The Genetic Diversity of Horse Native Breeds in Russia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. DNA Extraction
2.3. Genotyping DNA Samples
2.4. Statistical Analysis and Visualization
3. Results
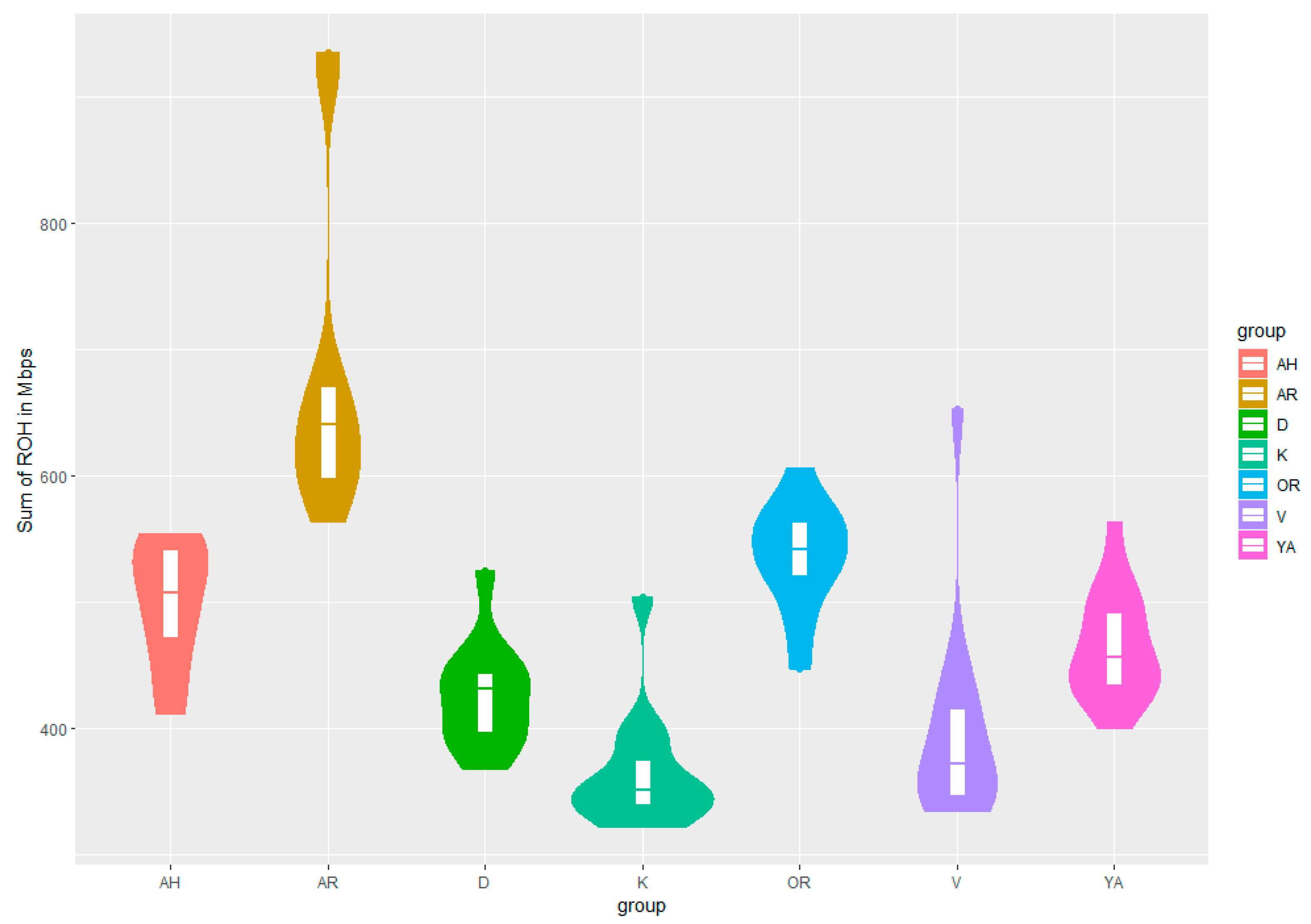
3.1. Genetic Diversity and Genomic Variability in Horse Breeds
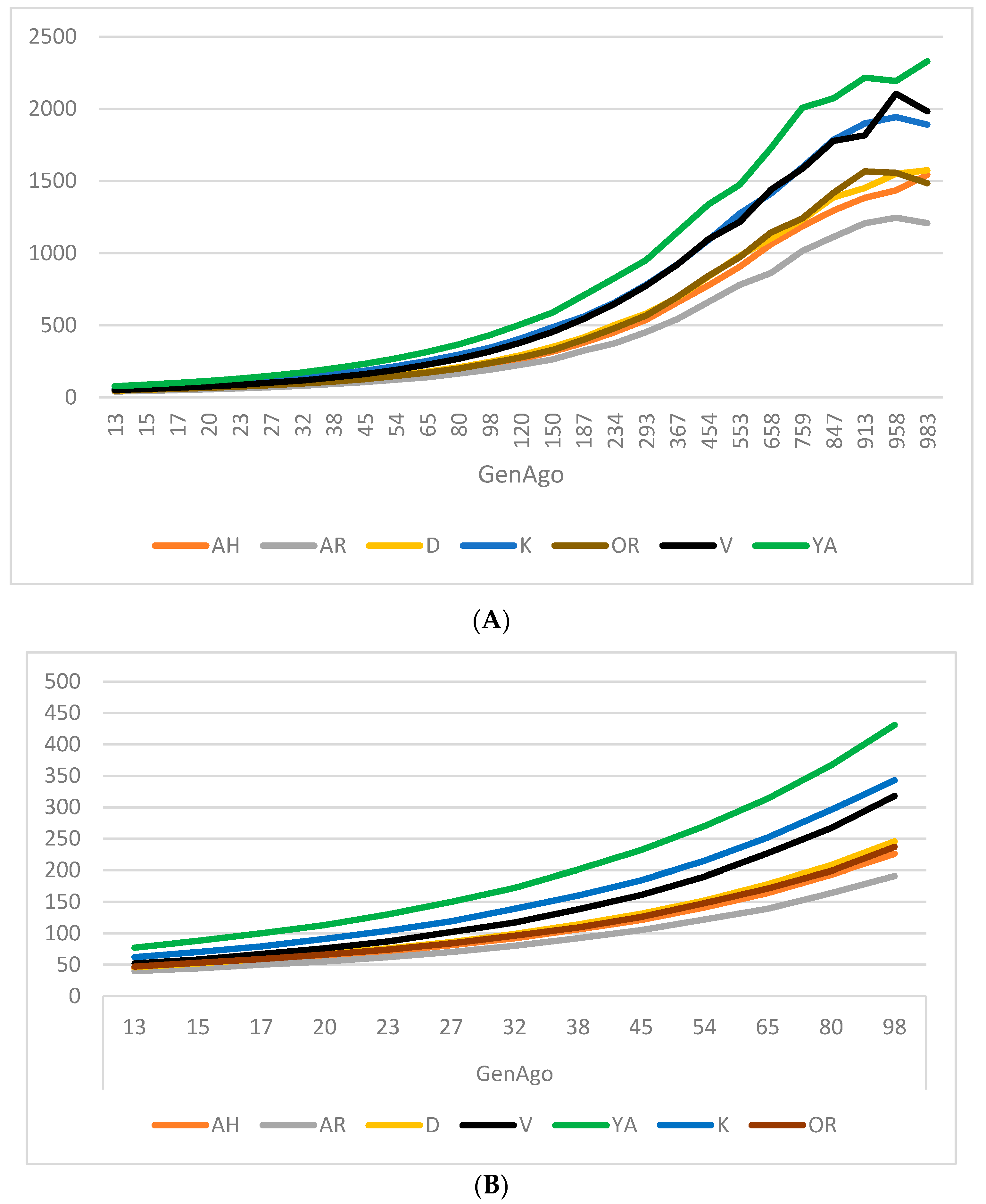
3.2. Effective Population Size
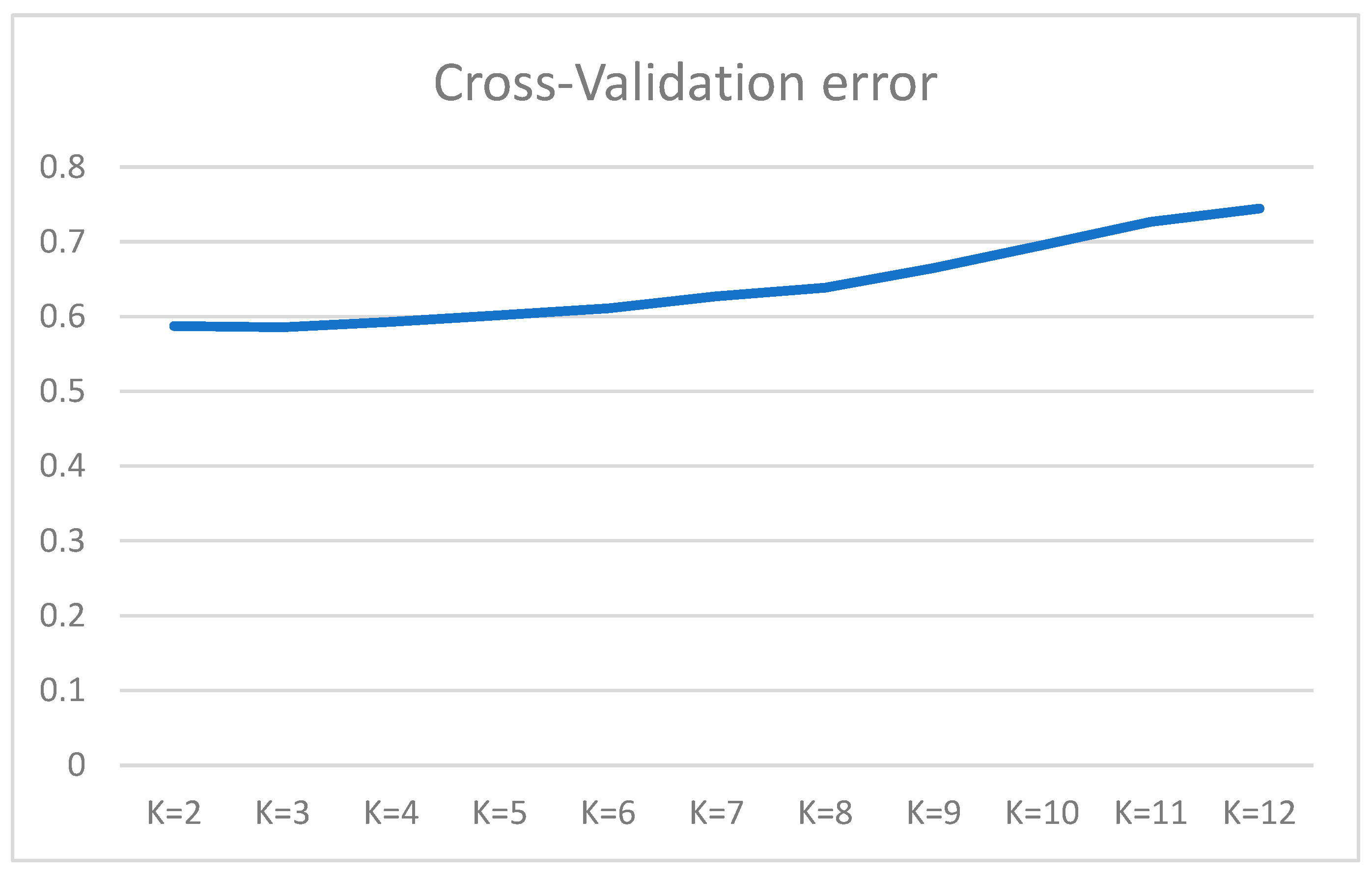
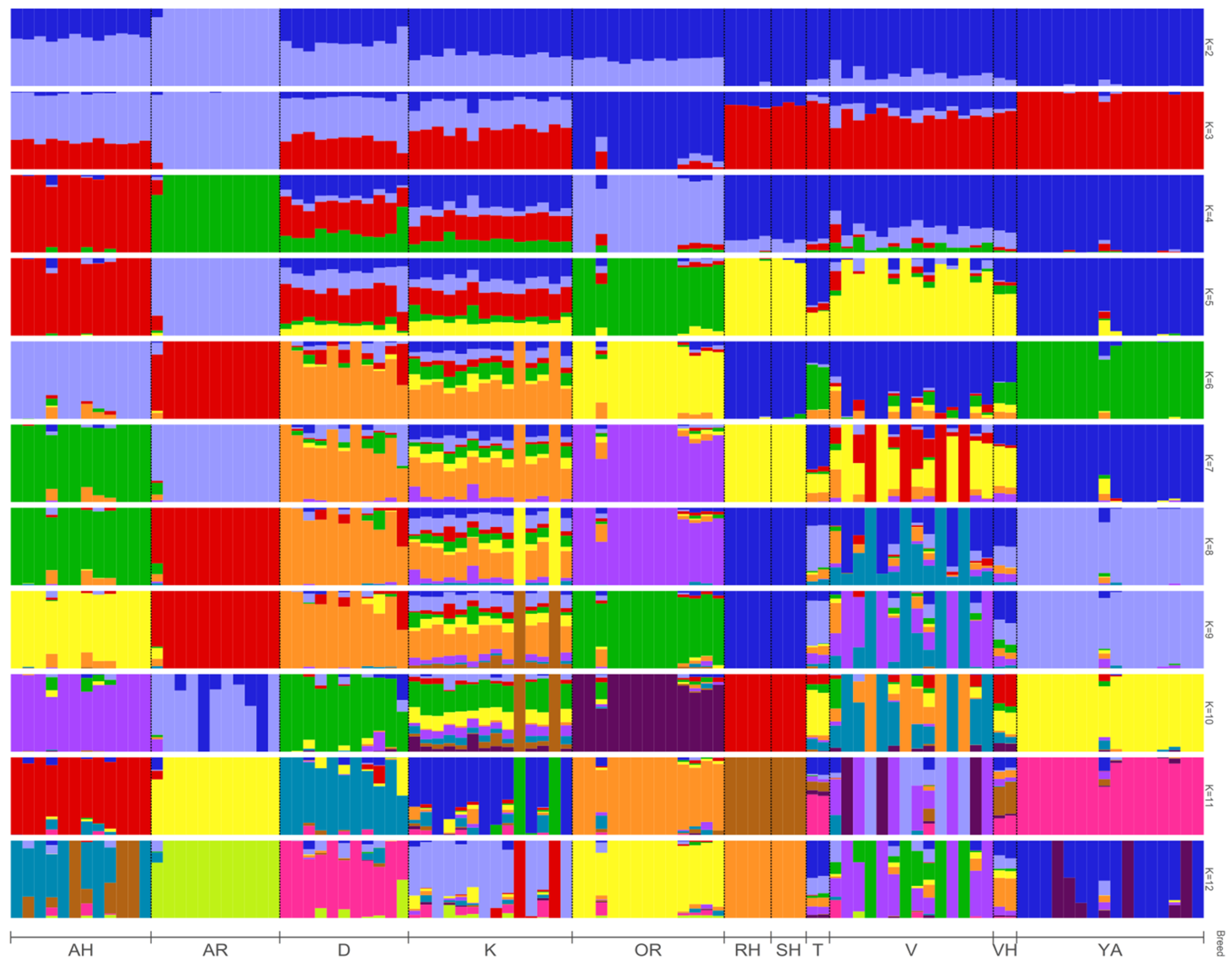
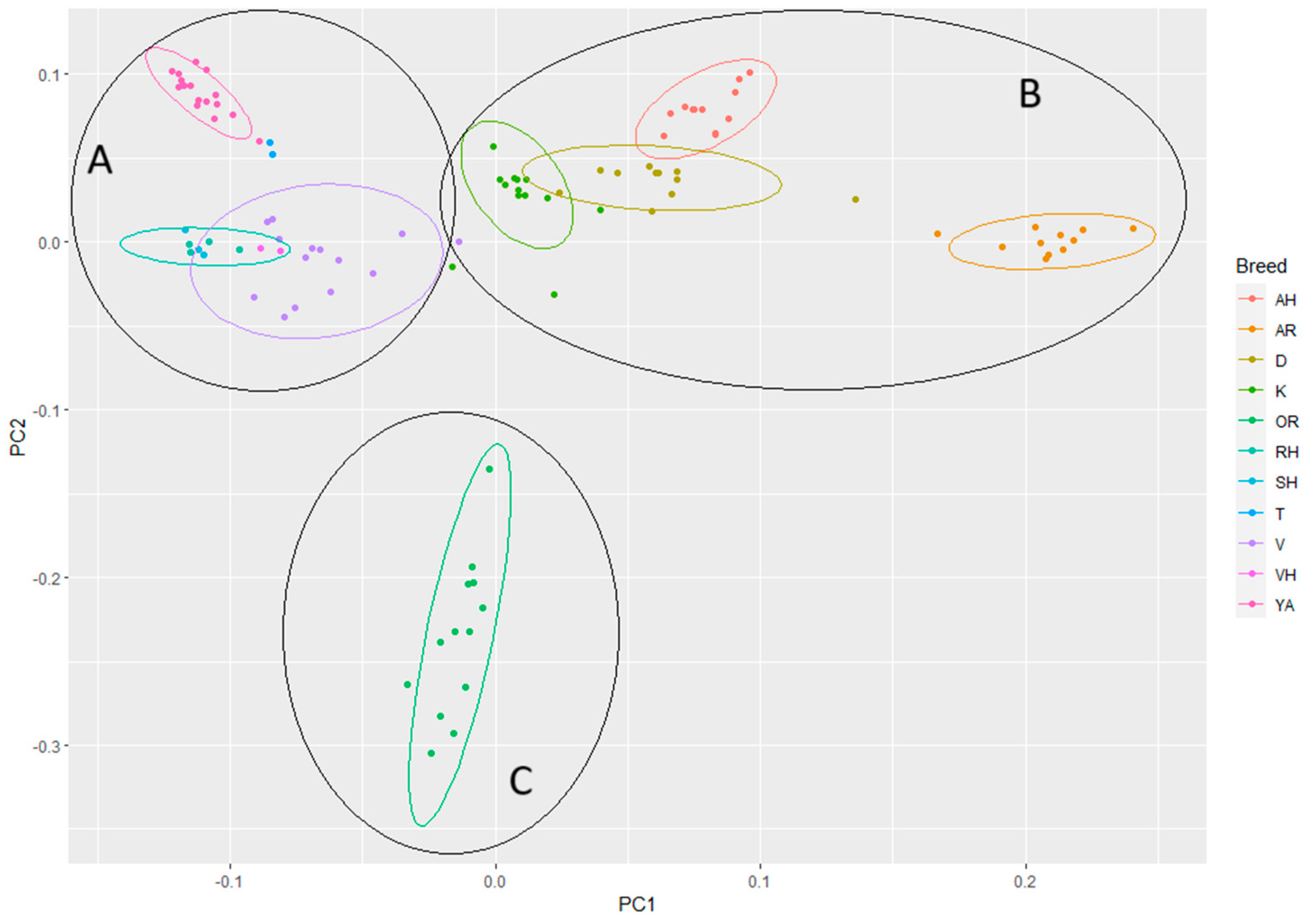
3.3. ADMIXTURE and PCA Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fu, M.; Li, Y. The Origin and Domestication History of Domestic Horses and the Domestication Characteristics of Breeds. Yi Chuan 2022, 44, 216–229. [Google Scholar] [PubMed]
- Capomaccio, S.; Ablondi, M.; Colombi, D.; Sartori, C.; Giontella, A.; Cappelli, K.; Mancin, E.; Asti, V.; Mantovani, R.; Sabbioni, A.; et al. Exploring the Italian Equine Gene Pool via High-Throughput Genotyping. Front. Genet. 2023, 14, 1099896. [Google Scholar] [CrossRef] [PubMed]
- Petrov, K.A.; Makhutova, O.N.; Gladyshev, M.I. Fatty Acid Composition of Yakut Horse Tissues. Dokl. Biochem. Biophys. 2020, 492, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Langlois, B. The history, ethnology and social importance of mare’s milk consumption in Central Asia. J. Life Sci. 2011, 5, 863–872. [Google Scholar]
- Librado, P.; Khan, N.; Fages, A.; Kusliy, M.A.; Suchan, T.; Tonasso-Calvière, L.; Schiavinato, S.; Alioglu, D.; Fromentier, A.; Perdereau, A.; et al. The Origins and Spread of Domestic Horses from the Western Eurasian Steppes. Nature 2021, 598, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Remer, V.; Bozlak, E.; Felkel, S.; Radovic, L.; Rigler, D.; Grilz-Seger, G.; Stefaniuk-Szmukier, M.; Bugno-Poniewierska, M.; Brooks, S.; Miller, D.C.; et al. Y-Chromosomal Insights into Breeding History and Sire Line Genealogies of Arabian Horses. Genes 2022, 13, 229. [Google Scholar] [CrossRef]
- Mongush, B.M.; Zaitsev, A.M.; Atroshchenko, M.M. Breeding Quality Control of Tuvan Sire-Stallions. Bull. KSAU 2022, 3, 87–92. [Google Scholar] [CrossRef]
- Dementieva, N.; Nikitkina, E.; Shcherbakov, Y.; Nikolaeva, O.; Mitrofanova, O.; Ryabova, A.; Atroshchenko, M.; Makhmutova, O.; Zaitsev, A. The Genetic Diversity of Stallions of Different Breeds in Russia. Genes 2023, 14, 1511. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Francis, R.M. Pophelper: An R Package and Web App to Analyse and Visualize Population Structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Pettersson, F.H.; Clarke, G.M.; Cardon, L.R.; Morris, A.P.; Zondervan, K.T. Data Quality Control in Genetic Case-Control Association Studies. Nat. Protoc. 2010, 5, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, F.; Cozzi, P.; Gaspa, G.; Marras, G. DetectRUNS: An R Package to Detect Runs of Homozygosity and Heterozygosity in Diploid Homozygosity and Heterozygosity in Diploid Genomes. 2019. Available online: https://cran.r-project.org/web/packages/detectRUNS/vignettes/detectRUNS.vignette.html (accessed on 24 October 2023).
- Miraglia, N.; Salimei, E.; Fantuz, F. Equine Milk Production and Valorization of Marginal Areas—A Review. Animals 2020, 10, 353. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, M.F.; Nocelli, F.; Pasquini, M. Meat quality and intramuscular fatty acid composition of Catria Horse. Anim. Sci. J. 2017, 88, 1107–1112. [Google Scholar] [CrossRef]
- Orlando, L. Ancient Genomes Reveal Unexpected Horse Domestication and Management Dynamics. BioEssays News Rev. Mol. Cell. Dev. Biol. 2020, 42, e1900164. [Google Scholar] [CrossRef] [PubMed]
- Khrabrova, L.A.; Blohina, N.V.; Belousova, N.F.; Cothran, E.G. Estimation of the genealogical structure of Vyatskaya horse breed (Equus ferus caballus) using DNA analysis. Russ. J. Genet. 2022, 58, 462–466. [Google Scholar] [CrossRef]
- Librado, P.; Der Sarkissian, C.; Ermini, L.; Schubert, M.; Jónsson, H.; Albrechtsen, A.; Fumagalli, M.; Yang, M.A.; Gamba, C.; Seguin-Orlando, A.; et al. Tracking the origins of Yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proc. Natl. Acad. Sci. USA 2015, 112, E6889–E6897. [Google Scholar] [CrossRef] [PubMed]
- Sild, E.; Värv, S.; Kaart, T.; Kantanen, J.; Popov, R.; Viinalass, H. Maternal and paternal genetic variation in Estonian local horse breeds in the context of geographically adjacent and distant Eurasian breeds. Anim. Genet. 2019, 50, 757–760. [Google Scholar] [CrossRef]
- Mancin, E.; Ablondi, M.; Mantovani, R.; Pigozzi, G.; Sabbioni, A.; Sartori, C. Genetic Variability in the Italian Heavy Draught Horse from Pedigree Data and Genomic Information. Animals 2020, 10, 1310. [Google Scholar] [CrossRef]
- Csizmár, N.; Mihók, S.; Jávor, A.; Kusza, S. Genetic analysis of the Hungarian draft horse population using partial mitochondrial DNA D-loop sequencing. PeerJ 2018, 6, e4198. [Google Scholar] [CrossRef]
- Chen, C.; Zhu, B.; Tang, X.; Chen, B.; Liu, M.; Gao, N.; Li, S.; Gu, J. Genome-Wide Assessment of Runs of Homozygosity by Whole-Genome Sequencing in Diverse Horse Breeds Worldwide. Genes 2023, 14, 1211. [Google Scholar] [CrossRef] [PubMed]
- Grilz-Seger, G.; Neuditschko, M.; Ricard, A.; Velie, B.; Lindgren, G.; Mesarič, M.; Cotman, M.; Horna, M.; Dobretsberger, M.; Brem, G.; et al. Genome-Wide Homozygosity Patterns and Evidence for Selection in a Set of European and near Eastern Horse Breeds. Genes 2019, 10, 491. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Shi, J.; Liu, T.; Zhang, Y.; Zhang, Q.; Liu, Z.; Wang, J.; Cheng, S. Genome-wide single-nucleotide polymorphism data and mitochondrial hypervariable region 1 nucleotide sequence reveal the origin of the Akhal-Teke horse. Anim. Biosci. 2023, 36, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Bailey, E.; Petersen, J.L.; Kalbfleisch, T.S. Genetics of Thoroughbred Racehorse Performance. Annu. Rev. Anim. Biosci. 2022, 10, 131–150. [Google Scholar] [CrossRef]
- Khaudov, A.B.D.; Duduev, A.S.; Kokov, Z.A.; Amshokov, K.K.; Zhekamukhov, M.K.; Zaitsev, A.M.; Reissmann, M. Genetic analysis of maternal and paternal lineages in kabardian horses by uniparental molecular markers. Open Vet. J. 2018, 8, 40–46. [Google Scholar] [CrossRef]
- Khrabrova, L.A.; Nikolaeva, A.A.; Blokhina, N.V. Features of the matrilineal structure of the don horse breed formed in the central area of domestication. Horse Breed. Equest. Sports 2022, 1, 60291. [Google Scholar] [CrossRef]

| Breed | Number of Samples | Ho | He | FIS |
|---|---|---|---|---|
| AH | 12 | 0.329 ± 0.001 | 0.310 ± 0.001 | −0.05 ± 0.001 |
| AR | 11 | 0.303 ± 0.001 | 0.294 ± 0.001 | −0.027 ± 0.001 |
| D | 11 | 0.342 ± 0.001 | 0.325 ± 0.001 | −0.048 ± 0.001 |
| K | 14 | 0.342 ± 0.001 | 0.328 ± 0.001 | −0.039 ± 0.001 |
| OR | 13 | 0.309 ± 0.001 | 0.294 ± 0.001 | −0.043 ± 0.001 |
| V | 14 | 0.321 ± 0.001 | 0.302 ± 0.001 | −0.054 ± 0.001 |
| YA | 16 | 0.298 ± 0.001 | 0.288 ± 0.001 | −0.030 ± 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atroshchenko, M.; Dementieva, N.; Shcherbakov, Y.; Nikolaeva, O.; Azovtseva, A.; Ryabova, A.; Nikitkina, E.; Makhmutova, O.; Datsyshin, A.; Zakharov, V.; et al. The Genetic Diversity of Horse Native Breeds in Russia. Genes 2023, 14, 2148. https://doi.org/10.3390/genes14122148
Atroshchenko M, Dementieva N, Shcherbakov Y, Nikolaeva O, Azovtseva A, Ryabova A, Nikitkina E, Makhmutova O, Datsyshin A, Zakharov V, et al. The Genetic Diversity of Horse Native Breeds in Russia. Genes. 2023; 14(12):2148. https://doi.org/10.3390/genes14122148
Chicago/Turabian StyleAtroshchenko, Mikhail, Natalia Dementieva, Yuri Shcherbakov, Olga Nikolaeva, Anastasiia Azovtseva, Anna Ryabova, Elena Nikitkina, Oksana Makhmutova, Andrey Datsyshin, Viktor Zakharov, and et al. 2023. "The Genetic Diversity of Horse Native Breeds in Russia" Genes 14, no. 12: 2148. https://doi.org/10.3390/genes14122148
APA StyleAtroshchenko, M., Dementieva, N., Shcherbakov, Y., Nikolaeva, O., Azovtseva, A., Ryabova, A., Nikitkina, E., Makhmutova, O., Datsyshin, A., Zakharov, V., & Zaitsev, A. (2023). The Genetic Diversity of Horse Native Breeds in Russia. Genes, 14(12), 2148. https://doi.org/10.3390/genes14122148







