Transcriptome Profiling after Early Spinal Cord Injury in the Axolotl and Its Comparison with Rodent Animal Models through RNA-Seq Data Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Pre-Processing
2.3. Differential Gene Expression Analysis
2.4. Gene Ontology Analysis
2.5. Code Availability
3. Results and Discussion
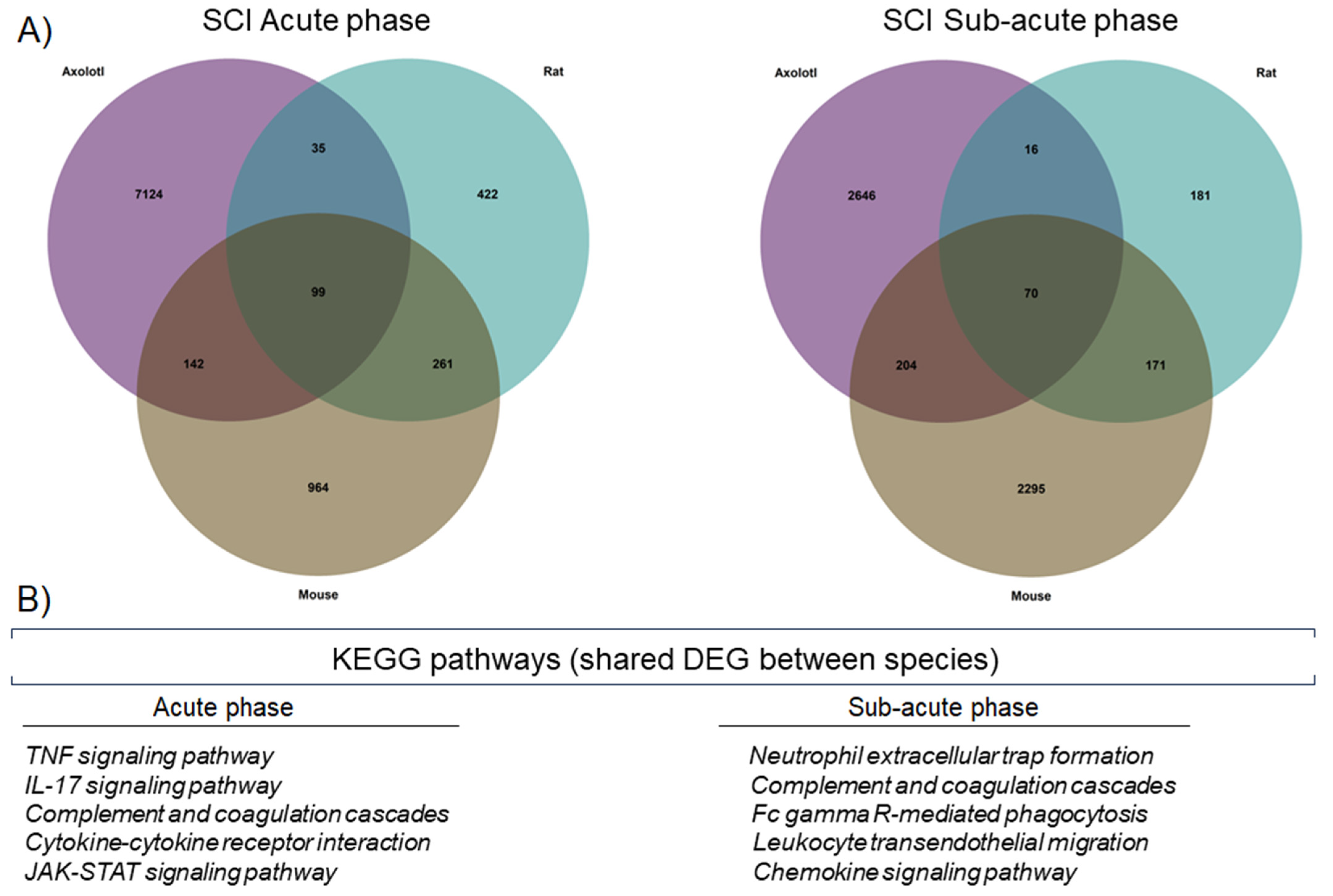
3.1. Changes in the Global Gene Expression after Traumatic SCI Show Large Differences between Axolotls and Rodents
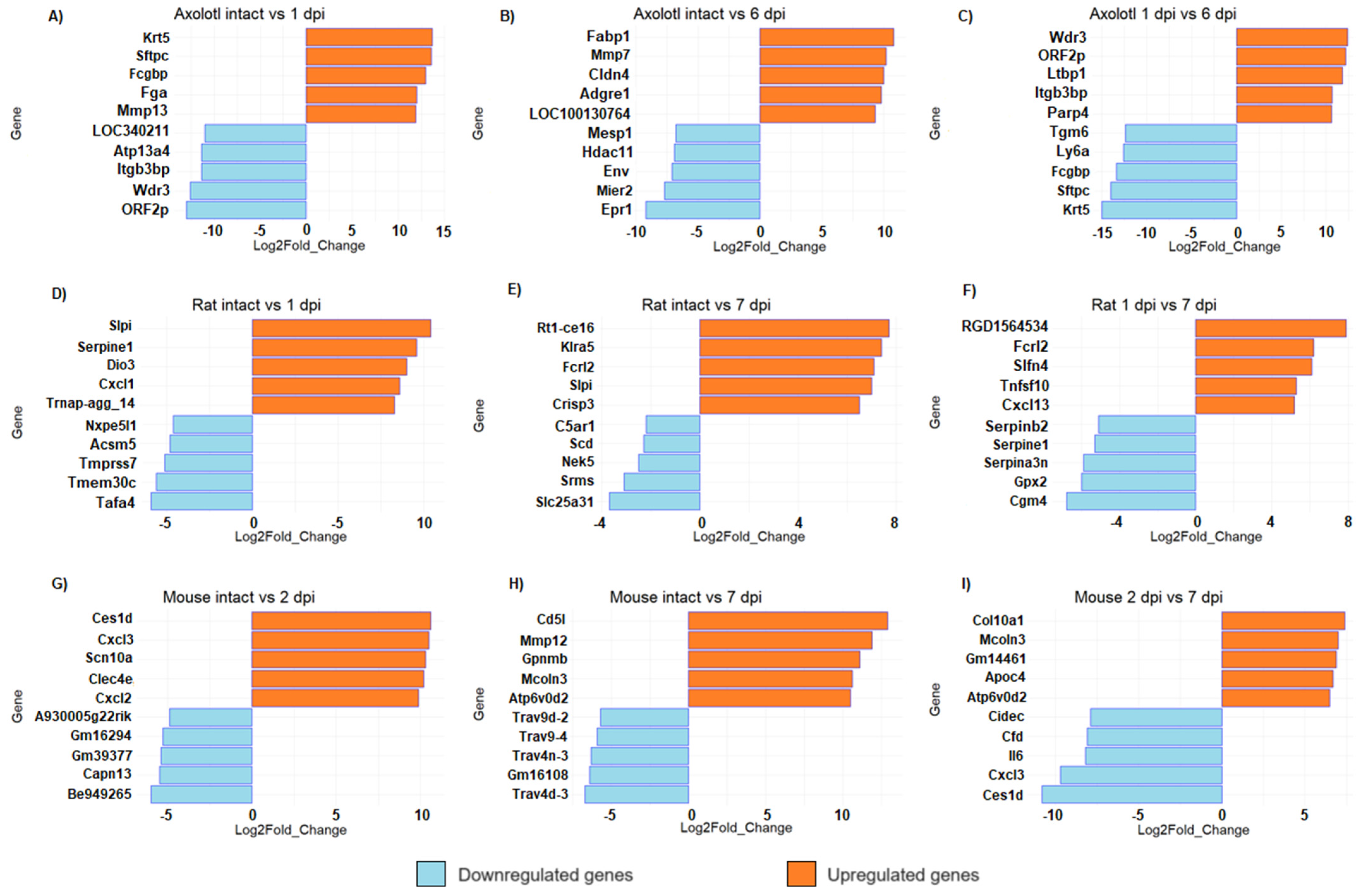
3.2. Top Regulated Genes in Axolotl and Rodents in the Acute and Sub-Acute Stages of SCI
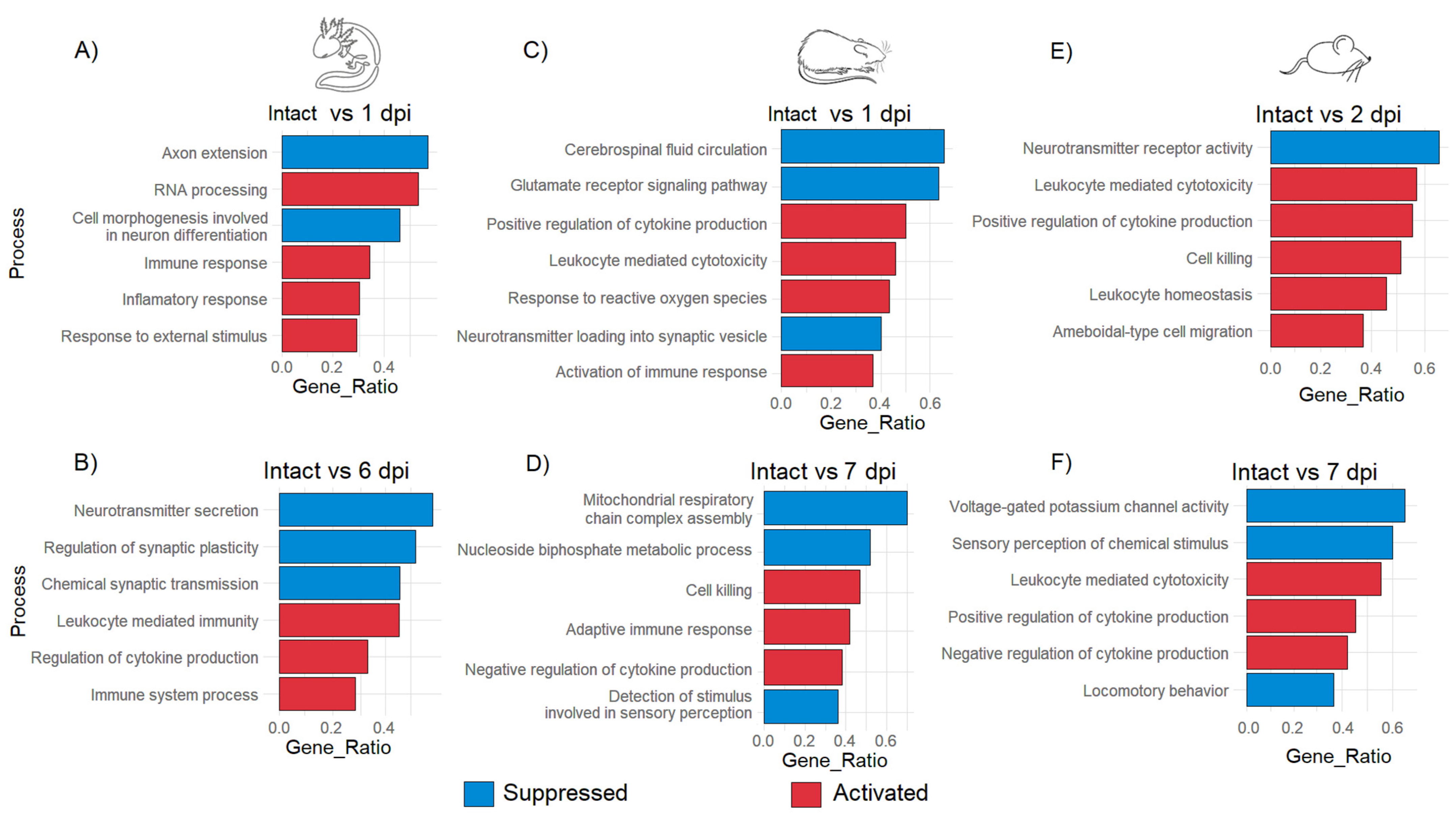
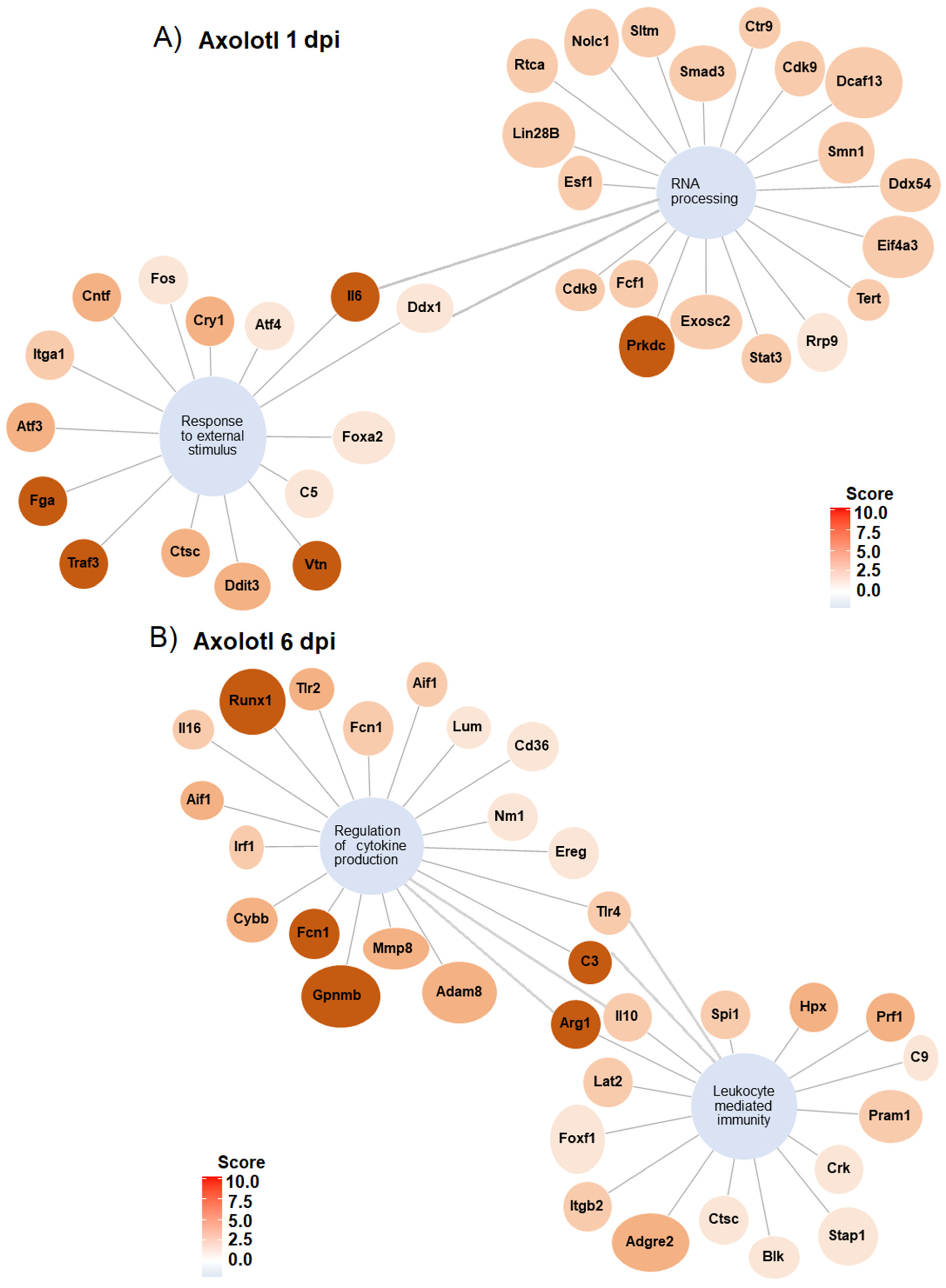
3.3. Gene Ontology Analysis Shows Differences and Similarities in Biological Processes between Axolotl and Rodents after Traumatic SCI
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ahuja, C.S.; Wilson, J.R.; Nori, S.; Kotter, M.R.N.; Druschel, C.; Curt, A.; Fehlings, M.G. Traumatic spinal cord injury. Nat. Rev. Dis. Primers 2017, 3, 17018. [Google Scholar] [CrossRef]
- Kooijmans, H.; Post, M.W.M.; Stam, H.J.; van der Woude, L.H.V.; Spijkerman, D.C.M.; Snoek, G.J.; Bongers-Janssen, H.M.H.; van Koppenhagen, C.F.; Twisk, J.W.; Bussmann, J.B.J. Effectiveness of a Self-Management Intervention to Promote an Active Lifestyle in Persons With Long-Term Spinal Cord Injury: The HABITS Randomized Clinical Trial. Neurorehabilit. Neural Repair 2017, 31, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.; Dyck, S.M.; Karimi-Abdolrezaee, S. Traumatic Spinal Cord Injury: An Overview of Pathophysiology, Models and Acute Injury Mechanisms. Front. Neurol. 2019, 10, 282. [Google Scholar] [CrossRef] [PubMed]
- Müller-Jensen, L.; Ploner, C.J.; Kroneberg, D.; Schmidt, W.U. Clinical Presentation and Causes of Non-Traumatic Spinal Cord Injury: An Observational Study in Emergency Patients. Front. Neurol. 2021, 12, 701927. [Google Scholar] [CrossRef]
- Yang, T.; Dai, Y.; Chen, G.; Cui, S. Dissecting the Dual Role of the Glial Scar and Scar-Forming Astrocytes in Spinal Cord Injury. Front. Cell. Neurosci. 2020, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Fauss, G.N.K.; Hudson, K.E.; Grau, J.W. Role of Descending Serotonergic Fibers in the Development of Pathophysiology after Spinal Cord Injury (SCI): Contribution to Chronic Pain, Spasticity, and Autonomic Dysreflexia. Biology 2022, 11, 234. [Google Scholar] [CrossRef]
- Wang, T.Y.; Park, C.; Zhang, H.; Rahimpour, S.; Murphy, K.R.; Goodwin, C.R.; Karikari, I.O.; Than, K.D.; Shaffrey, C.I.; Foster, N.; et al. Management of Acute Traumatic Spinal Cord Injury: A Review of the Literature. Front. Surg. 2021, 8, 698736. [Google Scholar] [CrossRef]
- Kumar, R.; Lim, J.; Mekary, R.A.; Rattani, A.; Dewan, M.C.; Sharif, S.Y.; Osorio-Fonseca, E.; Park, K.B. Traumatic Spinal Injury: Global Epidemiology and Worldwide Volume. World Neurosurg. 2018, 113, e345–e363. [Google Scholar] [CrossRef]
- Sharif-Alhoseini, M.; Khormali, M.; Rezaei, M.; Safdarian, M.; Hajighadery, A.; Khalatbari, M.M.; Safdarian, M.; Meknatkhah, S.; Rezvan, M.; Chalangari, M.; et al. Animal Models of Spinal Cord Injury: A Systematic Review. Spinal Cord 2017, 55, 714–721. [Google Scholar] [CrossRef]
- Amamoto, R.; Huerta, V.G.L.; Takahashi, E.; Dai, G.; Grant, A.K.; Fu, Z.; Arlotta, P. Adult axolotls can regenerate original neuronal diversity in response to brain injury. eLife 2016, 5, e13998. [Google Scholar] [CrossRef] [PubMed]
- Mchedlishvili, L.; Epperlein, H.H.; Telzerow, A.; Tanaka, E.M. A Clonal Analysis of Neural Progenitors during Axolotl Spinal Cord Regeneration Reveals Evidence for Both Spatially Restricted and Multipotent Progenitors. Development 2007, 134, 2083–2093. [Google Scholar] [CrossRef]
- Haas, B.J.; Whited, J.L. Advances in Decoding Axolotl Limb Regeneration. Trends Genet. 2017, 33, 553–565. [Google Scholar] [CrossRef]
- Sibai, M.; Parlayan, C.; Tuğlu, P.; Öztürk, G.; Demircan, T. Integrative Analysis of Axolotl Gene Expression Data from Regenerative and Wound Healing Limb Tissues. Sci. Rep. 2019, 9, 20280. [Google Scholar] [CrossRef]
- Leinonen, R.; Akhtar, R.; Birney, E.; Bower, L.; Cerdeno-Tarraga, A.; Cheng, Y.; Cleland, I.; Faruque, N.; Goodgame, N.; Gibson, R.; et al. The European Nucleotide Archive. Nucleic Acids Res. 2011, 39, D28–D31. [Google Scholar] [CrossRef] [PubMed]
- Nowoshilow, S.; Schloissnig, S.; Fei, J.-F.; Dahl, A.; Pang, A.W.C.; Pippel, M.; Winkler, S.; Hastie, A.R.; Young, G.; Roscito, J.G.; et al. The Axolotl Genome and the Evolution of Key Tissue Formation Regulators. Nature 2018, 554, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Z.; Zhu, Z.; Liang, Z.; Zuo, X.; Ju, C.; Song, Z.; Li, X.; Hu, X.; Wang, Z. Photobiomodulation Promotes Repair Following Spinal Cord Injury by Regulating the Transformation of A1/A2 Reactive Astrocytes. Front. Neurosci. 2021, 15, 768262. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Deng, S.; Lu, H.; Zheng, Y.; Yang, G.; Kim, D.; Cao, Q.; Wu, J.Q. RNA-Seq Characterization of Spinal Cord Injury Transcriptome in Acute/Subacute Phases: A Resource for Understanding the Pathology at the Systems Level. PLoS ONE 2013, 8, e72567. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Schloissnig, S.; Kawaguchi, A.; Nowoshilow, S.; Falcon, F.; Otsuki, L.; Tardivo, P.; Timoshevskaya, N.; Keinath, M.C.; Smith, J.J.; Voss, S.R.; et al. The Giant Axolotl Genome Uncovers the Evolution, Scaling, and Transcriptional Control of Complex Gene Loci. Proc. Natl. Acad. Sci. USA 2021, 118, e2017176118. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R Package Rsubread Is Easier, Faster, Cheaper and Better for Alignment and Quantification of RNA Sequencing Reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [PubMed]
- Saremi, B.; Gusmag, F.; Distl, O.; Schaarschmidt, F.; Metzger, J.; Becker, S.; Jung, K. A Comparison of Strategies for Generating Artificial Replicates in RNA-Seq Experiments. Sci. Rep. 2022, 12, 7170. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lv, J.; Huang, Y.-F.; Hao, D.-J.; Liu, J.-J. Bioinformatics analyses of differentially expressed genes associated with spinal cord injury: A microarray-based analysis in a mouse model. Neural Regen. Res. 2019, 14, 1262. [Google Scholar] [PubMed]
- Sabin, K.Z.; Jiang, P.; Gearhart, M.D.; Stewart, R.; Echeverri, K. AP-1cFos/JunB/miR-200a Regulate the pro-Regenerative Glial Cell Response during Axolotl Spinal Cord Regeneration. Commun. Biol. 2019, 2, 91. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.-L.; Zhang, N.; Xie, X.-M.; Chen, Y.-J.; Wang, R.; Shen, L.; Zhou, J.-S.; Hu, J.-G.; Lü, H.-Z. Transcriptome Profile of Rat Genes in Injured Spinal Cord at Different Stages by RNA-Sequencing. BMC Genom. 2017, 18, 173. [Google Scholar] [CrossRef]
- Seymour, T.; Twigger, A.-J.; Kakulas, F. Pluripotency Genes and Their Functions in the Normal and Aberrant Breast and Brain. Int. J. Mol. Sci. 2015, 16, 27288–27301. [Google Scholar] [CrossRef]
- Hillary, R.F.; FitzGerald, U. A Lifetime of Stress: ATF6 in Development and Homeostasis. J. Biomed. Sci. 2018, 25, 48. [Google Scholar] [CrossRef]
- Haenzi, B.; Moon, L.D. The Function of FGFR1 Signalling in the Spinal Cord: Therapeutic Approaches Using FGFR1 Ligands after Spinal Cord Injury. Neural Plast. 2017, 2017, 2740768. [Google Scholar] [CrossRef]
- Tica, J.; Didangelos, A. Comparative Transcriptomics of Rat and Axolotl After Spinal Cord Injury Dissects Differences and Similarities in Inflammatory and Matrix Remodeling Gene Expression Patterns. Front. Neurosci. 2018, 12, 808. [Google Scholar] [CrossRef] [PubMed]
- Hellenbrand, D.J.; Quinn, C.M.; Piper, Z.J.; Morehouse, C.N.; Fixel, J.A.; Hanna, A.S. Inflammation after spinal cord injury: A review of the critical timeline of signaling cues and cellular infiltration. J. Neuroinflammation 2021, 18, 284. [Google Scholar] [CrossRef] [PubMed]
- Leibinger, M.; Zeitler, C.; Gobrecht, P.; Andreadaki, A.; Gisselmann, G.; Fischer, D. Transneuronal Delivery of Hyper-Interleukin-6 Enables Functional Recovery after Severe Spinal Cord Injury in Mice. Nat. Commun. 2021, 12, 391. [Google Scholar] [CrossRef] [PubMed]
- Tazaki, A.; Tanaka, E.M.; Fei, J.-F. Salamander Spinal Cord Regeneration: The Ultimate Positive Control in Vertebrate Spinal Cord Regeneration. Dev. Biol. 2017, 432, 63–71. [Google Scholar] [CrossRef]
- Randal Voss, S.; Murrugarra, D.; Jensen, T.B.; Monaghan, J.R. Transcriptional Correlates of Proximal-Distal Identify and Regeneration Timing in Axolotl Limbs. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2018, 208, 53–63. [Google Scholar] [CrossRef]
- Shechter, R.; Raposo, C.; London, A.; Sagi, I.; Schwartz, M. The Glial Scar-Monocyte Interplay: A Pivotal Resolution Phase in Spinal Cord Repair. PLoS ONE 2011, 6, e27969. [Google Scholar] [CrossRef]
- Ardeljan, D.; Wang, X.; Oghbaie, M.; Taylor, M.S.; Husband, D.; Deshpande, V.; Steranka, J.P.; Gorbounov, M.; Yang, W.R.; Sie, B.; et al. LINE-1 ORF2p expression is nearly imperceptible in human cancers. Mob. DNA 2019, 11, 1. [Google Scholar] [CrossRef]
- Kamizato, K.; Sato, S.; Shil, S.K.; Umaru, B.A.; Kagawa, Y.; Yamamoto, Y.; Ogata, M.; Yasumoto, Y.; Okuyama, Y.; Ishii, N.; et al. The Role of Fatty Acid Binding Protein 7 in Spinal Cord Astrocytes in a Mouse Model of Experimental Autoimmune Encephalomyelitis. Neuroscience 2019, 409, 120–129. [Google Scholar] [CrossRef]
- Arrildt, K.T.; Joseph, S.B.; Swanstrom, R. The HIV-1 env protein: A coat of many colors. Curr. HIV/AIDS Rep. 2012, 9, 52–63. [Google Scholar] [CrossRef]
- Pineau, I.; Sun, L.; Bastien, D.; Lacroix, S. Astrocytes Initiate Inflammation in the Injured Mouse Spinal Cord by Promoting the Entry of Neutrophils and Inflammatory Monocytes in an IL-1 Receptor/MyD88-Dependent Fashion. Brain Behav. Immun. 2010, 24, 540–553. [Google Scholar] [CrossRef]
- Hannila, S.S. Secretory Leukocyte Protease Inhibitor (SLPI): Emerging Roles in CNS Trauma and Repair. Neurosci. Rev. J. Bringing Neurobiol. Neurol. Psychiatry 2015, 21, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Ghasemlou, N.; Bouhy, D.; Yang, J.; López-Vales, R.; Haber, M.; Thuraisingam, T.; He, G.; Radzioch, D.; Ding, A.; David, S. Beneficial effects of secretory leukocyte protease inhibitor after spinal cord injury. Brain 2010, 133, 126–138. [Google Scholar] [CrossRef]
- Urso, M.L.; Chen, Y.; Scrimgeour, A.G.; Lee, P.C.; Lee, K.F.; Clarkson, P.M. Alterations in mRNA Expression and Protein Products Following Spinal Cord Injury in Humans. J. Physiol. 2007, 579, 877–892. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.E.A.; Rice, T.K.; Nuttall, R.K.; Edwards, D.R.; Zekki, H.; Rivest, S.; Yong, V.W. An Adverse Role for Matrix Metalloproteinase 12 after Spinal Cord Injury in Mice. J. Neurosci. 2003, 23, 10107–10115. [Google Scholar] [CrossRef] [PubMed]
- Demircan, T. Dissecting the Molecular Signature of Spinal Cord Regeneration in the Axolotl Model. Cureus 2020, 12, e7014. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.L.; Ge, X.; Xie, Z.; Zhou, Y.; Tsai, L.H. Control of activating transcription factor 4 (ATF4) persistence by multisite phosphorylation impacts cell cycle progression and neurogenesis. J. Biol. Chem. 2010, 285, 33324–33337. [Google Scholar] [CrossRef] [PubMed]
- Si, Z.-P.; Wang, G.; Han, S.-S.; Jin, Y.; Hu, Y.-X.; He, M.-Y.; Brand-Saberi, B.; Yang, X.; Liu, G.-S. CNTF and Nrf2 Are Coordinately Involved in Regulating Self-Renewal and Differentiation of Neural Stem Cell during Embryonic Development. iScience 2019, 19, 303–315. [Google Scholar] [CrossRef]
- Petrović, A.; Ban, J.; Ivaničić, M.; Tomljanović, I.; Mladinic, M. The Role of ATF3 in Neuronal Differentiation and Development of Neuronal Networks in Opossum Postnatal Cortical Cultures. Int. J. Mol. Sci. 2022, 23, 4964. [Google Scholar] [CrossRef]
- Ruzha, Y.; Ni, J.; Quan, Z.; Li, H.; Qing, H. Role of Vitronectin and Its Receptors in Neuronal Function and Neurodegenerative Diseases. Int. J. Mol. Sci. 2022, 23, 12387. [Google Scholar] [CrossRef]
- Yuan, X.; Xu, D. Telomerase Reverse Transcriptase (TERT) in Action: Cross-Talking with Epigenetics. Int. J. Mol. Sci. 2019, 20, 3338. [Google Scholar] [CrossRef]
- Tsai, S.L.; Baselga-Garriga, C.; Melton, D.A. Blastemal Progenitors Modulate Immune Signaling during Early Limb Regeneration. Development 2019, 146, dev169128. [Google Scholar] [CrossRef] [PubMed]
- Vieira, W.A.; Wells, K.M.; McCusker, C.D. Advancements to the Axolotl Model for Regeneration and Aging. Gerontology 2020, 66, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.A.; Burda, J.E.; Ren, Y.; Ao, Y.; O’Shea, T.M.; Kawaguchi, R.; Coppola, G.; Khakh, B.S.; Deming, T.J.; Sofroniew, M.V. Astrocyte scar formation aids central nervous system axon regeneration. Nature 2016, 532, 195–200. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, T.M.; Burda, J.E.; Sofroniew, M.V. Cell biology of spinal cord injury and repair. J. Clin. Investig. 2017, 127, 3259–3270. [Google Scholar] [CrossRef]
- Sousounis, K.; Athippozhy, A.T.; Voss, S.R.; Tsonis, P.A. Plasticity for axolotl lens regeneration is associated with age-related changes in gene expression. Regeneration 2014, 1, 47–57. [Google Scholar] [CrossRef]

| Species | Sequencing Platform | Type of Induced Injury | Samples | Reference |
|---|---|---|---|---|
| Axolotl | Illumina HiSeq 2500 paired end sequencing | The spinal cord was crushed at multiple levels, including the thoracic portion | Intact spinal cord, 1 dpi and 6 dpi | [15] |
| Rat | Illumina NovaSeq 6000 paired end sequencing | Lateral compression at the thoracic vertebrae T10 level | Intact spinal cord, 1 dpi and 7 dpi | [16] |
| Mouse | Illumina HiSeq 2000 paired end sequencing | Contusive injury at the thoracic vertebrae T9 level | Intact spinal cord, 2 dpi and 7 dpi | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Orozco, J.C.; Escobedo-Avila, I.; Velasco, I. Transcriptome Profiling after Early Spinal Cord Injury in the Axolotl and Its Comparison with Rodent Animal Models through RNA-Seq Data Analysis. Genes 2023, 14, 2189. https://doi.org/10.3390/genes14122189
González-Orozco JC, Escobedo-Avila I, Velasco I. Transcriptome Profiling after Early Spinal Cord Injury in the Axolotl and Its Comparison with Rodent Animal Models through RNA-Seq Data Analysis. Genes. 2023; 14(12):2189. https://doi.org/10.3390/genes14122189
Chicago/Turabian StyleGonzález-Orozco, Juan Carlos, Itzel Escobedo-Avila, and Iván Velasco. 2023. "Transcriptome Profiling after Early Spinal Cord Injury in the Axolotl and Its Comparison with Rodent Animal Models through RNA-Seq Data Analysis" Genes 14, no. 12: 2189. https://doi.org/10.3390/genes14122189
APA StyleGonzález-Orozco, J. C., Escobedo-Avila, I., & Velasco, I. (2023). Transcriptome Profiling after Early Spinal Cord Injury in the Axolotl and Its Comparison with Rodent Animal Models through RNA-Seq Data Analysis. Genes, 14(12), 2189. https://doi.org/10.3390/genes14122189






