I.; Westwood, S.; Maniguet, S.; Gillery, P. Overcoming challenges regarding reference materials and regulations that influence global standardization of medical laboratory testing results. Clin. Chem. Lab. Med. 2023, 61, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Van den Bruel, A.; Cleemput, I.; Aertgeerts, B.; Ramaekers, D.; Buntinx, F. The evaluation of diagnostic tests: Evidence on technical and diagnostic accuracy, impact on patient outcome and cost-effectiveness is needed. J. Clin. Epidemiol. 2007, 60, 1116–1122. [Google Scholar] [CrossRef] [PubMed]
- Dupouey, J.; Ninove, L.; Ferrier, V.; Py, O.; Gazin, C.; Thirion-Perrier, L.; De Lamballerie, X. Molecular Detection of Human Rhinoviruses in Respiratory Samples: A Comparison of Taqman Probe-, SYBR Green I- and BOXTO-Based Real-Time PCR Assays. Virol. J. 2014, 11, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Wang, W.; Bessaud, M.; Ren, P.; Sheng, J.; Yan, H.; Zhang, J.; Lin, X.; Wang, Y.; Delpeyroux, F.; et al. Evidence of Recombination and Genetic Diversity in Human Rhinoviruses in Children with Acute Respiratory Infection. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; McIntyre, C.; Savolainen-Kopra, C.; Tapparel, C.; Mackay, I.M.; Hovi, T. Proposals for the Classification of Human Rhinovirus Species C into Genotypically Assigned Types. J. Gen. Virol. 2010, 91, 2409–2419. [Google Scholar] [CrossRef] [PubMed]
- Esneau, C.; Bartlett, N.; Bochkov, Y.A. Rhinovirus Structure, Replication, and Classification; Elsevier Inc.: Amsterdam, The Netherlands, 2019; ISBN 9780128164174. [Google Scholar]
- Whale, A.S.; Devonshire, A.S.; Karlin-Neumann, G.; Regan, J.; Javier, L.; Cowen, S.; Fernandez-Gonzalez, A.; Jones, G.M.; Redshaw, N.; Beck, J.; et al. International Interlaboratory Digital PCR Study Demonstrating High Reproducibility for the Measurement of a Rare Sequence Variant. Anal. Chem. 2017, 89, 1724–1733. [Google Scholar] [CrossRef]
- Košir, A.B.; Divieto, C.; Pavšič, J.; Pavarelli, S.; Dobnik, D.; Dreo, T.; Bellotti, R.; Sassi, M.P.; Žel, J. Droplet Volume Variability as a Critical Factor for Accuracy of Absolute Quantification Using Droplet Digital PCR. Anal. Bioanal. Chem. 2017, 409, 6689–6697. [Google Scholar] [CrossRef]
- Pinheiro, L.B.; Coleman, V.A.; Hindson, C.M.; Herrmann, J.; Hindson, B.J.; Bhat, S.; Emslie, K.R. Evaluation of a Droplet Digital Polymerase Chain Reaction Format for DNA Copy Number Quantification. Anal. Chem. 2012, 84, 1003–1011. [Google Scholar] [CrossRef]




| Homogeneity | Value |
|---|---|
| Average | 1.6 × 105 copies/μL |
| Standard deviation | 1.7 × 104 copies/μL |
| Relative standard deviation | 10.76% |
| Relative standard uncertainty | 3.2% |
| Expanded uncertainty | 4.5 × 104 copies/μL |
| k (95% level of confidence) | 2.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, D.U.; Park, D.; Kim, I.-H.; Kim, S.; Yoo, H.M. Development of Human Rhinovirus RNA Reference Material Using Digital PCR. Genes 2023, 14, 2210. https://doi.org/10.3390/genes14122210
Ju DU, Park D, Kim I-H, Kim S, Yoo HM. Development of Human Rhinovirus RNA Reference Material Using Digital PCR. Genes. 2023; 14(12):2210. https://doi.org/10.3390/genes14122210
Chicago/Turabian StyleJu, Dong U, Dongju Park, Il-Hwan Kim, Seil Kim, and Hee Min Yoo. 2023. "Development of Human Rhinovirus RNA Reference Material Using Digital PCR" Genes 14, no. 12: 2210. https://doi.org/10.3390/genes14122210
APA StyleJu, D. U., Park, D., Kim, I.-H., Kim, S., & Yoo, H. M. (2023). Development of Human Rhinovirus RNA Reference Material Using Digital PCR. Genes, 14(12), 2210. https://doi.org/10.3390/genes14122210