Response of the Propylea japonica Microbiota to Treatment with Cry1B Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Reagents
2.3. Insect Sample Collection and Processing
2.4. DNA Extraction and PCR Amplification
2.5. Illumina Sequencing
2.6. Sequencing Data Processing
2.7. Statistical Analysis
3. Results
3.1. Sequencing Data Assessment
3.2. Effects of Cry1B on P. japonica Microbial Community Diversity
3.3. Effects of Cry1B on P. japonica Microbiota at the Phylum Level
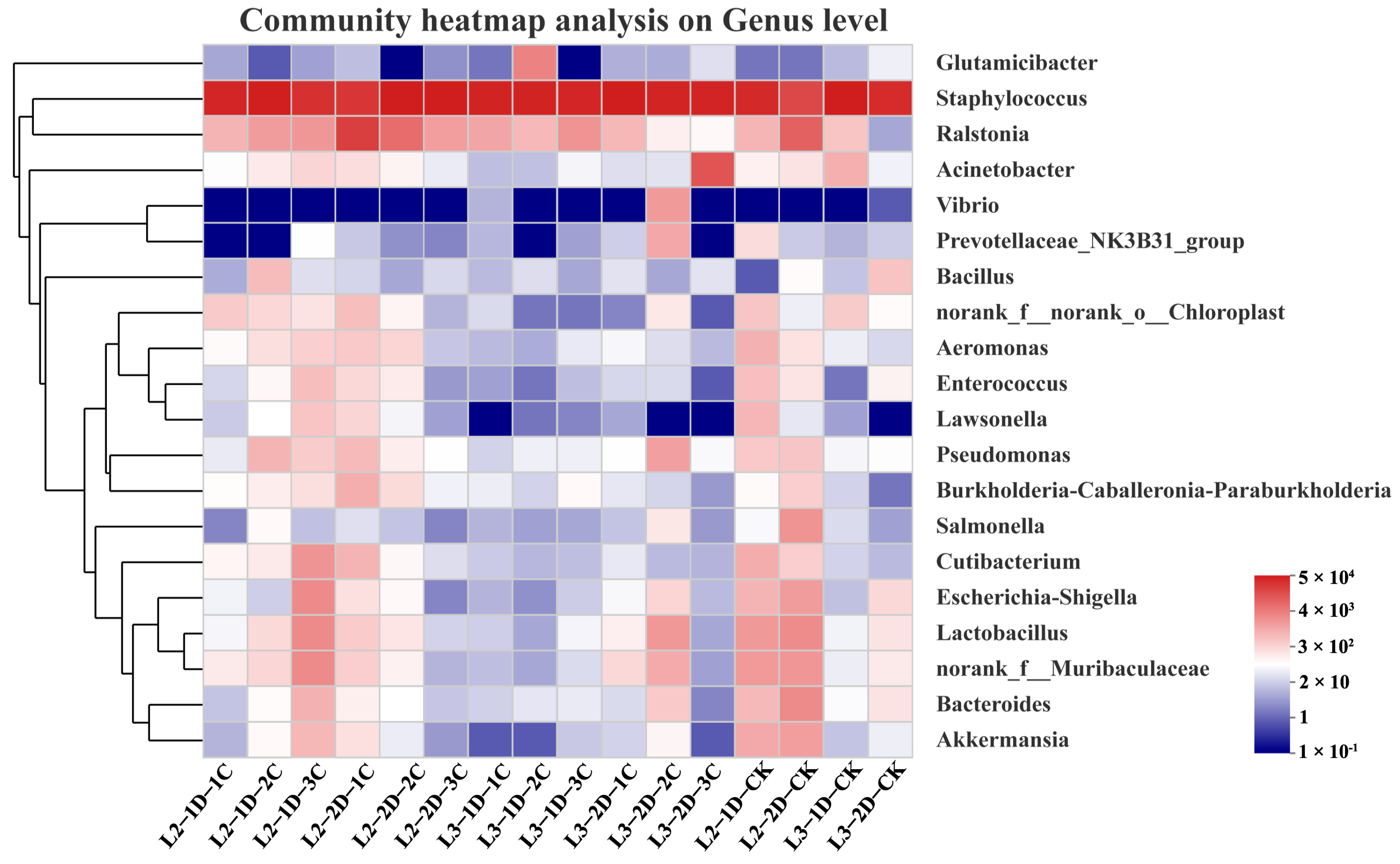
3.4. Effects of Cry1B Treatment on P. japonica Microbiota at the Genus Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tilgam:, J.; Kumar, K.; Jayaswal, D.; Choudhury, S.; Kumar, A.; Jayaswall, K.; Saxena, A.K. Success of microbial genes based transgenic crops: Bt and beyond Bt. Mol. Biol. Rep. 2021, 48, 8111–8122. [Google Scholar] [CrossRef]
- Řezbová, H.; Škubna, O. The Role of Transgenic Crops in the Future of Global Food and Feed. AGRIS-Line Pap. Econ. Inform. 2012, 4, 49–60. [Google Scholar]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef]
- Bigler, F.; Candolfi, M.P. Moving Through the Tiered and Methodological Framework for NonTarget Arthropod Risk Assessment of Transgenic Insecticidal Crops. In Proceedings of the 9th International Symposium on the Biosafety of Genetically Modified Organisms, Jeju Island, Republic of Korea, 24–29 September 2006. [Google Scholar]
- Marvier, M. Improving risk assessment for nontarget safety of transgenic crops. Ecol. Appl. 2002, 12, 1119–1124. [Google Scholar] [CrossRef]
- Romeis, J.; Meissle, M.; Alvarez-Alfageme, F.; Bigler, F.; Bohan, D.A.; Devos, Y.; Malone, L.A.; Pons, X.; Rauschen, S. Potential use of an arthropod database to support the non-target risk assessment and monitoring of transgenic plants. Transgenic Res. 2014, 23, 995–1013. [Google Scholar] [CrossRef]
- Romeis, J.; Meissle, M. Non-target risk assessment of Bt crops—Cry protein uptake by aphids. J. Appl. Entomol. 2011, 135, 1–6. [Google Scholar] [CrossRef]
- Waseem, M.; Khan, M.B.; Ahmad, F.; Farooq, S.; Id, M.H. The influence of transgenic (Bt) and non- transgenic (non-Bt) cotton mulches on weed dynamics, soil properties and productivity of different winter crops. PLoS ONE 2020, 15, e0238716. [Google Scholar]
- Tayyib, M.; Sohail, A.; Murtaza, A.; Jamil, F.F. Efficacy of some new-chemistry inseticides for controlling the sucking insect peats and mites on cotton. Pak. Entomol. 2005, 27, 63–66. [Google Scholar]
- Chen, Y.; Li, Y.; Zhou, M.; Cai, Z.; Tambel, L.I.M.; Zhang, X.; Chen, Y.; Chen, D. Nitrogen deficit decreases seed Cry1Ac endotoxin expression in Bt transgenic cotton. Plant Physiol. Biochem. 2019, 141, 114–121. [Google Scholar] [CrossRef]
- Bravo, A.; Gill, S.S.; Soberón, M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 2007, 49, 423–435. [Google Scholar] [CrossRef]
- Glare, T.R.; O’Callaghan, M. Bacillus thuringiensis: Biology, Ecology and Safety; Wiley: Hoboken, NJ, USA, 2000. [Google Scholar]
- Azizoglu, U.; Jouzani, G.S.; Yilmaz, N.; Baz, E.; Ozkok, D. Genetically modified entomopathogenic bacteria, recent developments, benefits and impacts: A review. Sci. Total Environ. 2020, 734, 139169. [Google Scholar] [CrossRef]
- Schnepf, E.; Crickmore, N.; Van Rie, J.; Lereclus, D.; Baum, J.; Feitelson, J.; Zeigler, D.R.; Dean, D.H. Bacillus thuringiensis and Its Pesticidal Crystal Proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 775–806. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, Y.; Zaheeruddin, F.; Bakhsh, K.; Anjum, M.B.; Ahmad, M. Impact of Bt. cotton varieties on productivity: Evidence from district Vehari, Pakistan. J. Agric. Soc. Sci. 2012, 8, 109–111. [Google Scholar]
- Lu, Y.; Wu, K.; Jiang, Y.; Guo, Y.; Desneux, N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 2012, 487, 362–365. [Google Scholar] [CrossRef]
- Chen, Y.; Lai, F.X.; Sun, Y.Q.; Hong, L.Y.; Tian, J.C.; Zhang, Z.T.; Qiang, F.U. Cry1Ab rice does not impact biological characters and functional response of Cyrtorhinus lividipennis preying on Nilaparvata lugens eggs. J. Integr. Agric. 2015, 14, 2011–2018. [Google Scholar] [CrossRef]
- Yang, H.; Peng, Y.; Tian, J.; Wang, J.; Hu, J.; Song, Q.; Wang, Z. Review: Biosafety assessment of Bt rice and other Bt crops using spiders as example for non-target arthropods in China. Plant Cell Rep. 2017, 36, 505–517. [Google Scholar] [CrossRef]
- Li, Y.; Diao, F.; Zhu, X.; Wang, L.; Zhang, K.; Li, D.; Ji, J.; Niu, L.; Gao, X.; Luo, J.; et al. Transgenic cotton expressing Cry1B protein has no adverse effect on predatory insect Propylea Japonica. Ecotoxicol. Environ. Saf. 2022, 245, 114088. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; Moran, N.A. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2010, 2, 708–718. [Google Scholar] [CrossRef]
- Dillon, R.J.; Vennard, C.T.; Buckling, A.; Charnley, A.K. Diversity of locust gut bacteria protects against pathogen invasion. Ecol. Lett. 2010, 8, 1291–1298. [Google Scholar] [CrossRef]
- Sharon, G.; Segal, D.; Ringo, J.M.; Hefetz, A.; Zilber-Rosenberg, L.; Rosenberg, E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2010, 107, 20051–20056. [Google Scholar] [CrossRef]
- Brand, J.M.; Bracke, J.W.; Markovetz, A.J.; Wood, D.L.; Browne, L.E. Production of verbenol pheromone by a bacterium isolated from bark beetles. Nature 1975, 254, 136–137. [Google Scholar] [CrossRef]
- Gaugler, R. Entomopathogenic Nematology; CABI Publishing: Wallingford, UK, 2002. [Google Scholar]
- König, H. Bacillus species in the intestine of termites and other soil invertebrates. J. Appl. Microbiol. 2006, 101, 620–627. [Google Scholar] [CrossRef]
- Wenzel, M.; Schönig, I.; Berchtold, M.; Kämpfer, P.; König, H. Aerobic and facultatively anaerobic cellulolytic bacteria from the gut of the termite Zootermopsis angusticollis. J. Appl. Microbiol. 2002, 92, 32–40. [Google Scholar] [CrossRef]
- Zhu, F.J. Effects of Bt Proteins Cry1Ab, Cry1F and Cry3B on Earthworm and Intestinal Microorganisms; Shandong Agricultural University: Taian, China, 2018. [Google Scholar]
- Wu, L.K. Studies of Biosafety of Bt Proteins on Propylea Japonica Using the Transcriptome and 16s Sequencing Technology; Chinese Academy of Agricultural Sciences: Beijing, China, 2018. [Google Scholar]
- Singh, A.K.; Dubey, S.K. Current trends in Bt crops and their fate on associated microbial community dynamics: A review. Protoplasma 2016, 253, 663–681. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, X.; Chen, X.; Romeis, J.R.; Yin, X.; Peng, Y. Consumption of Bt rice pollen containing Cry1C or Cry2A does not pose a risk to Propylea japonica (Thunberg) (Coleoptera: Coccinellidae). Sci. Rep. 2015, 5, 7679. [Google Scholar] [CrossRef]
- Romeis, J.; Hellmich, R.L.; Candolfi, M.P.; Carstens, K.; Schrijver, A.D.; Gatehouse, A.; Herman, R.A.; Huesing, J.E.; Mclean, M.A.; Raybould, A. Recommendations for the design of laboratory studies on non-target arthropods for risk assessment of genetically engineered plants. Transgenic Res. 2011, 20, 1–22. [Google Scholar] [CrossRef]
- Khan, I.A.; Wan, F.H. Life table of Propylea japonica Thunberg (Coleoptera, Coccinellidae) fed on Bemisia tabaci (Gennadius) (Homoptera, Aleyrodidae) biotype B prey. Sarhad. J. Agric. 2008, 24, 261–267. [Google Scholar]
- Zhang, L.; Li, S.; Luo, J.; Du, P.; Wu, L.; Li, Y.; Zhu, X.; Wang, L.; Zhang, S.; Cui, J. Chromosome-level genome assembly of the predator Propylea japonica to understand its tolerance to insecticides and high temperatures. Mol. Ecol. Resour. 2020, 20, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Shelton, A.M.; Zhao, J.Z.; Roush, R.T. Economic, ecological, food safety, and social consequences of the deployment of bt transgenic plants. Annu. Rev. Entomol. 2002, 47, 845–881. [Google Scholar] [CrossRef]
- Alvarez-Alfageme, F.; Bigler, F.; Romeis, J. Laboratory toxicity studies demonstrate no adverse effects of Cry1Ab and Cry3Bb1 to larvae of Adalia bipunctata (Coleoptera: Coccinellidae): The importance of study design. Transgenic Res. 2011, 20, 467–479. [Google Scholar] [CrossRef]
- Knight, K.; Head, G.; Rogers, J. Season-long expression of Cry1Ac and Cry2Ab proteins in Bollgard II cotton in Australia. Crop Prot. 2013, 44, 50–58. [Google Scholar] [CrossRef]
- Meissle, M.; Romeis, J. Transfer of Cry1Ac and Cry2Ab proteins from genetically engineered Bt cotton to herbivores and predators. Insect Sci. 2018, 25, 823–832. [Google Scholar] [CrossRef]
- Ramirez-Romero, R.; Desneux, N.; Chaufaux, J.; Kaiser, L. Bt-maize effects on biological parameters of the non-target aphid Sitobion avenae (Homoptera : Aphididae) and Cry1Ab toxin detection. Pestic. Biochem. Physiol. 2008, 91, 110–115. [Google Scholar] [CrossRef]
- Ali, I.; Zhang, S.; Iqbal, M.; Ejaz, S.; Cui, J.J. Trypsinized Cry1Fa and Vip3Aa have no detrimental effects on the adult green lacewing Chrysopa pallens (Neuroptera: Chrysopidae). Appl. Entomol. Zool. 2017, 52, 321–327. [Google Scholar] [CrossRef]
- Tian, J.C.; Yao, J.; Long, L.P.; Romeis, J.; Shelton, A.M. Bt crops benefit natural enemies to control non-target pests. Sci. Rep. 2015, 5, 16636. [Google Scholar] [CrossRef] [PubMed]
- Kremer, R.J.; Means, N.E. Glyphosate and glyphosate-resistant crop interactions with rhizosphere microorganisms. Eur. J. Agron. 2009, 31, 153–161. [Google Scholar] [CrossRef]
- Dong, L.; Meng, Y.; Wang, J.; Sun, G. Effects of Transgenic Bt + CpTI cotton on the abundance and diversity of rhizosphere ammonia oxidizing bacteria and archaea. J. Environ. Biol. 2016, 37, 881–888. [Google Scholar] [PubMed]
- Zhang, S.; Luo, J.; Jiang, W.; Wu, L.; Zhang, L.; Ji, J.; Wang, L.; Ma, Y.; Cui, J. Response of the bacterial community of Propylea japonica (Thunberg) to Cry2Ab protein. Environ. Pollut. 2019, 254, 113063. [Google Scholar] [CrossRef]
- Zhao, C.; Wu, L.; Luo, J.; Niu, L.; Wang, C.; Zhu, X.; Wang, L.; Zhao, P.; Zhang, S.; Cui, J. Bt, Not a Threat to Propylea japonica. Front. Physiol. 2020, 11, 758. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.; Song, X.; Yang, X.; Han, L.; Romeis, J.; Li, Y. Unintended changes in transgenic maize cause no nontarget effects. Plants People Plane 2022, 4, 313–415. [Google Scholar] [CrossRef]
- Niu, L.; Ma, Y.; Mannakkara, A.; Zhao, Y.; Ma, W.; Lei, C.; Chen, L. Impact of single and stacked insect-resistant Bt-cotton on the honey bee and silkworm. PLoS ONE 2013, 8, e72988. [Google Scholar]
- Xiong, H.; Li, D.; Liang, G.H.; Wang, Z.; Wei, B.Y. Effect of Bt protein on intestinal bacterial diversity of the Pardosa pseudoannulata. J. Hunan Agric. Univ. 2018, 44, 430–434. [Google Scholar]
- Han, L.; Jiang, X.; Peng, Y. Potential resistance management for the sustainable use of insect-resistant genetically modified corn and rice in China. Curr. Opin. Insect Sci. 2016, 15, 139–143. [Google Scholar] [CrossRef]
- Jiang, W.Y.; Dai, P.L.; Zhang, Y.J.; Zhou, T.; Lin, Y.; Shu, C.L.; Zhang, J. Effect of transgenic cotton with Cry1Ac gene on intestinal baterial community of Apis mellifera ligustica. Chin. J. Appl. Environ. Biol. 2010, 16, 211–215. [Google Scholar] [CrossRef]
- Romeis, J.; Meissle, M.; Bigler, F. Transgenic crops expressing Bacillus thuringiensis toxins and biological control. Nat. Biotechnol. 2006, 24, 63–71. [Google Scholar] [CrossRef]
- Crotti, E.; Balloi, A.; Hamdi, C.; Sansonno, L.; Marzorati, M.; Gonella, E.; Favia, G.; Cherif, A.; Bandi, C.; Alma, A. Microbial symbionts: A resource for the management of insect-related problems. Microb. Biotechnol. 2012, 5, 307–317. [Google Scholar] [CrossRef]
- Tsuchida, T.; Koga, R.; Horikawa, M.; Tsunoda, T.; Maoka, T.; Matsumoto, S.; Simon, J.C.; Fukatsu, T. Symbiotic bacterium modifies aphid body color. Science 2010, 330, 1102–1104. [Google Scholar] [CrossRef]
- Zhang, W.; He, Z.; Deng, X.; Yin, Y. Variety and distribution of intestinal flora of Apriona germari (hope) larvae. J. Southwest Agric. Univ. 2004, 26, 169–172. [Google Scholar]
- Jiang, W.Y.; Liang, G.M.; Lin, Y.; Shu, C.L.; Song, F.P.; Zhang, J. Comparison of midgut bacterial community between Bt resistant and sensitive Helicoverpa armiger. J. Microbiol. 2010, 50, 828–834. [Google Scholar]
- Dai, N.J. Diversity and Synergistic Effect with BT of the Intestine Bacterial Community of Plutella Xylostella Lavae; Huazhong Agricultural University: Wuhan, China, 2014. [Google Scholar]
- Behar, A.; Jurkevitch, E.; Yuval, B. Bringing back the fruit into fruit fly-bacteria interactions. Mol. Ecol. 2008, 17, 1375–1386. [Google Scholar] [CrossRef]
- Zhang, Q.P.; Cai, B.; Wang, X.; Liu, J.; Lin, R.; Hua, H.; Zhang, C.; Yi, X.; Song, Q.; Ji, J.; et al. Manipulation of Gut Symbionts for Improving the Sterile Insect Technique: Quality Parameters of Bactrocera dorsalis (Diptera: Tephritidae) Genetic Sexing Strain Males After Feeding on Bacteria-Enriched Diets. J. Econ. Entomol. 2021, 114, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, S.X.; Zhang, K.; Zhang, W.; Liu, R.; Zhang, R.L.; Zhang, Z. Enterobacter hormaechei in the intestines of housefly larvae promotes host growth by inhibiting harmful intestinal bacteria. Parasit Vectors 2021, 14, 598. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, A.; Habineza, P.; Ji, T.L.; Hou, Y.M.; Shi, Z.H. Intestinal Microbiota Confer Protection by Priming the Immune System of Red Palm Weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front. Physiol. 2019, 10, 1303. [Google Scholar] [CrossRef]
- Habineza, P.; Muhammad, A.; Ji, T.L.; Xiao, R.; Yin, X.Y.; Hou, Y.M.; Shi, Z.H. The Promoting Effect of Gut Microbiota on Growth and Development of Red Palm Weevil, Rhynchophorus ferrugineus (Olivier) (Coleoptera: Dryophthoridae) by Modulating Its Nutritional Metabolism. Front. Microbiol. 2019, 10, 1212. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.A.; Yoon, J.H.; Ryu, J.H.; Lee, W.J. Drosophila Microbiome Modulates Host Developmental and Metabolic Homeostasis via Insulin Signaling. Science 2011, 334, 670–674. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.H.; Berdy, B.M.; Velasquez, O.; Jovanovic, N.; Alkhalifa, S.; Minbiole, K.P.C.; Brucker, R.M. Changes in Microbiome Confer Multigenerational Host Resistance after Sub-toxic Pesticide Exposure. Cell Host Microbe 2020, 27, 213–224.e7. [Google Scholar] [CrossRef]
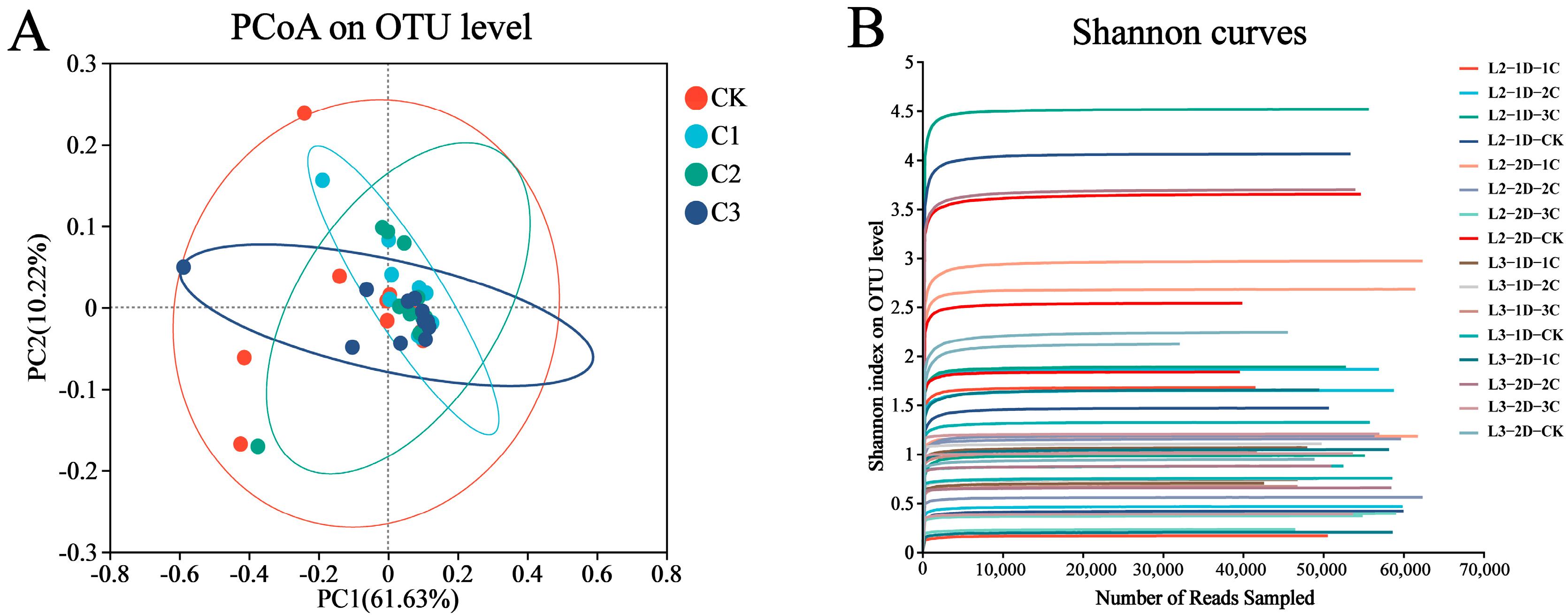
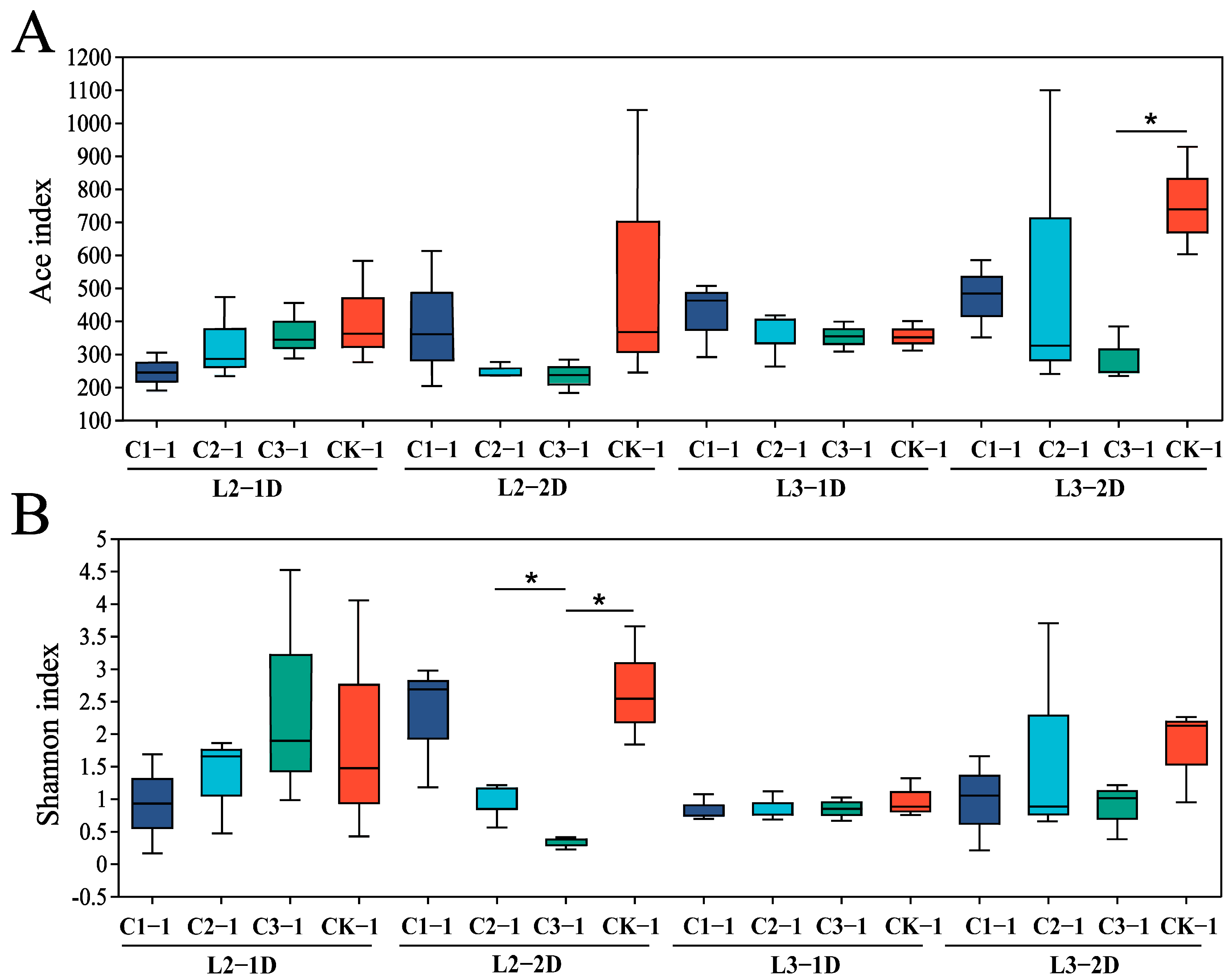

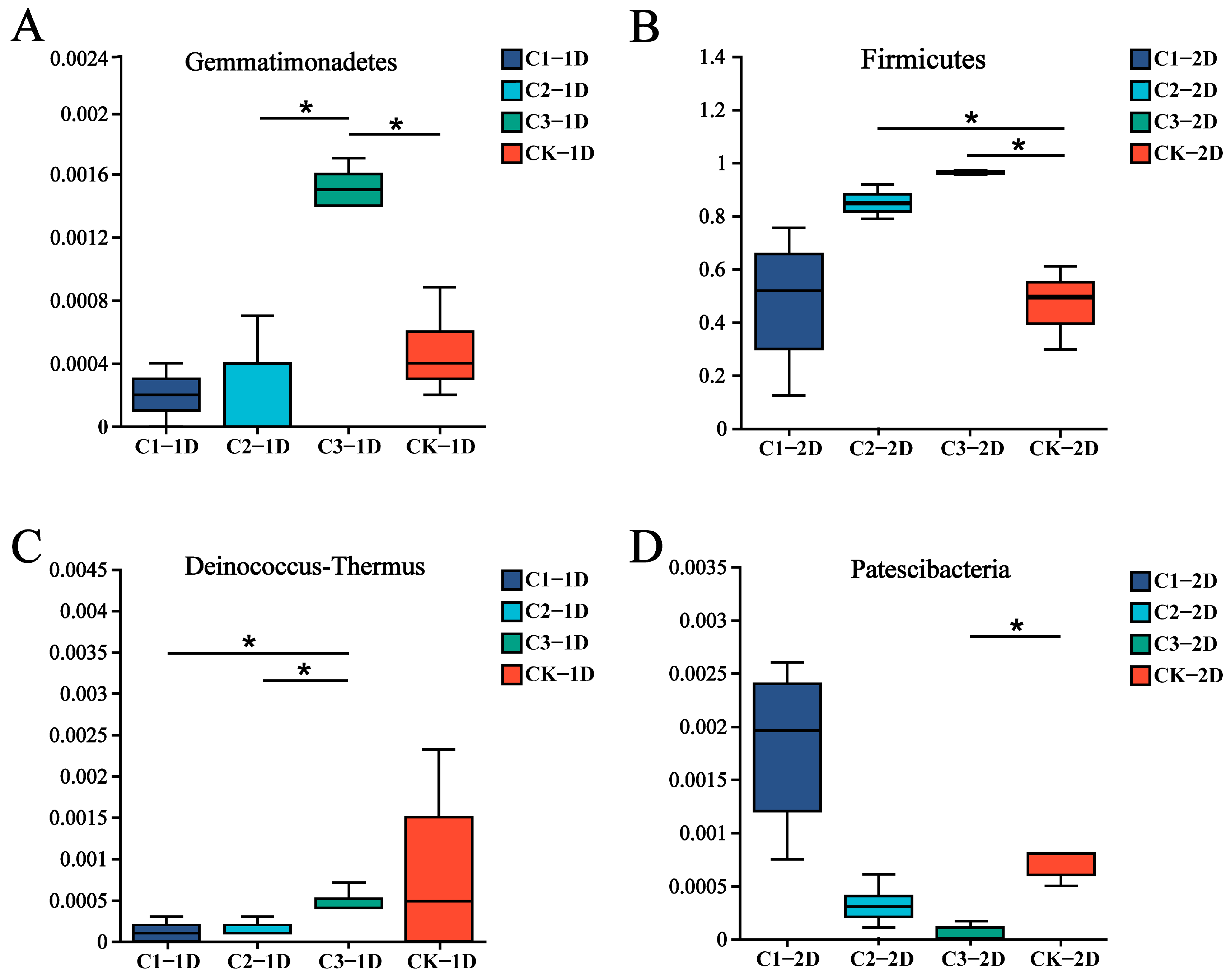
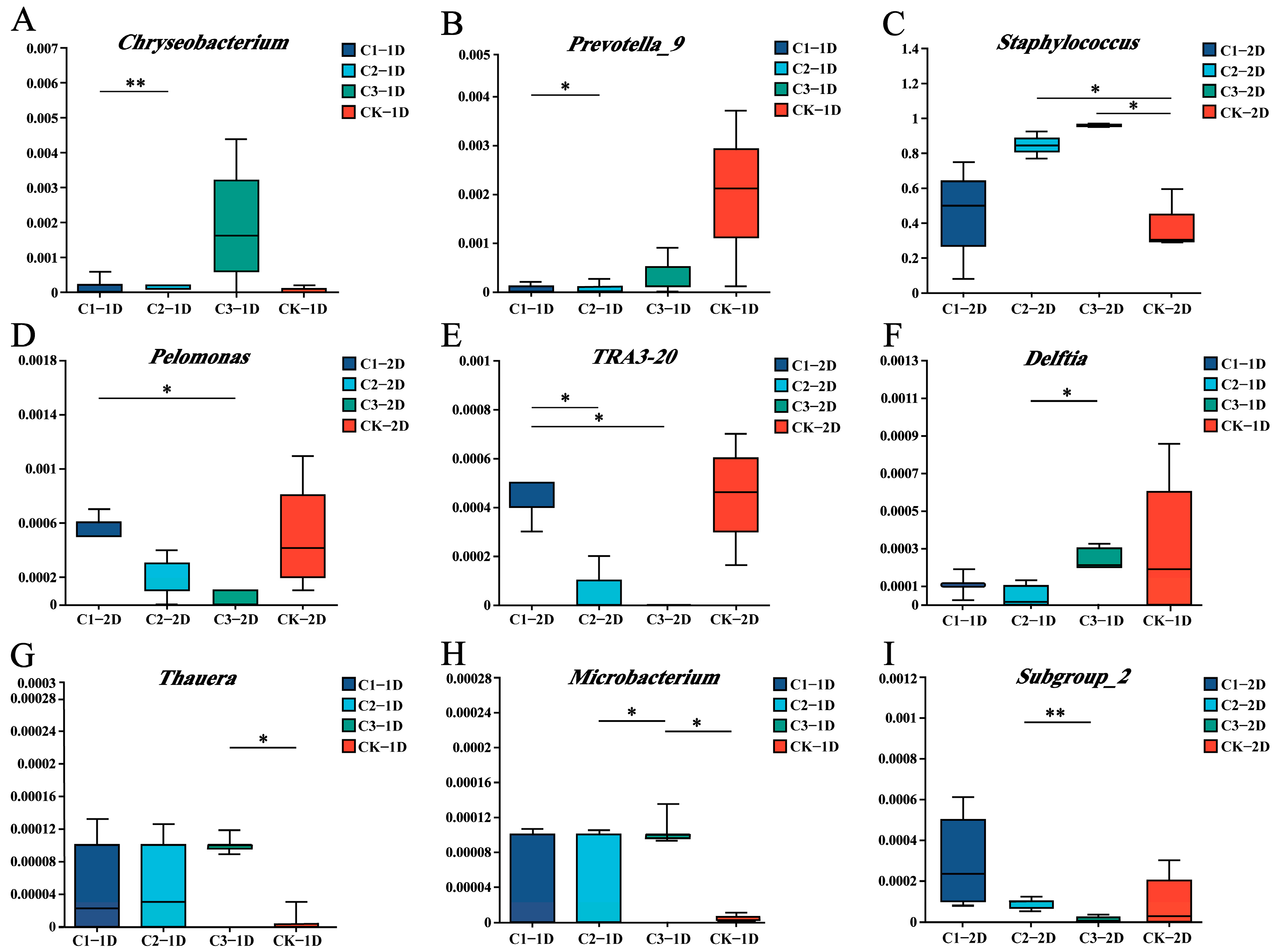
| Sample Name | Original Tags | High Quality Tags | OUTs | α Diversity Index | ||||
|---|---|---|---|---|---|---|---|---|
| Shannon | Simpson | Ace | Chao | Coverage | ||||
| L2_1D_C1 | 66,516 | 66,516 | 346 | 2.9537 | 0.2055 | 593.04 | 600.54 | 0.9991 |
| L2_1D_C2 | 61,032 | 60,538 | 331 | 1.0529 | 0.6660 | 340.31 | 342.44 | 0.9981 |
| L2_1D_C3 | 60,905 | 59,015 | 394 | 2.7374 | 0.3999 | 363.07 | 362.32 | 0.9991 |
| L2_1D_CK | 60,664 | 57,962 | 458 | 0.9368 | 0.7562 | 305.00 | 301.59 | 0.9984 |
| L2_2D_C1 | 63,288 | 64,417 | 600 | 2.6984 | 0.3527 | 337.38 | 350.43 | 0.9995 |
| L2_2D_C2 | 60,854 | 60,854 | 236 | 0.9639 | 0.6477 | 245.19 | 245.53 | 0.9986 |
| L2_2D_C3 | 54,419 | 54,419 | 191 | 0.3289 | 0.9000 | 232.71 | 234.11 | 0.9979 |
| L2_2D_CK | 48,682 | 48,682 | 532 | 2.6801 | 0.2359 | 575.59 | 563.47 | 0.9968 |
| L3_1D_C1 | 46,717 | 46,717 | 290 | 0.8338 | 0.6178 | 402.64 | 351.45 | 0.9968 |
| L3_1D_C2 | 48,830 | 48,830 | 249 | 0.8679 | 0.5559 | 404.96 | 345.06 | 0.9965 |
| L3_1D_C3 | 45,021 | 45,021 | 297 | 0.8438 | 0.6575 | 339.16 | 322.41 | 0.9973 |
| L3_1D_CK | 56,730 | 56,730 | 281 | 0.9826 | 0.5119 | 351.04 | 336.39 | 0.9971 |
| L3_2D_C1 | 57,147 | 57,147 | 406 | 0.9641 | 0.6492 | 456.31 | 432.79 | 0.9966 |
| L3_2D_C2 | 57,909 | 57,909 | 451 | 1.7358 | 0.4001 | 576.14 | 487.87 | 0.9963 |
| L3_2D_C3 | 55,863 | 55,863 | 153 | 0.8632 | 0.5460 | 308.40 | 211.03 | 0.9980 |
| L3_2D_CK | 43,716 | 43,716 | 662 | 1.7741 | 0.4718 | 746.87 | 738.36 | 0.9954 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diao, F.; Li, Y.; Gao, X.; Luo, J.; Zhu, X.; Wang, L.; Zhang, K.; Li, D.; Ji, J.; Cui, J. Response of the Propylea japonica Microbiota to Treatment with Cry1B Protein. Genes 2023, 14, 2008. https://doi.org/10.3390/genes14112008
Diao F, Li Y, Gao X, Luo J, Zhu X, Wang L, Zhang K, Li D, Ji J, Cui J. Response of the Propylea japonica Microbiota to Treatment with Cry1B Protein. Genes. 2023; 14(11):2008. https://doi.org/10.3390/genes14112008
Chicago/Turabian StyleDiao, Fengchao, Yarong Li, Xueke Gao, Junyu Luo, Xiangzhen Zhu, Li Wang, Kaixin Zhang, Dongyang Li, Jichao Ji, and Jinjie Cui. 2023. "Response of the Propylea japonica Microbiota to Treatment with Cry1B Protein" Genes 14, no. 11: 2008. https://doi.org/10.3390/genes14112008
APA StyleDiao, F., Li, Y., Gao, X., Luo, J., Zhu, X., Wang, L., Zhang, K., Li, D., Ji, J., & Cui, J. (2023). Response of the Propylea japonica Microbiota to Treatment with Cry1B Protein. Genes, 14(11), 2008. https://doi.org/10.3390/genes14112008






