The Pivotal Role of Noncoding RNAs in Flowering Time Regulation
Abstract
1. Introduction
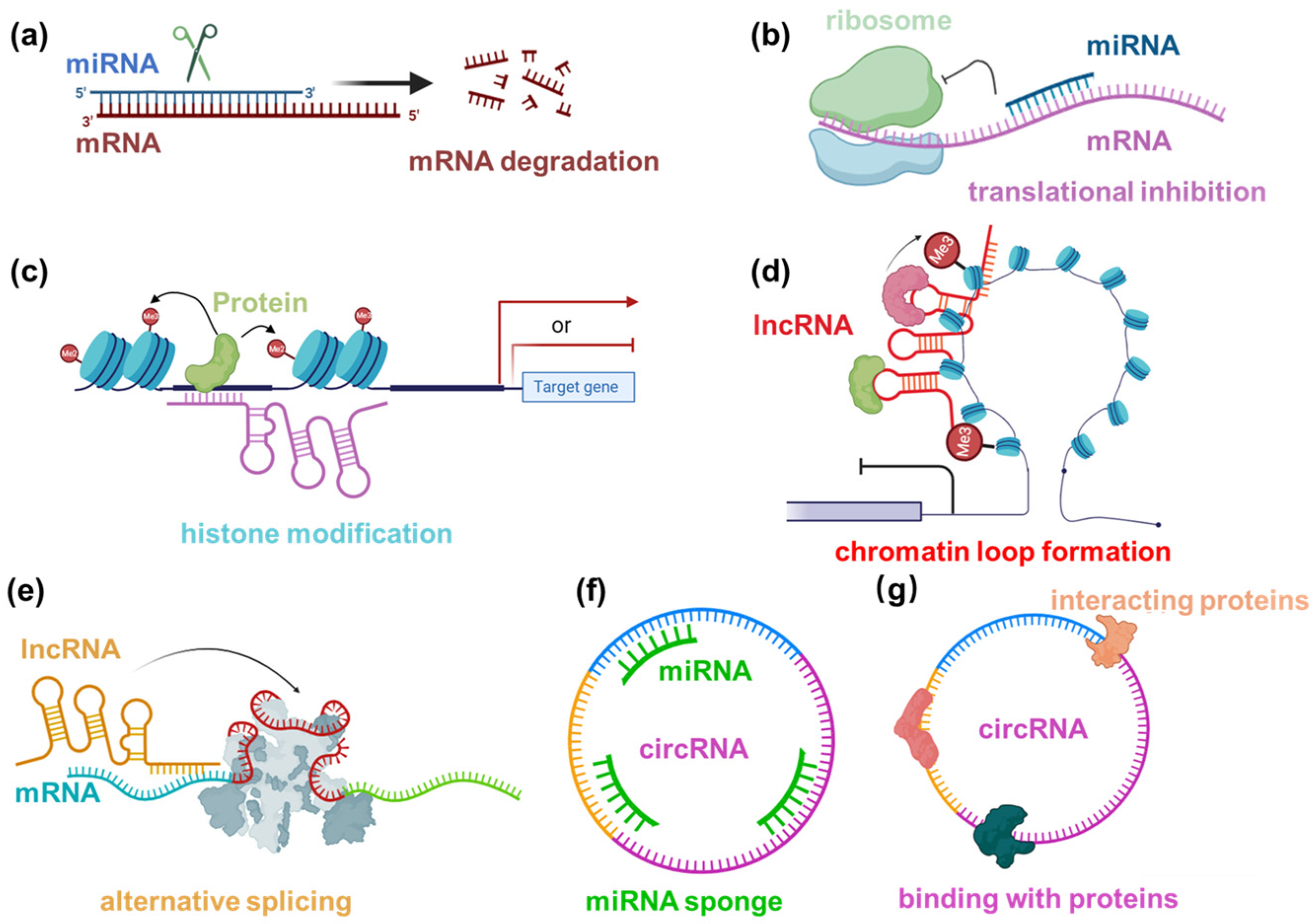
2. Classification of Noncoding RNAs and Their General Mechanisms
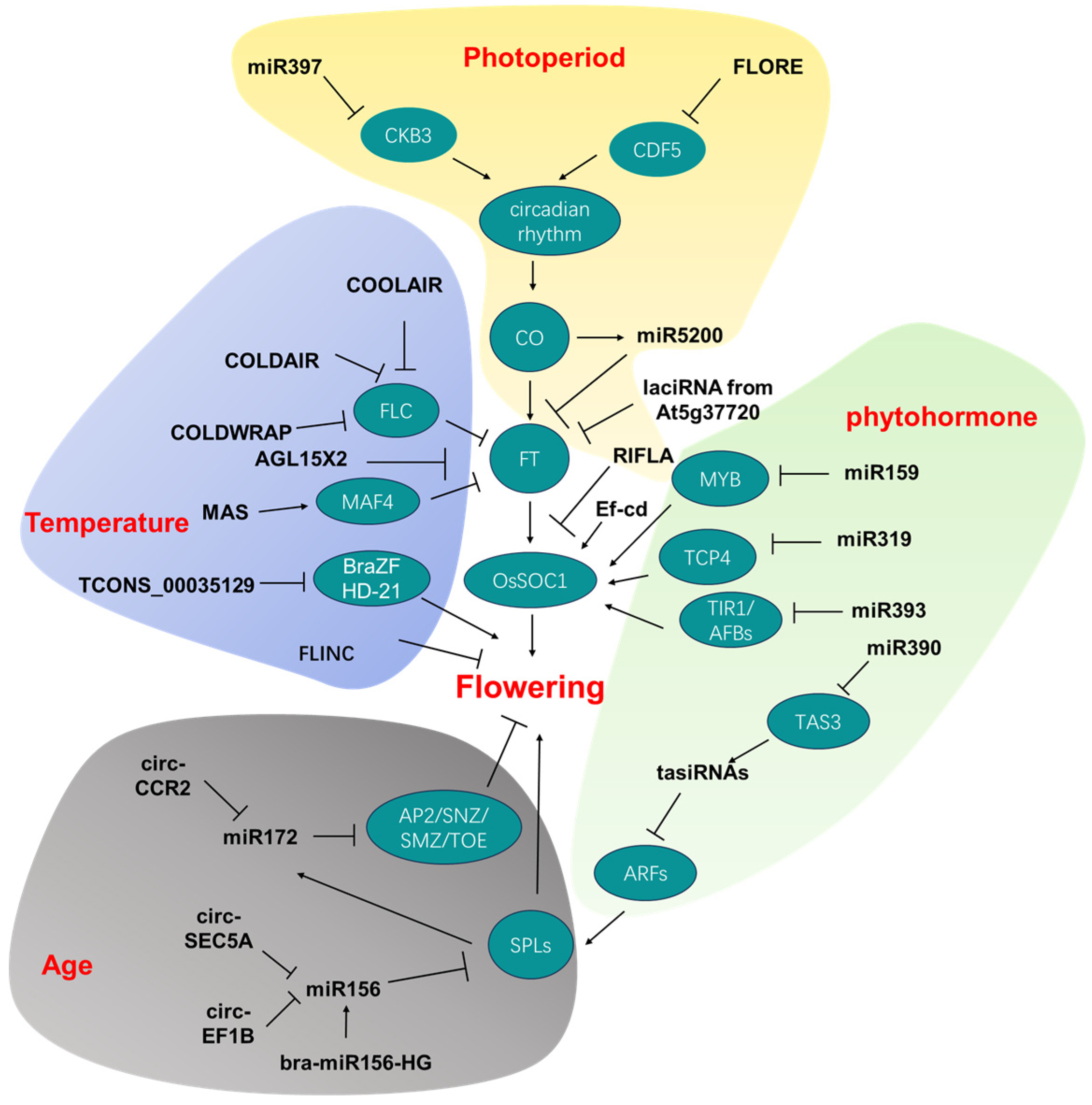
3. Noncoding RNAs Involved in Diverse Flowering Pathways
3.1. Photoperiodic Pathway
3.2. Autonomous Pathway and Vernalization Pathway
3.3. Aging Pathway
3.4. Phytohormone-Related Pathways
4. A Glimpse of Noncoding RNA in Flowering Adaption
5. Strategies for Noncoding RNA Manipulation in Crop Flowering Control
6. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, M.R.G.; Ai, X.-Y.; Zhang, J.-Z. Genetic regulation of flowering time in annual and perennial plants. WIREs RNA 2014, 5, 347–359. [Google Scholar] [CrossRef]
- Turck, F.; Fornara, F.; Coupland, G. Regulation and Identity of Florigen: FLOWERING LOCUS T Moves Center Stage. Annu. Rev. Plant Biol. 2008, 59, 573–594. [Google Scholar] [CrossRef]
- Zhang, Y.-C.; Yuan, C.; Chen, Y.-Q. Noncoding RNAs and their roles in regulating the agronomic traits of crops. Fundam. Res. 2023, 3, 718–726. [Google Scholar] [CrossRef]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Das, S.S.; Karmakar, P.; Nandi, A.K.; Sanan-Mishra, N. Small RNA mediated regulation of seed germination. Front. Plant Sci. 2015, 6, 828. [Google Scholar] [CrossRef]
- Gleeson, M.; Constantin, M.; Carroll, B.J.; Mitter, N. MicroRNAs as regulators of adventitious root development. J. Plant Biochem. Biotechnol. 2014, 23, 339–347. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, L.; Chen, G.; Shi, T. Identifying and Characterizing the Circular RNAs during the Lifespan of Arabidopsis Leaves. Front. Plant Sci. 2017, 8, 1278. [Google Scholar] [CrossRef]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yang, J.; Mathioni, S.M.; Yu, J.; Shen, J.; Yang, X.; Wang, L.; Zhang, Q.; Cai, Z.; Xu, C.; et al. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. USA 2016, 113, 15144–15149. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-q.; Wang, J.; Wu, Y.-y.; Li, D.-w.; Allan, A.C.; Yin, X.-r. Genome-wide analysis of coding and non-coding RNA reveals a conserved miR164-NAC regulatory pathway for fruit ripening. New Phytol. 2020, 225, 1618–1634. [Google Scholar] [CrossRef]
- Zhang, J.-P.; Yu, Y.; Feng, Y.-Z.; Zhou, Y.-F.; Zhang, F.; Yang, Y.-W.; Lei, M.-Q.; Zhang, Y.-C.; Chen, Y.-Q. MiR408 Regulates Grain Yield and Photosynthesis via a Phytocyanin Protein. Plant Physiol. 2017, 175, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-C.; Yu, Y.; Wang, C.-Y.; Li, Z.-Y.; Liu, Q.; Xu, J.; Liao, J.-Y.; Wang, X.-J.; Qu, L.-H.; Chen, F.; et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat. Biotechnol. 2013, 31, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Wen, X.; Mudunuri, S.; Varma, G.P.S.; Sablok, G. Diff isomiRs: Large-scale detection of differential isomiRs for understanding non-coding regulated stress omics in plants. Sci. Rep. 2019, 9, 1406. [Google Scholar] [CrossRef] [PubMed]
- Summanwar, A.; Basu, U.; Rahman, H.; Kav, N.N.V. Non-coding RNAs as emerging targets for crop improvement. Plant Sci. 2020, 297, 110521. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Chen, X.; Chen, Y. Plant Noncoding RNAs: Hidden Players in Development and Stress Responses. Annu. Rev. Cell Dev. Biol. 2019, 35, 407–431. [Google Scholar] [CrossRef]
- Liu, T.-T.; Zhu, D.; Chen, W.; Deng, W.; He, H.; He, G.; Bai, B.; Qi, Y.; Chen, R.; Deng, X.W. A Global Identification and Analysis of Small Nucleolar RNAs and Possible Intermediate-Sized Non-Coding RNAs in Oryza sativa. Mol. Plant 2013, 6, 830–846. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Deng, W.; Fan, X.; Liu, T.-T.; He, G.; Chen, R.; Terzaghi, W.; Zhu, D.; Deng, X.W. Genomic Features and Regulatory Roles of Intermediate-Sized Non-Coding RNAs in Arabidopsis. Mol. Plant 2014, 7, 514–527. [Google Scholar] [CrossRef]
- Rani, V.; Sengar, R.S. Biogenesis and mechanisms of microRNA-mediated gene regulation. Biotechnol. Bioeng. 2022, 119, 685–692. [Google Scholar] [CrossRef]
- Li, Z.; Xu, R.; Li, N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr. Metab. 2018, 15, 68. [Google Scholar] [CrossRef]
- Bologna, N.G.; Iselin, R.; Abriata, L.A.; Sarazin, A.; Pumplin, N.; Jay, F.; Grentzinger, T.; Dal Peraro, M.; Voinnet, O. Nucleo-cytosolic Shuttling of ARGONAUTE1 Prompts a Revised Model of the Plant MicroRNA Pathway. Mol. Cell 2018, 69, 709–719.e705. [Google Scholar] [CrossRef]
- Li, M.; Yu, B. Recent advances in the regulation of plant miRNA biogenesis. RNA Biol. 2021, 18, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Xu, M.; Willmann, M.R.; McCormick, K.; Hu, T.; Yang, L.; Starker, C.G.; Voytas, D.F.; Meyers, B.C.; Poethig, R.S. Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007337. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, Secondary, Small Interfering RNAs in Posttranscriptional Regulatory Networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Ma, X.; Wang, C.; Li, Q.; Meyers, B.C.; Springer, N.M.; Walbot, V. CHH DNA methylation increases at 24-PHAS loci depend on 24-nt phased small interfering RNAs in maize meiotic anthers. New Phytol. 2021, 229, 2984–2997. [Google Scholar] [CrossRef]
- Liu, Y.; Ke, L.; Wu, G.; Xu, Y.; Wu, X.; Xia, R.; Deng, X.; Xu, Q. miR3954 is a trigger of phasiRNAs that affects flowering time in citrus. Plant J. 2017, 92, 263–275. [Google Scholar] [CrossRef]
- Wierzbicki, A.T.; Blevins, T.; Swiezewski, S. Long noncoding RNAs in plants. Annu. Rev. Plant Biol. 2021, 72, 245–271. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Fang, X.; Zhu, D.; Dean, C. Autonomous Pathway: FLOWERING LOCUS C Repression through an Antisense-Mediated Chromatin-Silencing Mechanism. Plant Physiol. 2020, 182, 27–37. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-Mediated Epigenetic Silencing by a Long Intronic Noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Jin, Y.; Ivanov, M.; Dittrich, A.N.; Nelson, A.D.L.; Marquardt, S. LncRNA FLAIL affects alternative splicing and represses flowering in Arabidopsis. EMBO J. 2023, 42, e110921. [Google Scholar] [CrossRef]
- Quan, G.; Li, J. Circular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian Res. 2018, 11, 9. [Google Scholar] [CrossRef]
- Weirick, T.; Militello, G.; Uchida, S. Long Non-coding RNAs in Endothelial Biology. Front. Physiol. 2018, 9, 522. [Google Scholar] [CrossRef]
- Chu, Q.; Shen, E.; Ye, C.-Y.; Fan, L.; Zhu, Q.-H. Emerging Roles of Plant Circular RNAs. J. Plant Cell Dev. 2018, 1, 1–14. [Google Scholar] [CrossRef]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef]
- Fan, J.; Quan, W.; Li, G.-B.; Hu, X.-H.; Wang, Q.; Wang, H.; Li, X.-P.; Luo, X.; Feng, Q.; Hu, Z.-J.; et al. circRNAs Are Involved in the Rice-Magnaporthe oryzae Interaction. Plant Physiol. 2020, 182, 272–286. [Google Scholar] [CrossRef]
- Gao, Z.; Li, J.; Luo, M.; Li, H.; Chen, Q.; Wang, L.; Song, S.; Zhao, L.; Xu, W.; Zhang, C.; et al. Characterization and Cloning of Grape Circular RNAs Identified the Cold Resistance-Related Vv-circATS1. Plant Physiol. 2019, 180, 966–985. [Google Scholar] [CrossRef]
- Zhang, P.; Fan, Y.; Sun, X.; Chen, L.; Terzaghi, W.; Bucher, E.; Li, L.; Dai, M. A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J. 2019, 98, 697–713. [Google Scholar] [CrossRef]
- Babaei, S.; Singh, M.B.; Bhalla, P.L. Circular RNAs modulate the floral fate acquisition in soybean shoot apical meristem. BMC Plant Biol. 2023, 23, 322. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, Y.; Li, Z.; Wang, T.; Zhang, X.; Zheng, B. A lariat-derived circular RNA is required for plant development in Arabidopsis. Sci. China Life Sci. 2018, 61, 204–213. [Google Scholar] [CrossRef]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Song, Y.H.; Shim, J.S.; Kinmonth-Schultz, H.A.; Imaizumi, T. Photoperiodic Flowering: Time Measurement Mechanisms in Leaves. Annu. Rev. Plant Biol. 2015, 66, 441–464. [Google Scholar] [CrossRef]
- Feng, Y.-Z.; Yu, Y.; Zhou, Y.-F.; Yang, Y.-W.; Lei, M.-Q.; Lian, J.-P.; He, H.; Zhang, Y.-C.; Huang, W.; Chen, Y.-Q. A Natural Variant of miR397 Mediates a Feedback Loop in Circadian Rhythm. Plant Physiol. 2020, 182, 204–214. [Google Scholar] [CrossRef]
- Fornara, F.; Panigrahi, K.C.S.; Gissot, L.; Sauerbrunn, N.; Rühl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis DOF Transcription Factors Act Redundantly to Reduce CONSTANS Expression and Are Essential for a Photoperiodic Flowering Response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef]
- Henriques, R.; Wang, H.; Liu, J.; Boix, M.; Huang, L.-F.; Chua, N.-H. The antiphasic regulatory module comprising CDF5 and its antisense RNA FLORE links the circadian clock to photoperiodic flowering. New Phytol. 2017, 216, 854–867. [Google Scholar] [CrossRef]
- Shin, W.J.; Nam, A.H.; Kim, J.Y.; Kwak, J.S.; Song, J.T.; Seo, H.S. Intronic long noncoding RNA, RICE FLOWERING ASSOCIATED (RIFLA), regulates OsMADS56-mediated flowering in rice. Plant Sci. 2022, 320, 111278. [Google Scholar] [CrossRef]
- Wu, L.; Liu, D.; Wu, J.; Zhang, R.; Qin, Z.; Liu, D.; Li, A.; Fu, D.; Zhai, W.; Mao, L. Regulation of FLOWERING LOCUS T by a MicroRNA in Brachypodium distachyon. Plant Cell 2013, 25, 4363–4377. [Google Scholar] [CrossRef]
- Qi, P.-L.; Zhou, H.-R.; Zhao, Q.-Q.; Feng, C.; Ning, Y.-Q.; Su, Y.-N.; Cai, X.-W.; Yuan, D.-Y.; Zhang, Z.-C.; Su, X.-M.; et al. Characterization of an autonomous pathway complex that promotes flowering in Arabidopsis. Nucleic Acids Res. 2022, 50, 7380–7395. [Google Scholar] [CrossRef]
- Yan, Z.; Liang, D.; Liu, H.; Zheng, G. FLC: A key regulator of flowering time in Arabidopsis. Russ. J. Plant Physiol. 2010, 57, 166–174. [Google Scholar] [CrossRef]
- Tian, Y.; Zheng, H.; Zhang, F.; Wang, S.; Ji, X.; Xu, C.; He, Y.; Ding, Y. PRC2 recruitment and H3K27me3 deposition at FLC require FCA binding of COOLAIR. Sci. Adv. 2019, 5, eaau7246. [Google Scholar] [CrossRef]
- Csorba, T.; Questa, J.I.; Sun, Q.; Dean, C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl. Acad. Sci. USA 2014, 111, 16160–16165. [Google Scholar] [CrossRef]
- Kim, D.H.; Xi, Y.; Sung, S. Modular function of long noncoding RNA, COLDAIR, in the vernalization response. PLoS Genet. 2017, 13, e1006939. [Google Scholar] [CrossRef]
- Kim, D.-H.; Sung, S. Vernalization-Triggered Intragenic Chromatin Loop Formation by Long Noncoding RNAs. Dev. Cell 2017, 40, 302–312.e304. [Google Scholar] [CrossRef]
- Yang, J.; Cao, Y.; Ma, L. Co-Transcriptional RNA Processing in Plants: Exploring from the Perspective of Polyadenylation. Int. J. Mol. Sci. 2021, 22, 3300. [Google Scholar] [CrossRef]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef]
- Liu, T.; Wu, P.; Wang, Q.; Wang, W.; Zhang, C.; Sun, F.; Liu, Z.; Li, Y.; Hou, X. Comparative transcriptome discovery and elucidation of the mechanism of long noncoding RNAs during vernalization in Brassica rapa. Plant Growth Regul. 2018, 85, 27–39. [Google Scholar] [CrossRef]
- Liang, N.; Cheng, D.; Cui, J.; Dai, C.; Luo, C.; Liu, T.; Li, J. Vernalisation mediated LncRNA-like gene expression in Beta vulgaris. Funct. Plant Biol. 2017, 44, 720–726. [Google Scholar] [CrossRef]
- Severing, E.; Faino, L.; Jamge, S.; Busscher, M.; Kuijer-Zhang, Y.; Bellinazzo, F.; Busscher-Lange, J.; Fernández, V.; Angenent, G.C.; Immink, R.G.H.; et al. Arabidopsis thaliana ambient temperature responsive lncRNAs. BMC Plant Biol. 2018, 18, 145. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef]
- Zhou, Y.; Myat, A.A.; Liang, C.; Meng, Z.; Guo, S.; Wei, Y.; Sun, G.; Wang, Y.; Zhang, R. Insights Into MicroRNA-Mediated Regulation of Flowering Time in Cotton Through Small RNA Sequencing. Front. Plant Sci. 2022, 13, 761244. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, Y.-Q.; Gao, X.; Jia, G.-X. Analysis of miRNA-mediated regulation of flowering induction in Lilium × formolongi. BMC Plant Biol. 2021, 21, 190. [Google Scholar] [CrossRef]
- Chuck, G.; Cigan, A.M.; Saeteurn, K.; Hake, S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 2007, 39, 544–549. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, J.; Zhou, C. The role of miR156 in developmental transitions in Nicotiana tabacum. Sci. China Life Sci. 2015, 58, 253–260. [Google Scholar] [CrossRef][Green Version]
- Cho, S.H.; Coruh, C.; Axtell, M.J. miR156 and miR390 Regulate tasiRNA Accumulation and Developmental Timing in Physcomitrella patens. Plant Cell 2012, 24, 4837–4849. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. To Bloom or Not to Bloom: Role of MicroRNAs in Plant Flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef]
- Jung, J.-H.; Seo, Y.-H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.-H.; Park, C.-M. The GIGANTEA-Regulated MicroRNA172 Mediates Photoperiodic Flowering Independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef]
- Aukerman, M.J.; Sakai, H. Regulation of Flowering Time and Floral Organ Identity by a MicroRNA and Its APETALA2-Like Target Genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific Effects of MicroRNAs on the Plant Transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Zhou, D.; Zhao, S.; Zhou, H.; Chen, J.; Huang, L. A lncRNA bra-miR156HG regulates flowering time and leaf morphology as a precursor of miR156 in Brassica campestris and Arabidopsis thaliana. Plant Sci. 2023, 337, 111889. [Google Scholar] [CrossRef]
- Yang, R.; Li, P.; Mei, H.; Wang, D.; Sun, J.; Yang, C.; Hao, L.; Cao, S.; Chu, C.; Hu, S.; et al. Fine-Tuning of MiR528 Accumulation Modulates Flowering Time in Rice. Mol. Plant 2019, 12, 1103–1113. [Google Scholar] [CrossRef]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, M.; Inukai, Y.; Ueguchi-Tanaka, M.; Itoh, H.; Izawa, T.; Kobayashi, Y.; Hattori, T.; Miyao, A.; Hirochika, H.; Ashikari, M.; et al. Loss-of-Function Mutations of the Rice GAMYB Gene Impair α-Amylase Expression in Aleurone and Flower Development. Plant Cell 2004, 16, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Montgomery, T.A.; Howell, M.D.; Allen, E.; Dvorak, S.K.; Alexander, A.L.; Carrington, J.C. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA Affects Developmental Timing and Patterning in Arabidopsis. Curr. Biol. 2006, 16, 939–944. [Google Scholar] [CrossRef]
- O’Neill, D.P.; Ross, J.J. Auxin Regulation of the Gibberellin Pathway in Pea. Plant Physiol. 2002, 130, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Harberd, N.P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 2003, 421, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 Downregulation via OsmiR393 Overexpression Leads to More Tillers, Early Flowering and Less Tolerance to Salt and Drought in Rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef]
- Domagalska, M.A.; Schomburg, F.M.; Amasino, R.M.; Vierstra, R.D.; Nagy, F.; Davis, S.J. Attenuation of brassinosteroid signaling enhances FLC expression and delays flowering. Development 2007, 134, 2841–2850. [Google Scholar] [CrossRef]
- Li, Z.; Ou, Y.; Zhang, Z.; Li, J.; He, Y. Brassinosteroid Signaling Recruits Histone 3 Lysine-27 Demethylation Activity to FLOWERING LOCUS C Chromatin to Inhibit the Floral Transition in Arabidopsis. Mol. Plant 2018, 11, 1135–1146. [Google Scholar] [CrossRef]
- Tadege, M.; Sheldon, C.C.; Helliwell, C.A.; Upadhyaya, N.M.; Dennis, E.S.; Peacock, W.J. Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 2003, 1, 361–369. [Google Scholar] [CrossRef]
- Wang, H.; Li, Y.; Chern, M.; Zhu, Y.; Zhang, L.-L.; Lu, J.-H.; Li, X.-P.; Dang, W.-Q.; Ma, X.-C.; Yang, Z.-R.; et al. Suppression of rice miR168 improves yield, flowering time and immunity. Nat. Plants 2021, 7, 129–136. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L. The Critical Role of miRNAs in Regulation of Flowering Time and Flower Development. Genes 2020, 11, 319. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Zhang, F.; Wang, H.; Wang, W.; Zhao, F.; Li, Z.; Sun, C.; Chen, F.; Xu, F.; Chang, S.; et al. Ef-cd locus shortens rice maturity duration without yield penalty. Proc. Natl. Acad. Sci. USA 2019, 116, 18717–18722. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.-H.; Helliwell, C.A. Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 2011, 62, 487–495. [Google Scholar] [CrossRef]
- Millar, A.A.; Lohe, A.; Wong, G. Biology and Function of miR159 in Plants. Plants 2019, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Javier, F.P.; Detlef, W. Specific Regulation of TCP genes by miR319. bioRxiv 2019, 747790. [Google Scholar] [CrossRef]
- McKeown, M.; Schubert, M.; Preston, J.C.; Fjellheim, S. Evolution of the miR5200-FLOWERING LOCUS T flowering time regulon in the temperate grass subfamily Pooideae. Mol. Phylogenetics Evol. 2017, 114, 111–121. [Google Scholar] [CrossRef]
- Bernardi, Y.; Ponso, M.A.; Belén, F.; Vegetti, A.C.; Dotto, M.C. MicroRNA miR394 regulates flowering time in Arabidopsis thaliana. Plant Cell Rep. 2022, 41, 1375–1388. [Google Scholar] [CrossRef]
- Xu, M.Y.; Zhang, L.; Li, W.W.; Hu, X.L.; Wang, M.-B.; Fan, Y.L.; Zhang, C.Y.; Wang, L. Stress-induced early flowering is mediated by miR169 in Arabidopsis thaliana. J. Exp. Bot. 2014, 65, 89–101. [Google Scholar] [CrossRef]
- Kim, W.; Ahn, H.J.; Chiou, T.-J.; Ahn, J.H. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol. Cells 2011, 32, 83–88. [Google Scholar] [CrossRef]
- Gramzow, L.; Theißen, G. Plant miRNA Conservation and Evolution. In Plant MicroRNAs: Methods and Protocols; de Folter, S., Ed.; Springer: New York, NY, USA, 2019; pp. 41–50. [Google Scholar]
- Van Valen, L. Foundations of Macroecology 19. A New Evolutionary Law (1973). In Classic Papers with Commentaries; Smith, F.A., Gittleman, J.L., Brown, J.H., Eds.; University of Chicago Press: Chicago, IL, USA, 2014; pp. 284–314. [Google Scholar]
- Zhao, Y.; Lu, G.-A.; Yang, H.; Lin, P.; Liufu, Z.; Tang, T.; Xu, J. Run or Die in the Evolution of New MicroRNAs—Testing the Red Queen Hypothesis on De Novo New Genes. Mol. Biol. Evol. 2021, 38, 1544–1553. [Google Scholar] [CrossRef]
- Deng, P.; Liu, S.; Nie, X.; Weining, S.; Wu, L. Conservation analysis of long non-coding RNAs in plants. Sci. China Life Sci. 2018, 61, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Lee, J.H.; Kim, W.; Jung, H.S.; Huijser, P.; Ahn, J.H. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 Module Regulates Ambient Temperature-Responsive Flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012, 159, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Small, I. RNAi for revealing and engineering plant gene functions. Curr. Opin. Biotechnol. 2007, 18, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.-Z.; Yu, Y. Experimental Strategies for Studying the Function of Plant CircRNAs. In Plant Circular RNAs: Methods and Protocols; Vaschetto, L.M., Ed.; Springer: New York, NY, USA, 2021; pp. 21–33. [Google Scholar]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Liu, T.; Tan, J.; Zhang, Y.; Zheng, Z.; Wang, B.; Zhou, D.; Xie, X.; Guo, M.; Liu, Y.-G.; et al. PhieCBEs: Plant High-Efficiency Cytidine Base Editors with Expanded Target Range. Mol. Plant 2020, 13, 1666–1669. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Zeng, D.; Zhao, Y.; Wang, Y.; Liu, T.; Li, S.; Xue, Y.; Luo, Y.; Xie, X.; Chen, L.; et al. PhieABEs: A PAM-less/free high-efficiency adenine base editor toolbox with wide target scope in plants. Plant Biotechnol. J. 2022, 20, 934–943. [Google Scholar] [CrossRef]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef]
- Lin, Q.; Jin, S.; Zong, Y.; Yu, H.; Zhu, Z.; Liu, G.; Kou, L.; Wang, Y.; Qiu, J.-L.; Li, J.; et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat. Biotechnol. 2021, 39, 923–927. [Google Scholar] [CrossRef]
- Zhu, L.; Ow, D.W.; Dong, Z. Transfer RNA-derived small RNAs in plants. Sci. China Life Sci. 2018, 61, 155–161. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, C.; Wu, T.; Liu, R.; Mao, W.; Gan, X.; Lu, X.; Liu, Y.; Wan, L.; Xu, B.; et al. Current status and perspectives of non-coding RNA and phase separation interactions. BioSci. Trends 2022, 16, 330–345. [Google Scholar] [CrossRef]
- Alberti, S. Phase separation in biology. Curr. Biol. 2017, 27, R1097–R1102. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Chen, M.; Niu, J.; Wang, L.; Li, Y.; Fang, X.; Li, P.; Qi, Y. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat. Cell Biol. 2021, 23, 32–39. [Google Scholar] [CrossRef] [PubMed]
| Species | Categories | Names | Targets | Functions | Reference |
|---|---|---|---|---|---|
| Arabidopsis/rice/maize/tomato, etc. | miRNA | miR156 | SPLs | delays flowering | [59,60] |
| Arabidopsis | miRNA | miR172 | AP2/SMZ/SNZ /TOE | accelerates flowering | [83] |
| rice | miRNA | miR528 | OsRFI2 | promotes flowering under long-day conditions | [69] |
| Arabidopsis | miRNA | miR159 | MYB | delays flowering in short day | [84] |
| Arabidopsis | miRNA | miR319 | TCP | delays flowering in short day | [85] |
| B. distachyon | miRNA | miR5200 | FT | promotes flowering under short-day conditions and inhibits flowering under long-day conditions | [86] |
| rice | miRNA | miR393 | OsTIR1/OsAFB2 | increases tiller number and promotes heading | [76] |
| rice | miRNA | miR168 | AGO1 | suppression of miR168 shortens flowering time | [80] |
| Arabidopsis | miRNA | miR397 | CKB3 | delays flowering time by modulating the circadian clock | [42] |
| Arabidopsis | miRNA | miR394a/ miR394b | LCR | loss-of-function showed early flowering with decreased branching and lower seed production | [87] |
| Arabidopsis | miRNA | miR390 | ARF3/ARF4 | delays flowering time | [63] |
| citrus | miRNA | miR3954 | NAC | facilitates flowering time | [7] |
| Arabidopsis | miRNA | miR169 | NF-YA | promotes early flowering through the abiotic stress response | [88] |
| Arabidopsis | miRNA | miR399 | PHO2 | promotes flowering at normal temperature | [89] |
| Arabidopsis | lncRNA | COOLAIR | FLC | promotes flowering | [8] |
| Arabidopsis | lncRNA | COLDAIR | FLC | promotes flowering | [51] |
| Arabidopsis | lncRNA | COLDWRAP | FLC | promotes flowering | [52] |
| B. rapa | lncRNA | TCONS_00035129 | BraZF-HD21 | promotes flowering | [55] |
| sugar beet | lncRNA | AGL15X2 | BvFT1 | promotes reproductive growth upon vernalization | [56] |
| Arabidopsis | lncRNA | FLINC | not clear | regulates temperature-mediated flowering | [57] |
| Arabidopsis | lncRNA | FLORE | FT | promotes flowering in a circadian-dependent manner | [44] |
| rice | lncRNA | Ef-cd | OsSOC1 | shortens maturity duration with no yield penalty | [82] |
| Arabidopsis | lncRNA | FLAIL | LAC8 | represses flowering | [30] |
| rice | lncRNA | RIFLA | OsMADS56 | promotes flowering | [45] |
| Soybean | circRNA | circ-CCR2, circ-SEC5A, circ-EF1B | miR172, miR156 | induces the floral meristem | [38] |
| Arabidopsis | circRNA | laciRNA from At5g37720 | FT | delays flowering | [39] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Zhu, Q.-F.; Li, W.-Y.; Chen, P.; Xue, J.; Yu, Y.; Feng, Y.-Z. The Pivotal Role of Noncoding RNAs in Flowering Time Regulation. Genes 2023, 14, 2114. https://doi.org/10.3390/genes14122114
Liu Y, Zhu Q-F, Li W-Y, Chen P, Xue J, Yu Y, Feng Y-Z. The Pivotal Role of Noncoding RNAs in Flowering Time Regulation. Genes. 2023; 14(12):2114. https://doi.org/10.3390/genes14122114
Chicago/Turabian StyleLiu, Yun, Qing-Feng Zhu, Wen-Yan Li, Pei Chen, Jiao Xue, Yang Yu, and Yan-Zhao Feng. 2023. "The Pivotal Role of Noncoding RNAs in Flowering Time Regulation" Genes 14, no. 12: 2114. https://doi.org/10.3390/genes14122114
APA StyleLiu, Y., Zhu, Q.-F., Li, W.-Y., Chen, P., Xue, J., Yu, Y., & Feng, Y.-Z. (2023). The Pivotal Role of Noncoding RNAs in Flowering Time Regulation. Genes, 14(12), 2114. https://doi.org/10.3390/genes14122114






