The Identification of the Banana Endogenous Reference Gene MaSPS1 and the Construction of Qualitative and Quantitative PCR Detection Methods
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. DNA Extraction and Purification
2.3. Primers and Probes
2.4. MaSPS1 Sequence Analysis
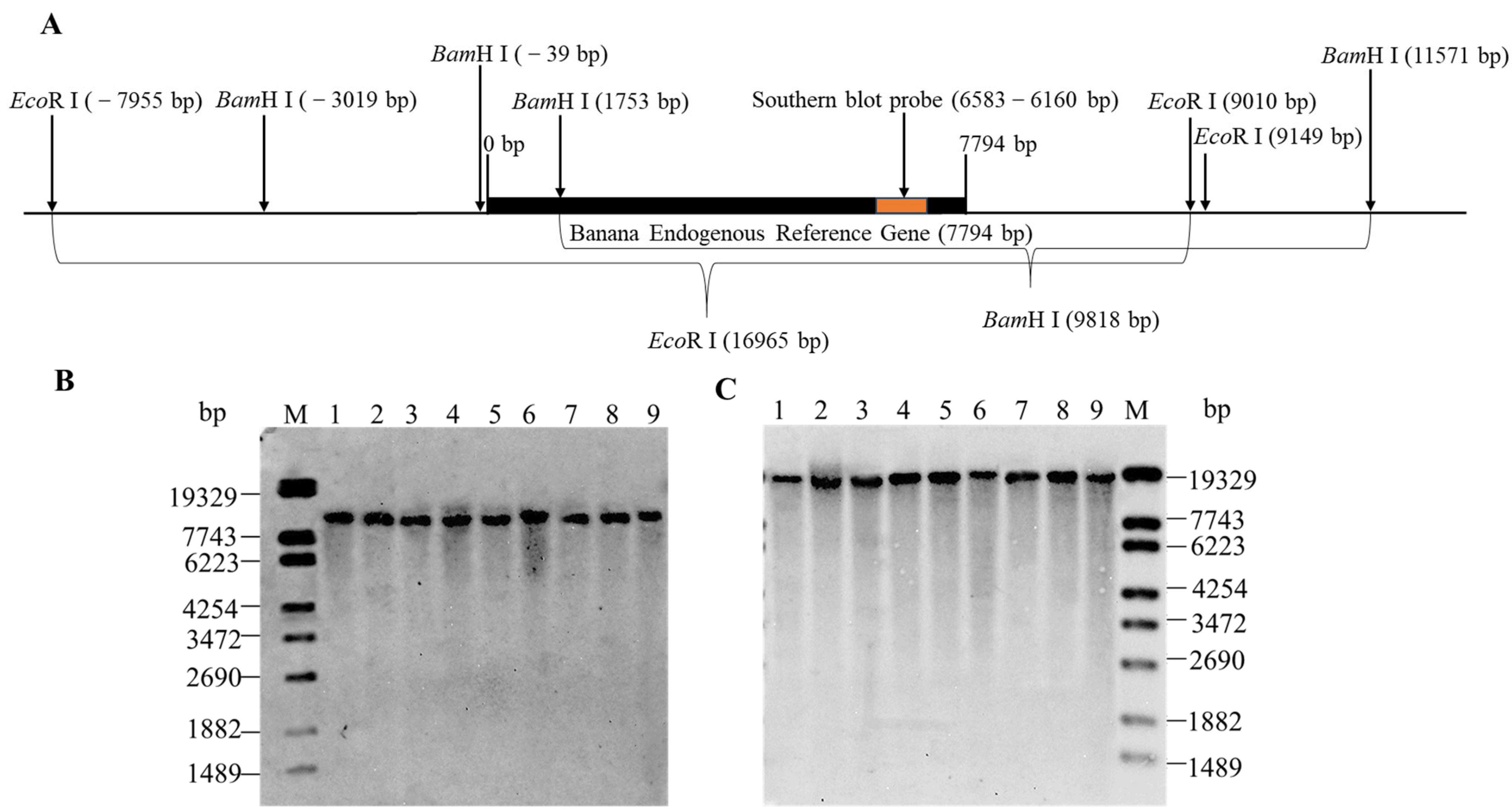
2.5. MaSPS1 Genome Copy Number Analysis
2.6. PCR Amplification Methods
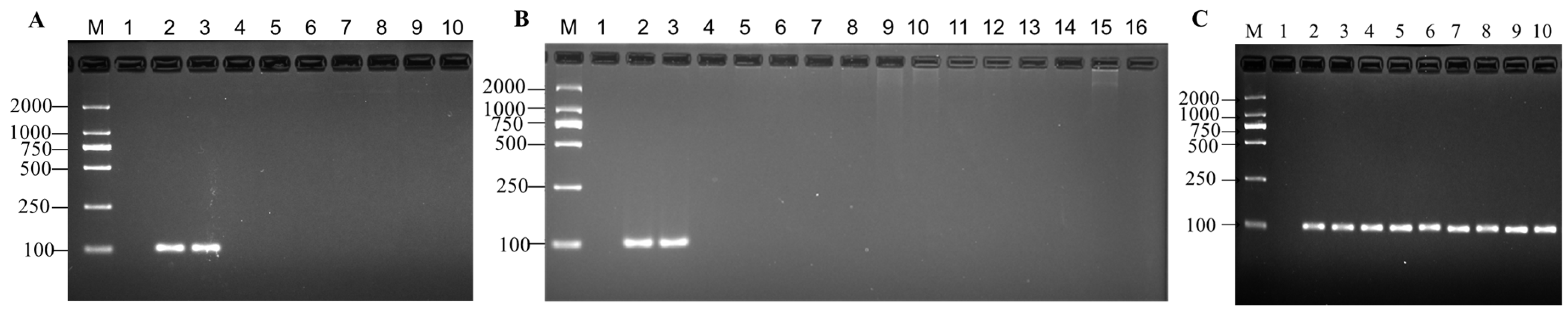
2.7. Specificity Detection of MaSPS1
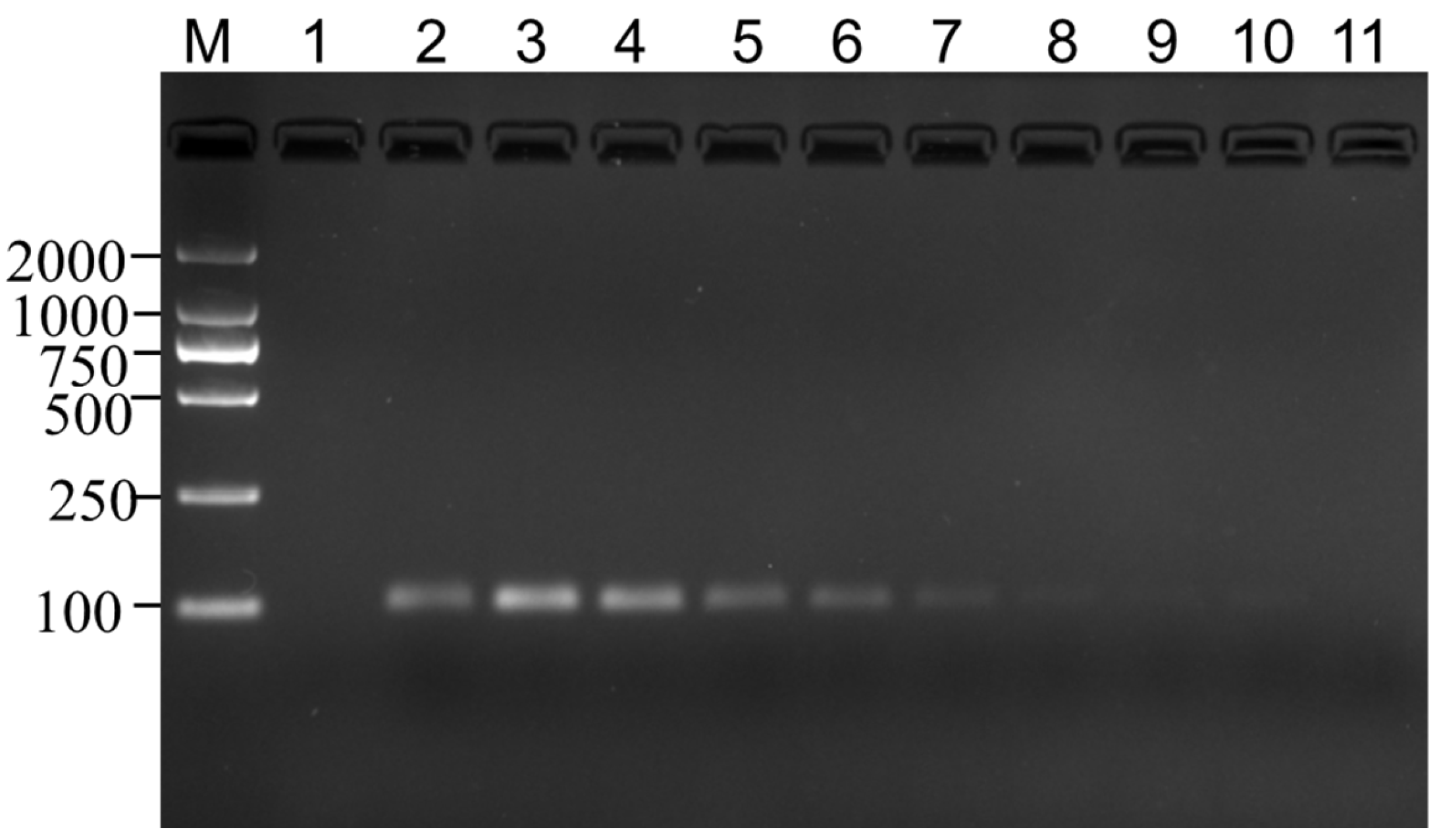
2.8. Limit of Detection Test for MaSPS1
2.9. Standard Curve Construction and Limit of Quantification Test for qPCR
3. Results
3.1. Sequence Analysis of MaSPS1
3.2. Specificity Test for MaSPS1
3.3. Determination of the Copy Number of MaSPS1 Using Southern Blot Analysis
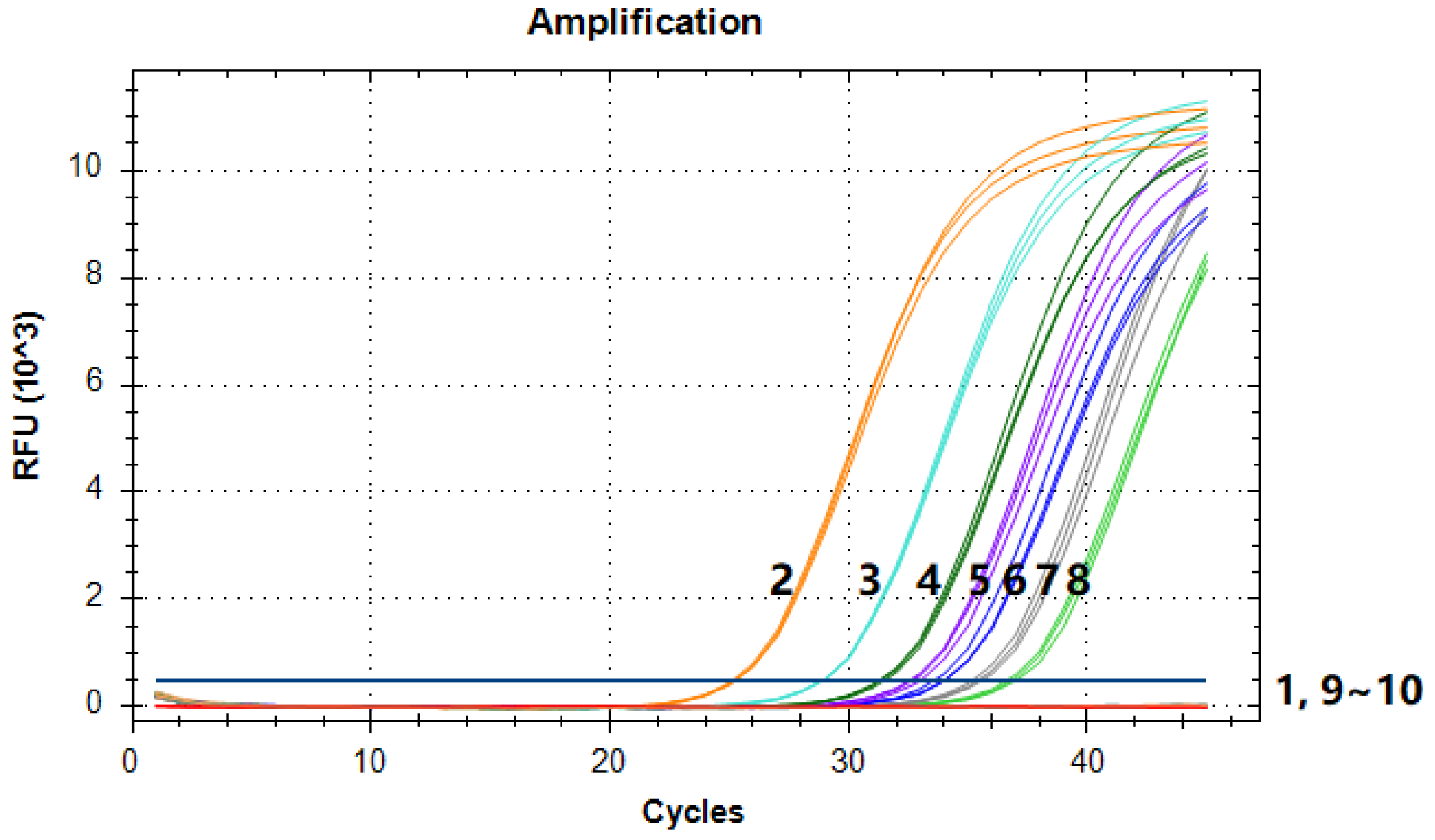
3.4. Limit of Detection Test
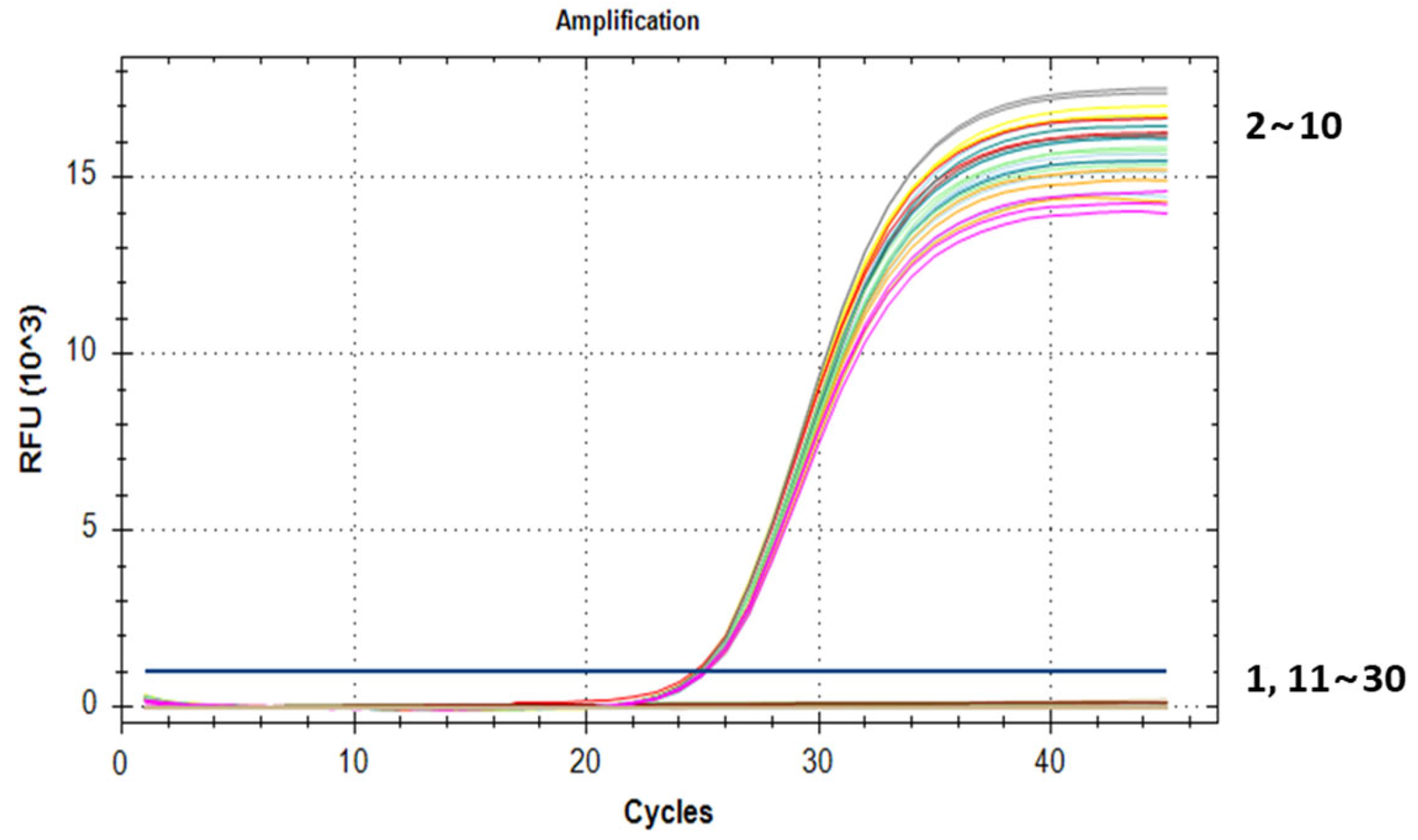
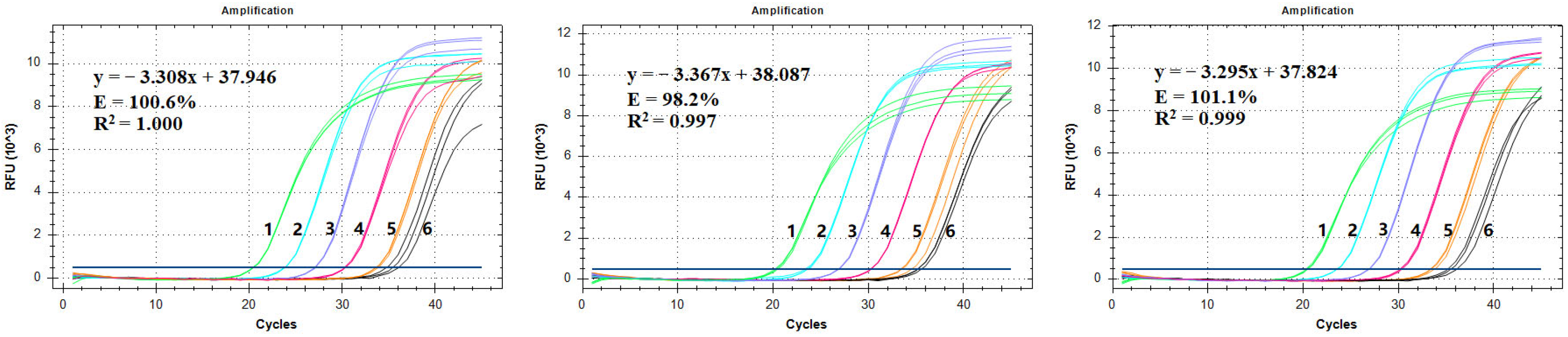
3.5. Construction of a Standard Curve and the LOQ Test
3.6. Repeatability and Stability Tests for the qPCR Method
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Aurore, G.; Parfait, B.; Fahrasmane, L. Banana, raw materials for making processed food products. Trends Food Sci. Technol. 2009, 20, 78–91. [Google Scholar] [CrossRef]
- Wang, Z.; Miao, H.X.; Liu, J.H.; Xu, B.Y.; Yao, X.M.; Xu, C.Y.; Zhao, S.C.; Fang, X.D.; Jia, C.H.; Wang, J.Y.; et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nat. Plants 2019, 5, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Marín, D.H.; Romero, R.A.; Guzmán, M.; Sutton, T.B. Black Sigatoka: An Increasing Threat to Banana Cultivation. Plant Dis. 2003, 87, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Stokstad, E. GM banana shows promise against deadly fungus strain. Science 2017, 358, 979. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.; Ntui, V.O.; Tripathi, J.N. CRISPR/Cas9-based genome editing of banana for disease resistance. Curr. Opin. Plant Biol. 2020, 56, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Namukwaya, B.; Tripathi, L.; Tripathi, J.N.; Arinaitwe, G.; Mukasa, S.B.; Tushemereirwe, W.K. Transgenic banana expressing Pflp gene confers enhanced resistance to Xanthomonas wilt disease. Transgenic Res. 2012, 21, 855–865. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, J.H.; Jia, C.H.; Hu, W.; Song, S.; Xu, B.Y.; Jin, Z.Q. Overexpression of an banana aquaporin gene MaPIP1;1 enhances tolerance to multiple abiotic stresses in transgenic banana and analysis of its interacting transcription factors. Front. Plant Sci. 2021, 25, 780544. [Google Scholar] [CrossRef]
- Wang, C.; Jiang, L.X.; Rao, J.; Liu, Y.N.; Yang, L.T.; Zhang, D.B. Evaluation of four genes in rice for their suitability as endogenous reference standards in quantitative PCR. J. Agric. Food Chem. 2010, 58, 11543–11547. [Google Scholar] [CrossRef]
- Chaouachi, M.; Giancola, S.; Romaniuk, M.; Laval, V.; Bertheau, Y.; Brunel, D. A strategy for designing multi-taxa specific reference gene systems. Example of application--ppi Phosphofructokinase (ppi-PPF) used for the detection and quantification of three taxa: Maize (Zea mays); cotton (Gossypium hirsutum) and rice (Oryza sativa). J. Agric. Food Chem. 2007, 55, 8003–8010. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Martinelli, L. Development of a real-time PCR method based on duplo target plasmids for determining an unexpected genetically modified soybean intermix with feed components. J. Agric. Food Chem. 2007, 55, 1264–1273. [Google Scholar] [CrossRef]
- Guo, J.C.; Yang, L.T.; Liu, X.; Zhang, H.B.; Qian, B.J.; Zhang, D.B. Applicability of the chymopapain gene used as endogenous reference gene for transgenic huanong no. 1 papaya detection. J. Agric. Food Chem. 2009, 57, 6502–6509. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.T.; Huang, W.S.; Ren, J.A.; Ma, X.L.; Ge, Y.Q.; Chen, Y. Verification and applicability of endogenous reference genes for quantifying GM rice by digital PCR. Anal. Biochem. 2019, 15, 113442. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.G.; Lu, S.; Zhou, H.; Wu, X.J.; Zhuang, C.X.; Liu, Y.G.; Mei, M.T. Mapping of the rice (Oryza sativa L.) thermo-sensitive genic male sterile gene tms5 with EST and SSR markers. Chin. Sci. Bull. 2006, 51, 417–420. [Google Scholar] [CrossRef]
- International Service for the Acquisition of Agri-Biotech Applications (ISAAA). Available online: https://www.isaaa.org/resources/publications/briefs/55/default.asp (accessed on 8 October 2023).
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Sharma, P. Recent advances in cucumber (Cucumis sativus L.). J. Hortic. Sci. Biotechnol. 2021, 97, 3–23. [Google Scholar] [CrossRef]
- Marette, S.; Disdier, A.C.; Beghin, J.C. A comparison of EU and US consumers’ willingness to pay for gene-edited food: Evidence from apples. Appetite 2021, 159, 105064. [Google Scholar] [CrossRef] [PubMed]
- Weng, H.; Pan, A.H.; Yang, L.T.; Zhang, C.M.; Liu, Z.L.; Zhang, D.B. Estimating number of transgene copies in transgenic rapeseed by real-time PCR assay with HMG I/Y as an endogenous reference gene. Plant Mol. Biol. 2004, 22, 289–300. [Google Scholar] [CrossRef]
- Yang, L.T.; Guo, J.C.; Pan, A.H.; Zhang, H.B.; Zhang, K.W.; Wang, Z.M.; Zhang, D.B. Event-specific quantitative detection of nine genetically modified maizes using one novel standard reference molecule. J. Agric. Food Chem. 2007, 55, 15–24. [Google Scholar] [CrossRef]
- Xu, W.T.; Bai, W.B.; Guo, F.; Luo, Y.B.; Yuan, Y.F.; Huang, K.L. A papaya-specific gene; papain; used as an endogenous reference gene in qualitative and real-time quantitative PCR detection of transgenic papayas. Eur. Food Res. Technol. 2008, 228, 301–309. [Google Scholar] [CrossRef]
- Debode, F.; Marien, A.; Janssen, E.; Berben, G. Design of multiplex calibrant plasmids; their use in GMO detection and the limit of their applicability for quantitative purposes owing to competition effects. Anal. Bioanal. Chem. 2010, 396, 2151–2164. [Google Scholar] [CrossRef]
- Artico, S.; Nardeli, S.M.; Neto, O.B.O.; Grossi-de-Sa, M.F.; Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutumfor accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 2010, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Mandaci, M.; Cakir, O.; Turgut Kara, N.; Meriç, S.; Ari, S. Detection of genetically modified organisms in soy products sold in Turkish market. Food Sci. Technol. 2014, 34, 717–722. [Google Scholar] [CrossRef]
- Yuan, J.Q.; Chang, H.; Zhao, J.H.; Tang, Z.W.; Shi, Z.Y.; Wang, J.D. Detection of transgenic components in animal feeds on Shanxi markets. Sheng Wu Gong Cheng Xue Bao 2016, 32, 1576–1589. [Google Scholar] [PubMed]
- Jiang, L.X.; Yang, L.T.; Rao, J.; Guo, J.C.; Wang, S.; Liu, J.; Lee, S.H.; Zhang, D.B. Development and in-house validation of the event-specific qualitative and quantitative PCR detection methods for genetically modified cotton MON15985. J. Sci. Food Agric. 2010, 90, 402–408. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, D.B.; Yang, L.T. Development of event-specific PCR detection methods for genetically modified tomato Huafan No. 1. J. Sci. Food Agric. 2013, 93, 652–660. [Google Scholar] [CrossRef]

| Purpose | Primers/Probe Name | Sequence (5′–3′) | Amplicon (bp) |
|---|---|---|---|
| Sequencing analysis | SPS-1F | CAAACAACGGGAAGCCTTTTTC | 1170 |
| SPS-1R | CCTTTTCGCCTTCTGATAGTTC | ||
| Southern blot probe | SPS-2F | CTGATAATTTGTTCGTAGACC | 423 |
| SPS-2R | CCGAATCTTGCATAACGTCTAA | ||
| PCR amplification | SPS-QF | CATGGCTCATTTATCTCAAACTAA | 104 |
| SPS-QR | CCGAATCTTGCATAACGTCTAA | ||
| SPS-QP | FAM-GGACCTTGTTCCATTGTTCCTTCTGTATGG-BHQ1 |
| Plant Species | Ct Value | Mean Ct | SD | RSD | ||
|---|---|---|---|---|---|---|
| Repeat 1 | Repeat 2 | Repeat 3 | ||||
| Blank control | N/A | N/A | N/A | |||
| Malaccensis | 25.03 | 24.77 | 24.58 | 24.79 | 0.23 | 0.91% |
| Brazilian banana | 25.20 | 25.10 | 25.15 | 25.15 | 0.05 | 0.20% |
| FHIA-04 | 25.05 | 25.11 | 24.96 | 25.04 | 0.08 | 0.30% |
| French Sombre | 25.12 | 25.03 | 25.04 | 25.06 | 0.05 | 0.20% |
| FHIA-21 | 25.05 | 24.97 | 24.94 | 24.99 | 0.06 | 0.23% |
| Musa ABB Pisang Awak | 25.16 | 25.00 | 25.07 | 25.08 | 0.08 | 0.32% |
| M. acuminata | 24.70 | 25.08 | 24.79 | 24.86 | 0.20 | 0.80% |
| CB5 | 24.99 | 24.98 | 25.11 | 25.03 | 0.07 | 0.29% |
| Guangxi hongjiao | 24.92 | 24.76 | 24.81 | 24.83 | 0.08 | 0.33% |
| Rapeseed | N/A | N/A | N/A | |||
| Rice | N/A | N/A | N/A | |||
| Papaya | N/A | N/A | N/A | |||
| Maize | N/A | N/A | N/A | |||
| Soybean | N/A | N/A | N/A | |||
| Cotton | N/A | N/A | N/A | |||
| Wheat | N/A | N/A | N/A | |||
| Mango | N/A | N/A | N/A | |||
| Grapefruit | N/A | N/A | N/A | |||
| Green jujube | N/A | N/A | N/A | |||
| Lemon | N/A | N/A | N/A | |||
| Orange | N/A | N/A | N/A | |||
| Jackfruit | N/A | N/A | N/A | |||
| Watermelon | N/A | N/A | N/A | |||
| Wax apple | N/A | N/A | N/A | |||
| Pear | N/A | N/A | N/A | |||
| Grape | N/A | N/A | N/A | |||
| Peach | N/A | N/A | N/A | |||
| Cantaloupe | N/A | N/A | N/A | |||
| Strawberry | N/A | N/A | N/A | |||
| DNA Concentration | Signal Rate (Positive Signals) | Copy Number | |||
|---|---|---|---|---|---|
| ng/reaction | Copies/reaction | Mean | SD | RSD | |
| 5.70 × 100 | 10,000 | 3/3 | 9351.2 | 353.12 | 3.78% |
| 5.70 × 10−1 | 1000 | 3/3 | 987.3 | 14.93 | 1.51% |
| 1.14 × 10−1 | 200 | 3/3 | 231.5 | 14.71 | 6.36% |
| 5.70 × 10−2 | 100 | 3/3 | 101.9 | 14.88 | 14.59% |
| 2.85 × 10−2 | 50 | 3/3 | 51.7 | 8.29 | 16.03% |
| 1.14 × 10−2 | 20 | 3/3 | 20.8 | 2.48 | 11.94% |
| 5.70 × 10−3 | 10 | 3/3 | 8.8 | 1.06 | 12.07% |
| 2.85 × 10−3 | 5 | 0/3 | N/A | ||
| 1.43 × 10−3 | 2.5 | 0/3 | N/A | ||
| DNA Concentration | Ct Value | Copy Number | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ng/reaction | Copies/ reaction | Repeat 1 | Repeat 2 | Repeat 3 | Mean | SD | RSD | Mean | SD | RSD |
| 1.14 × 102 | 200,000 | 20.39 | 20.34 | 20.40 | 20.38 | 0.03 | 0.16% | 204,983.7 | 4613.07 | 2.25% |
| 1.14 × 101 | 20,000 | 23.74 | 23.76 | 23.82 | 23.77 | 0.04 | 0.18% | 19,269.5 | 554.91 | 2.88% |
| 1.14 × 100 | 2000 | 27.09 | 27.06 | 27.02 | 27.06 | 0.04 | 0.13% | 1959.8 | 47.99 | 2.45% |
| 1.14 × 10−1 | 200 | 30.30 | 30.27 | 30.20 | 30.26 | 0.05 | 0.17% | 211.3 | 7.60 | 3.59% |
| 1.14 × 10−2 | 20 | 33.60 | 33.57 | 33.85 | 33.67 | 0.15 | 0.46% | 19.7 | 2.04 | 10.37% |
| 5.70 × 10−3 | 10 | 36.04 | 34.73 | 35.39 | 35.39 | 0.66 | 1.85% | 6.4 | 2.83 | 44.52% |
| Test Number | Amplification Efficiency | R2 | Slope | Y-Intercept |
|---|---|---|---|---|
| Test 1 | 100.6% | 1.000 | −3.308 | 37.946 |
| Test 2 | 98.2% | 0.997 | −3.367 | 38.087 |
| Test 3 | 101.1% | 0.999 | −3.295 | 37.824 |
| DNA Concentration | Test 1 Copy Number | Test 2 Copy Number | Test 3 Copy Number | ||||
|---|---|---|---|---|---|---|---|
| ng/reaction | Copies/ reaction | Mean | Bias | Mean | Bias | Mean | Bias |
| 1.14 × 102 | 200,000 | 204,755.5 | 2.4% | 185,916.0 | 7.0% | 193,029.0 | 3.5% |
| 1.14 × 101 | 20,000 | 19,252.3 | 3.7% | 21,091.1 | 5.5% | 21,280.5 | 6.4% |
| 1.14 × 100 | 2000 | 1958.4 | 2.1% | 2107.2 | 5.4% | 1996.4 | 0.2% |
| 1.14 × 10−1 | 200 | 211.2 | 5.6% | 210.7 | 5.4% | 195.5 | 2.2% |
| 1.14 × 10−2 | 20 | 19.6 | 1.8% | 19.6 | 2.2% | 20.3 | 1.4% |
| 5.70 × 10−3 | 10 | 6.4 | 36.4% | 6.7 | 33.0% | 4.8 | 51.9% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Lin, Y.; Yang, W.; Pan, Z.; Chen, W.; Yao, J.; Sheng, O.; Zhou, L.; Jiang, D. The Identification of the Banana Endogenous Reference Gene MaSPS1 and the Construction of Qualitative and Quantitative PCR Detection Methods. Genes 2023, 14, 2116. https://doi.org/10.3390/genes14122116
Zhu L, Lin Y, Yang W, Pan Z, Chen W, Yao J, Sheng O, Zhou L, Jiang D. The Identification of the Banana Endogenous Reference Gene MaSPS1 and the Construction of Qualitative and Quantitative PCR Detection Methods. Genes. 2023; 14(12):2116. https://doi.org/10.3390/genes14122116
Chicago/Turabian StyleZhu, Lili, Ying Lin, Wenli Yang, Zhiwen Pan, Weiting Chen, Juan Yao, Ou Sheng, Lingyan Zhou, and Dagang Jiang. 2023. "The Identification of the Banana Endogenous Reference Gene MaSPS1 and the Construction of Qualitative and Quantitative PCR Detection Methods" Genes 14, no. 12: 2116. https://doi.org/10.3390/genes14122116
APA StyleZhu, L., Lin, Y., Yang, W., Pan, Z., Chen, W., Yao, J., Sheng, O., Zhou, L., & Jiang, D. (2023). The Identification of the Banana Endogenous Reference Gene MaSPS1 and the Construction of Qualitative and Quantitative PCR Detection Methods. Genes, 14(12), 2116. https://doi.org/10.3390/genes14122116







