Optical Genome Mapping Helps to Identify BCR::JAK2 Rearrangement Arising from Cryptic Complex Chromosomal Aberrations: A Case Report and Literature Review
Abstract
:1. Introduction
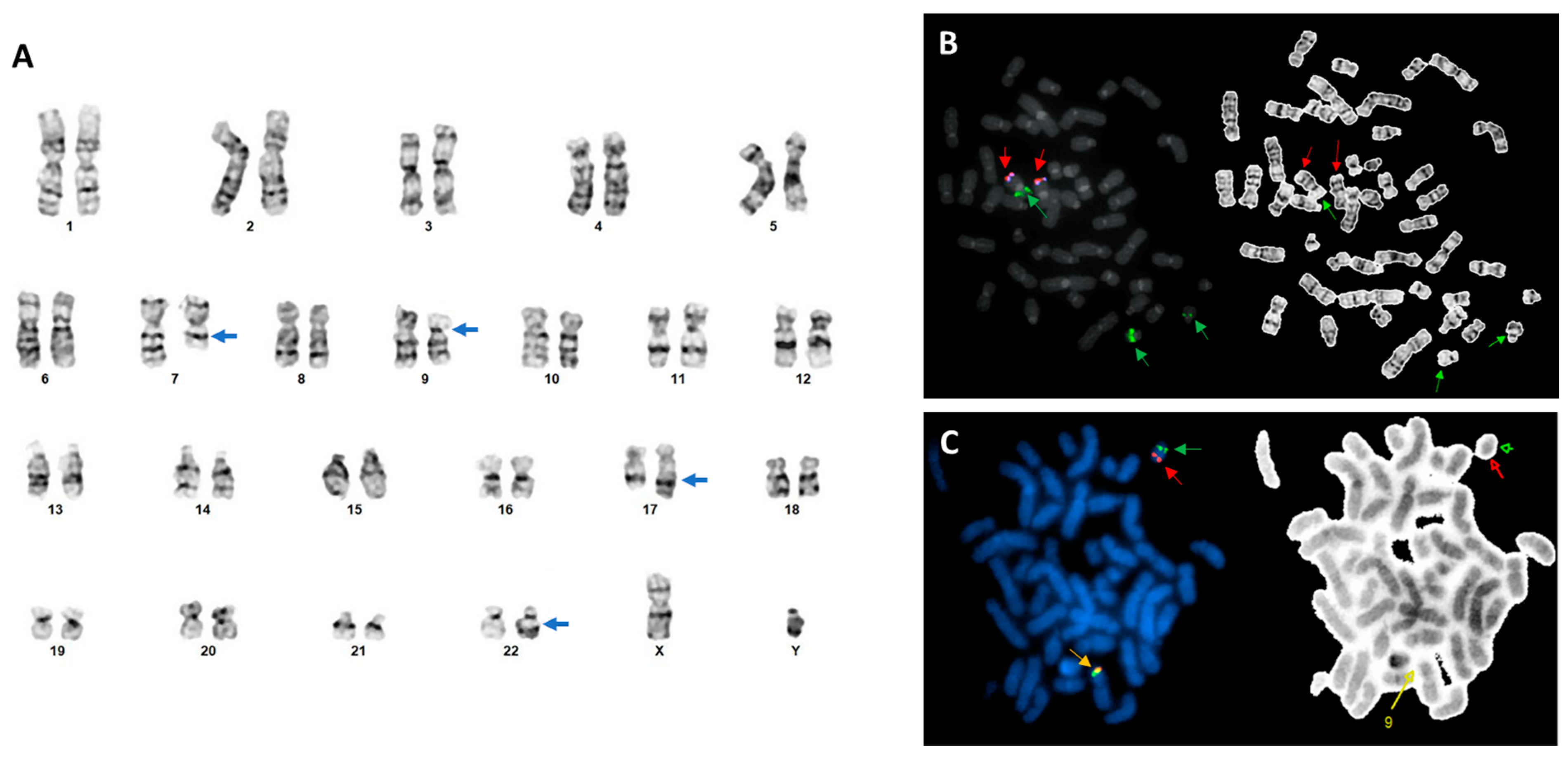
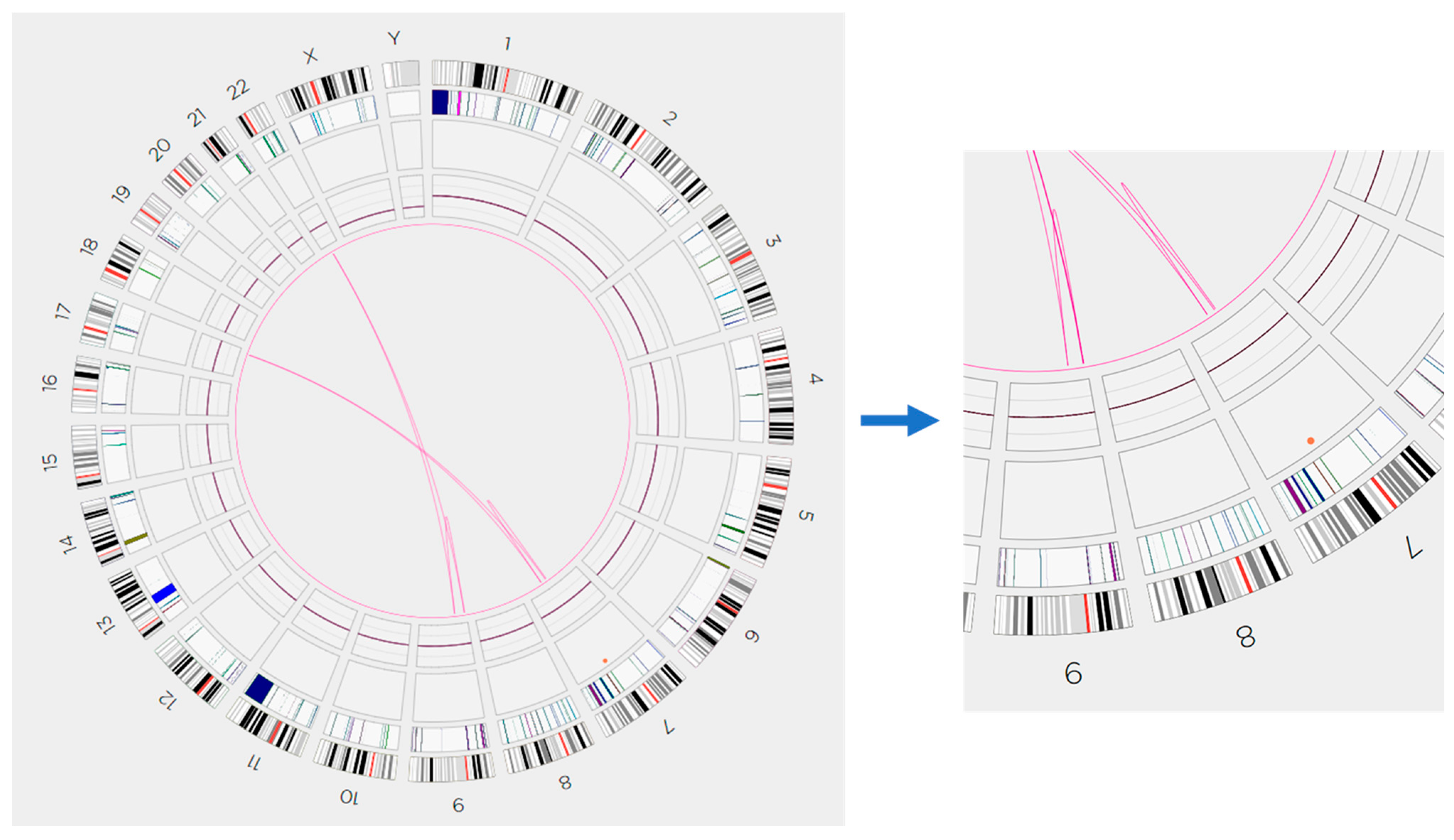
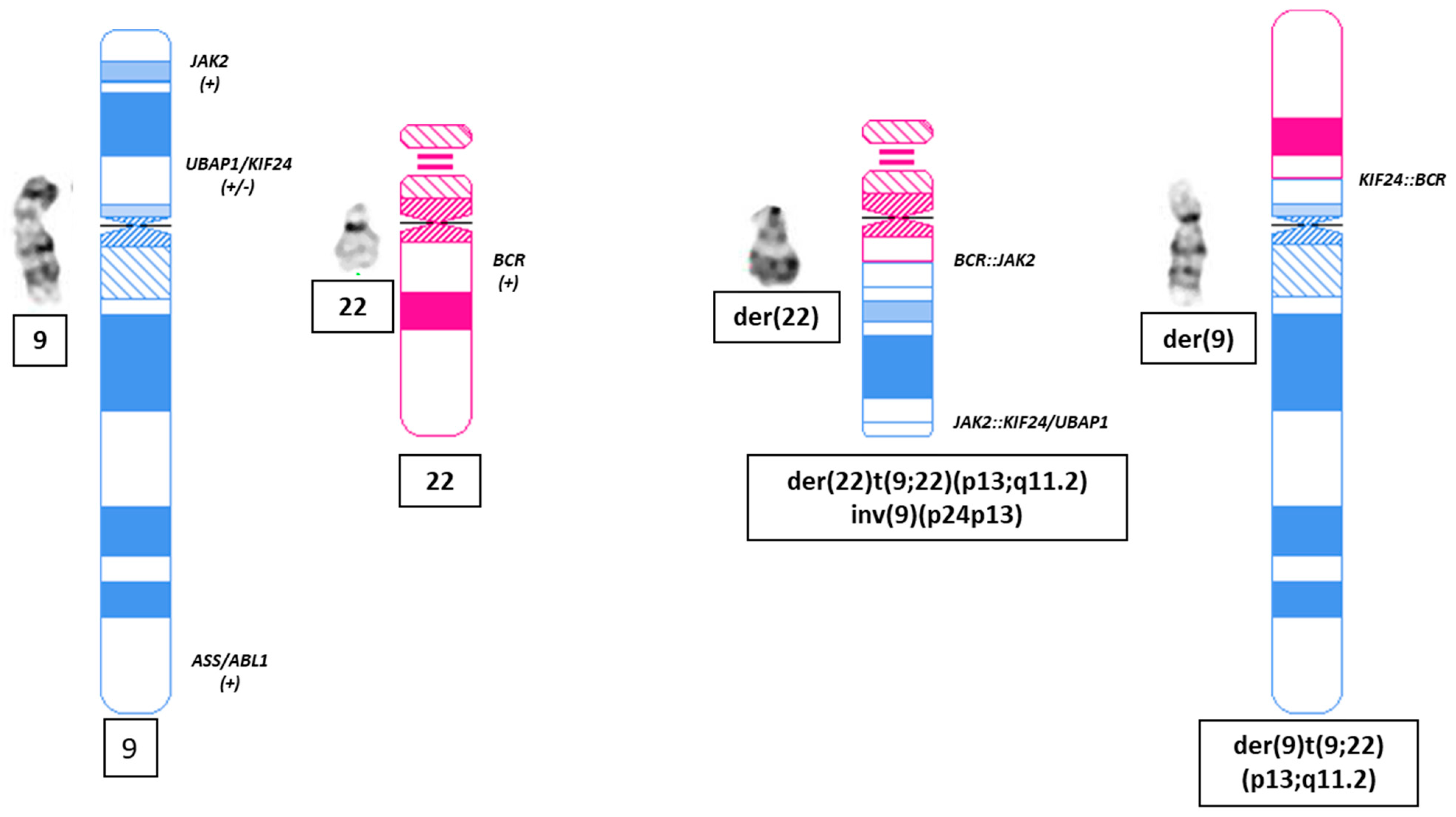
2. Case Report
3. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Griesinger, F.; Hennig, H.; Hillmer, F.; Podleschny, M.; Steffens, R.; Pies, A.; Wörmann, B.; Haase, D.; Bohlander, S.K. A BCR-JAK2 fusion gene as the result of a t(9;22)(p24;q11.2) translocation in a patient with a clinically typical chronic myeloid leukemia. Genes Chromosomes Cancer 2005, 44, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Lane, S.W.; Fairbairn, D.J.; McCarthy, C.; Nandini, A.; Perry-Keene, J.; Kennedy, G.A. Leukaemia cutis in atypical chronic myeloid leukaemia with a t(9;22)(p24;q11.2) leading to BCR-JAK2 fusion. Br. J. Haematol. 2008, 142, 503. [Google Scholar] [CrossRef] [PubMed]
- Impera, L.; Lonoce, A.; Fanfulla, D.A.; Moreilhon, C.; Legros, L.; Raynaud, S.; Storlazzi, C.T. Two alternatively spliced 5’BCR/3’JAK2 fusion transcripts in a myeloproliferative neoplasm with a three-way t(9;18;22)(p23;p11.3;q11.2) translocation. Cancer Genet. 2011, 204, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Elnaggar, M.M.; Agersborg, S.; Sahoo, T.; Girgin, A.; Ma, W.; Rakkhit, R.; Zorrilla, I.; Leal, A. BCR-JAK2 fusion as a result of a translocation (9;22)(p24;q11.2) in a patient with CML-like myeloproliferative disease. Mol. Cytogenet. 2012, 5, 23. [Google Scholar] [CrossRef]
- Xu, Y.; Yin, J.; Pan, J.; Wu, C.; Wang, Q.; Yao, H.; Wu, D.; Chen, S.; Sun, A. A BCR-JAK2 fusion gene from ins(22;9)(q11;p13p24) in a patient with atypical chronic myeloid leukemia. Leuk. Lymphoma 2013, 54, 2322–2324. [Google Scholar] [CrossRef]
- Chamseddine, A.N.; Etancelin, P.; Penther, D.; Parmentier, F.; Kuadjovi, C.; Camus, V.; Contentin, N.; Lenain, P.; Bastard, C.; Tilly, H.; et al. Transformation of an Unclassified Myeloproliferative Neoplasm with a Rare BCR-JAK2 Fusion Transcript Resulting from the Translocation (9;22)(p24;q11). Case Rep. Hematol. 2015, 2015, 252537. [Google Scholar] [CrossRef]
- Schwaab, J.; Knut, M.; Haferlach, C.; Metzgeroth, G.; Horny, H.P.; Chase, A.; Tapper, W.; Score, J.; Waghorn, K.; Naumann, N.; et al. Limited duration of complete remission on ruxolitinib in myeloid neoplasms with PCM1-JAK2 and BCR-JAK2 fusion genes. Ann. Hematol. 2015, 94, 233–238. [Google Scholar] [CrossRef]
- He, R.; Greipp, P.T.; Rangan, A.; Mai, M.; Chen, D.; Reichard, K.K.; Nelsen, L.L.; Pardanani, A.; Hanson, C.A.; Viswanatha, D.S. BCR-JAK2 fusion in a myeloproliferative neoplasm with associated eosinophilia. Cancer Genet. 2016, 209, 223–228. [Google Scholar] [CrossRef]
- Millett, R.; Aggarwal, A.; Tabbara, I.; Nassereddine, S. Chronic Myeloid Leukemia as Secondary Malignancy Following the Treatment of Hodgkin Lymphoma: A Case Series. Anticancer Res. 2019, 39, 4333–4335. [Google Scholar] [CrossRef]
- Snider, J.S.; Znoyko, I.; Lindsey, K.G.; Morse, J.; Baughn, L.B.; Hoppman, N.L.; Pitel, B.A.; Pearce, K.E.; Schandl, C.A.; Wolff, D.J. Integrated genomic analysis using chromosomal microarray, fluorescence in situ hybridization and mate pair analyses: Characterization of a cryptic t(9;22)(p24.1;q11.2)/BCR-JAK2 in myeloid/lymphoid neoplasm with eosinophilia. Cancer Genet. 2020, 246–247, 44–47. [Google Scholar] [CrossRef]
- Lap, C.J.; Nassereddine, S.; Liu, M.L.; Nava, V.E.; Aggarwal, A. Combined Ruxolitinib and Venetoclax Treatment in a Patient with a BCR-JAK2 Rearranged Myeloid Neoplasm. Case Rep. Hematol. 2021, 2021, 2348977. [Google Scholar] [CrossRef] [PubMed]
- Cirmena, G.; Aliano, S.; Fugazza, G.; Bruzzone, R.; Garuti, A.; Bocciardi, R.; Bacigalupo, A.; Ravazzolo, R.; Ballestrero, A.; Sessarego, M. A BCR-JAK2 fusion gene as the result of a t(9;22)(p24;q11) in a patient with acute myeloid leukemia. Cancer Genet. Cytogenet. 2008, 183, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Domínguez, Á.; Ortega, M.; Ormazábal, C.; Santos-Roncero, M.; Galán-Díez, M.; Steegmann, J.L.; Figuera, Á.; Arranz, E.; Vizmanos, J.L.; Bueren, J.A.; et al. Transforming and tumorigenic activity of JAK2 by fusion to BCR: Molecular mechanisms of action of a novel BCR-JAK2 tyrosine-kinase. PLoS ONE 2012, 7, e32451. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G.; Morin, R.D.; Zhang, J.; Hirst, M.; Zhao, Y.; Su, X.; Chen, S.C.; Payne-Turner, D.; Churchman, M.L.; Harvey, R.C.; et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell 2012, 22, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Duployez, N.; Nibourel, O.; Ducourneau, B.; Grardel, N.; Boyer, T.; Bories, C.; Darre, S.; Coiteux, V.; Berthon, C.; Preudhomme, C.; et al. Acquisition of genomic events leading to lymphoblastic transformation in a rare case of myeloproliferative neoplasm with BCR-JAK2 fusion transcript. Eur. J. Haematol. 2016, 97, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.A.; Hou, Y.; Roskin, K.M.; Arber, D.A.; Bangs, C.D.; Baughn, L.B.; Cherry, A.M.; Ewalt, M.D.; Fire, A.Z.; Fresard, L.; et al. Lymphoid blast transformation in an MPN with BCR-JAK2 treated with ruxolitinib: Putative mechanisms of resistance. Blood Adv. 2021, 5, 3492–3496. [Google Scholar] [CrossRef]
- Thakral, B.; Muzzafar, T.; Wang, S.A.; Medeiros, L.J. Myeloid neoplasm with eosinophilia and BCR-JAK2/t(9;22)(p24;q11.2) morphologically mimicking chronic myeloid leukemia. Ann. Diagn. Pathol. 2020, 44, 151405. [Google Scholar] [CrossRef]
- McGowan-Jordan, J.; Hastings, R.J.; Moore, S. ISCN 2020: An International System for Human Cytogenomic Nomenclature (2020); S.Karger AG: Basel, Switzerland, 2020. [Google Scholar]
- Yang, H.; Garcia-Manero, G.; Sasaki, K.; Montalban-Bravo, G.; Tang, Z.; Wei, Y.; Kadia, T.; Chien, K.; Rush, D.; Nguyen, H.; et al. High-resolution structural variant profiling of myelodysplastic syndromes by optical genome mapping uncovers cryptic aberrations of prognostic and therapeutic significance. Leukemia 2022, 36, 2306–2316. [Google Scholar] [CrossRef]
- Tirado, C.A.; Chen, W.; Huang, L.J.; Laborde, C.; Hiemenz, M.C.; Valdez, F.; Ho, K.; Winick, N.; Lou, Z.; Koduru, P. Novel JAK2 rearrangement resulting from a t(9;22)(p24;q11.2) in B-acute lymphoblastic leukemia. Leuk. Res. 2010, 34, 1674–1676. [Google Scholar] [CrossRef]
- Angelova, S.; Spassova, S.; Toshkov, S.; Shivarov, V. Chromosomal translocation t(9;22)(p24;q11) appears to be recurrently associated with myeloid malignancy with aggressive course. Leuk. Lymphoma 2011, 52, 1809–1810. [Google Scholar] [CrossRef]
- Bellesso, M.; Santucci, R.; Dias, D.F.; Centrone, R.; Elias, R.C. Atypical chronic myeloid leukemia with t(9;22)(p24,11.2), a BCR-JAK2 fusion gene. Rev. Bras. Hematol. Hemoter. 2013, 35, 218–219. [Google Scholar] [CrossRef] [PubMed]
- Kantarcioglu, B.; Kaygusuz-Atagunduz, I.; Uzay, A.; Toptas, T.; Tuglular, T.F.; Bayik, M. Myelodysplastic syndrome with t(9;22)(p24;q11.2), a BCR-JAK2 fusion: Case report and review of the literature. Int. J. Hematol. 2015, 102, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, P.; Zhang, D. Haemophagocytic syndrome triggered by acute lymphoblastic leukaemia with t(9;22)(p24; q11.2). J. Int. Med. Res. 2020, 48, 300060519874144. [Google Scholar] [CrossRef] [PubMed]
| SV Type | Chr Involved #1 | Chr Involved #2 | Breakpoint #1 (bp) | Breakpoint #1 (bp) | Confidence | VAF | Putative Gene Fusion | ISCN |
|---|---|---|---|---|---|---|---|---|
| Transl. | 9 | 22 | 34,271,272 | 23,219,177 | 0.98 | 0.48 | KIF24-BCR | ogm[GRCh38) t(9;22)(p13.3;q11.23) |
| Transl. | 9 | 22 | 5,079,902 | 23,205,817 | 0.99 | 0.5 | JAK2-BCR | ogm[GRCh38) t(9;22)(p24.1;q11.23) |
| intrachr_fusion | 9 | 22 | 5,077,416 | 34,246,297 | 0.99 | 0.47 | JAK2-UBAP1/KIF24 | ogm[GRCh38) fus(9;9)(p24.1;p13.3) |
| SV Type | Chr Involved #1 | Chr Involved #2 | Breakpoint #1 (bp) | Breakpoint #1 (bp) | Confidence | VAF | Putative Gene Fusion | ISCN |
|---|---|---|---|---|---|---|---|---|
| Transl. | 7 | 17 | 108,464,953 | 72,155,558 | 1 | 0.45 | ogm[GRCh38) t(7;17)(q31.1;q24.3) | |
| Transl. | 7 | 17 | 92,814,090 | 72,155,558 | 1 | 0.38 | CDK6-SOX9 | ogm[GRCh38) t(7;17)(q21.2;q24.3) |
| intrachr_fusion | 7 | 7 | 89,954,704 | 108,465,844 | 0.94 | 0.45 | ogm[GRCh38) fus(7;7)(q21.13;q31.1) |
| Year | Authors | Age/Sex | Diagnosis | Chromosomal Analysis | FISH Analysis | Confirmation of BCR::JAK2 | BCR::ABL1 Isoform(s) |
|---|---|---|---|---|---|---|---|
| 2005 | Griesinger et al. [1] | 63 yo, F | MPN-NOS | 46,XX,del(7)(q22q36)[21]/47,idem,t(9;22)(p24;q11.2),+19[5] | BCRx3 by BCR/ABL1 DF FISH | Yes. RT-PCR and Sanger Seq | E1::E19 |
| 2008 | Cirmena et al. [12] | 67 yo, F | AML * | 46,XX,t(9;22)(p24;q11) | BCRx3 by BCR/ABL1 DF FISH | Yes. RT-PCR and Sanger Seq | E14::E11 |
| 2008 | Lane et al. [2] | 44 yo, M | MPN-NOS | 46,XY,t(9;22)(p24;q11.2) [23]/46,XY[27] | BCR/ABL1 DF FISH-negative | Yes. RT-PCR and Sanger Seq | E1::E17 |
| 2011 | Impera et al. [3] | 54 yo, F | MPN-NOS | 46,XX,t(9;18;22)(p23;p11.3;q11.2)[18]/46,XX[2] | BCR/ABL1 FISH-negative, other intensive FISH studies | Yes. SNP array, RT-PCR, and Sanger Seq | E1::E15 and E1::E17 |
| 2012 | Cuesta-Dominguez et al. [13] | 55 yo, F | B-ALL | 49,XY,+X,+2,+4,−9,−11,+19,add(19)(q13),+20,−22,+mar[24]/46,XY[1] | BCR/JAK2 fusion FISH-positive | Yes. RT-PCR and Sanger seq | E1::E19 |
| 2012 | Elnaggar et al. [4] | 84 yo, M | MPN-NOS | 46,XX,t(9;22)(p24;q11.2) | BCRx3 by BCR/ABL1 DF FISH | Yes. RT-PCR and Sanger seq | E1::E19 |
| 2012 | Roberts et al. [14] | 2 yo, M | B-ALL | 47,XY,+2,del(2)(p23),t(3;22;9)(p12;q11.2; p24)[10]/46,XY[2] | n/a | Yes. WGS, RNA Seq, RT-PCR, and Sanger Seq | E1::E15 and E1::E17 |
| 2013 | Xu et al. [5] | 28 yo, M | MPN-NOS | ins(22;9)(q11;p13p24) | JAK2 BAP-positive | Yes. RT-PCR and Sanger seq | E1::E19 |
| 2015 | Chamseddine et al. [6] | 49 yo, M | MPN-NOS | 46,Y,t(9;22)(p24;q11)[12/20]/47,idem,+der(22)t(9;22)[2/20]/46,XY [6/20] | BCRx3 by BCR/ABL1 DF FISH; JAK2 BAP FISH-positive | Yes. RT-PCR and Sanger seq | E13::E17 |
| 2015 | Schwaab et al. [7] | ? | MPN-NOS | t(9;18)(p24;q12),der(18)ins(22)(BCR) | JAK2 BAP-positive | Yes. RT-PCR and Sanger seq | E1::E17 |
| 2016 | Duployez et al. [15] | 58 yo, M | B-ALL * | 47,XY,+8,t(9;22;15)(p24;q11;q21)(at MPN-U stage) | JAK2 BAP FISH-positive | Yes. RT-PCR and Sanger seq | E1::E17 |
| 2016 | He et al. [8] | 36 yo, F | MPN-NOS | Complex karyotype with multiple rearrangements involving chromosome 22, including a non-classical, unbalanced t(9;22) resulting in a derivative 9 from a t(9;22) and a dicentric rearrangement involving chromosomes 22 and 10. | BCRx3 by BCR/ABL1 DF FISH; JAK2 BAP FISH-positive | Yes. RT-PCR and Sanger seq | E1::E15 and E1::E17 |
| 2019 | Millett et al. [9] | 53 yo, M | MPN-NOS ** | n/a | JAK2 BAP-positive | Yes. RT-PCR | n/a |
| 2020 | Snider et al. [10] | 59 yo, M | Myeloid/lymphoid neoplasm with eosinophilia | 46,XY,t(3;21)(q21;q22)[10]/47,XY,+11[5]/46,XY[5] | JAK2 BAP-positive | Yes. Mate-pair seq | n/a |
| 2020 | Thakral et al. [17] | 31 yo, M | Myeloid neoplasm with eosinophilia | 46,XY,add(22)(q11.2)[13]/46,XY[7] | BCRx3 by BCR/ABL1 DF FISH | Yes. RNA seq | n/a |
| 2021 | Chen et al. [16] | 32 yo, M | B-ALL * | 46,XY,t(9;22)(p24;q11.2)[15]/46,XY[5] (with IKZF1 deletion) | BCR signal on der(9) by BCR/ABL1 DF FISH | Yes. WGS, RT-PCR, and Sanger Seq | E1::E19 |
| 2021 | Lap et al. [11] | 54 yo, M | MPN-NOS ** | 46,XY,del(6)(p21.2p24),t(22;9;11)(q11.2,p24,p11.2)[11]/46XY,del(9)[9] | BCRx3 by BCR/ABL1 DF FISH; JAK2 BAP FISH-positive | Yes. RT-PCR and Sanger seq | E1::E19 |
| 2010 | Tirado et al. [20] | 14 yo, M | B-ALL | 46,XY,t(9;22)(p24;q11.2)[14]/46,XY[6] | JAK2 BAP-positive | No | |
| 2011 | Angelova et al. [21] | 53 yo, M | MPN-NOS/MDS with Eos and baso | 47,XY,dup(3)(q25q27),+8,der(9)t(9;22)(p24;q11),dup(14)(q24q32),+14,−22[13]/47,XY,dup(1)(p13p22),dup(3)(q25q27),+8,add(8)(q24),der(9)t(9;22)(p24;q11),−22,+mar [6]/47,XY,dup(3)(q25q27),+8,t(9;22)(p24;q11),dup(14)(q24q32),+14,−21[1] | No | No | n/a |
| 2013 | Bellesso et al. [22] | 54 yo, M | MPN-NOS | 46,XY,t(9;22)(p24;q11.2) | BCR/ABL1 DF, BCR/PDGFR DF FISH-negative | No | n/a |
| 2015 | Kantarcioglu et al. [23] | 64 yo, F | MDS | 47,X,-X,+3,t(9;22)(p24;q11.2),+18[3]/46,XX[17] | BCR/ABL1 DF | No | |
| 2020 | Chen et al. [24] | 45 yo, M | B-ALL | 46,XY,t(9;22)(p24;q11.2)[9]/46,XY[2] | No | No JAK2-BCR and BCR-JAK2 fusion was detected by RT-PCR | |
| This study | Vanjari et al. | 39 yo, M | AML * | 46,XY,t(7;17)(q21;q24),der(9)t(9;22)(p13;q11.2),der(22)t(9;22)inv(9)(p13p24)[20] | BCRx3 by BCR/ABL1/ASS1 DF FISH; JAK2 BAP FISH-positive | Yes. OGM | E1:E17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanjari, N.; Tang, G.; Toruner, G.A.; Wang, W.; Thakral, B.; Zhao, M.; Dave, B.J.; Khoury, J.D.; Medeiros, L.J.; Tang, Z. Optical Genome Mapping Helps to Identify BCR::JAK2 Rearrangement Arising from Cryptic Complex Chromosomal Aberrations: A Case Report and Literature Review. Genes 2023, 14, 2188. https://doi.org/10.3390/genes14122188
Vanjari N, Tang G, Toruner GA, Wang W, Thakral B, Zhao M, Dave BJ, Khoury JD, Medeiros LJ, Tang Z. Optical Genome Mapping Helps to Identify BCR::JAK2 Rearrangement Arising from Cryptic Complex Chromosomal Aberrations: A Case Report and Literature Review. Genes. 2023; 14(12):2188. https://doi.org/10.3390/genes14122188
Chicago/Turabian StyleVanjari, Neelam, Guilin Tang, Gokce A. Toruner, Wei Wang, Beenu Thakral, Ming Zhao, Bhavana J. Dave, Joseph D. Khoury, L. Jeffrey Medeiros, and Zhenya Tang. 2023. "Optical Genome Mapping Helps to Identify BCR::JAK2 Rearrangement Arising from Cryptic Complex Chromosomal Aberrations: A Case Report and Literature Review" Genes 14, no. 12: 2188. https://doi.org/10.3390/genes14122188
APA StyleVanjari, N., Tang, G., Toruner, G. A., Wang, W., Thakral, B., Zhao, M., Dave, B. J., Khoury, J. D., Medeiros, L. J., & Tang, Z. (2023). Optical Genome Mapping Helps to Identify BCR::JAK2 Rearrangement Arising from Cryptic Complex Chromosomal Aberrations: A Case Report and Literature Review. Genes, 14(12), 2188. https://doi.org/10.3390/genes14122188









