Abstract
RNA sequencing makes it possible to uncover genetic mechanisms that underlie certain performance traits. In order to gain a deeper insight into the genetic background and biological processes involved in endurance performance in horses, the changes in the gene expression profiles induced by endurance runs over long (70 km) and short (15 km) distances in the blood of Kabardian horses (Equus caballus) were analyzed. For the long-distance runs, we identified 1484 up- and 691 downregulated genes, while after short-distance runs, only 13 up- and 8 downregulated genes (FC > |1.5|; p < 0.05) were found. These differentially expressed genes (DEGs) are involved in processes and pathways that are primarily related to stress response (interleukin production, activation of inflammatory system) but also to metabolism (carbohydrate catabolic process, lipid biosynthesis, NADP metabolic process). The most important genes involved in these processes therefore represent good candidates for the monitoring and evaluation of the performance of horses in order to avoid excessive demands when endurance performance is required, like ACOD1, CCL5, CD40LG, FOS, IL1R2, IL20RA, and IL22RA2, on the one hand, and, on the other hand, for assessing the suitability of a horse for endurance races, like GATA2, GYG1, HIF1A, MOGAT1, PFKFB3, PLIN5, SIK1, and STBD1.
1. Introduction
To date, numerous genomic polymorphisms have been discovered in horses that are associated with traits relevant to breeding. However, these are primarily mutations that are associated with monogenic phenotypic properties such as hereditary defects [1], coat color [2], or appearance [3]. However, there is a distinct lack of knowledge about the genetic basis of athletic performance, which is the focus of many breeders. Thanks to modern methods of molecular genetics and bioinformatics, initial successes have also been achieved here [4,5,6]. For example, the myostatin gene (MSTN), which is involved in the formation and development of muscle fibers, has been identified as a strong candidate gene for running ability [7,8]. Several mutations are known to be associated with race distance fitness [9,10] and body composition [11] in Thoroughbreds. Additional candidate genes have been found in Arabians [12] and Quarter Horses [13]. However, the sporting use of horses covers more areas than just racing performance. Good skills in jumping, dressage, Western riding, or mastering different gaits significantly expand the range of desired characteristics. Here, too, initial successes were achieved, which led, for example, to the identification of QTLs for jumping [14] or a mutation in the DMRT3 gene associated with gait [15].
However, all these genes found are mainly major genes in which individual polymorphisms in protein-coding regions have an above-average influence on the traits of interest. However, most properties are caused by complex metabolic processes in which not only numerous genes with their specific nucleotide sequences are involved, but which can also be modulated by environmental factors. Analyses using cDNA microarrays, RNA-seq, or qPCR are one way to obtain a deeper insight into the underlying biological processes and their genetic basis. By comparing gene expression in different states such as before and after specific exercises [16,17] and in different muscles [18,19], blood vs. muscle [20], or in various breeds [21,22] and hybrids [23], it may be possible to identify the genes involved in the biological context. Further investigations, in which analysis of eQTLs [24], noncoding RNAs [25,26], or changes in the methylation pattern [27,28,29] are in the foreground, deal with possible causes of expression changes, try to uncover alternative splice variants [17,18], selective isoform expression, or protein-/expression-changing SNPs in the genes involved [30,31,32], or focus on the combination of several omics levels [33,34,35] to better understand the relationships between genotype and phenotype as well as the influence of the environment. The final result is to determine the genetic influences on diseases and, conversely, the genetic background for good health, welfare, and performance [36,37,38].
Endurance rides have become increasingly popular in recent years, both for competition and for leisure. Endurance, which is defined as the ability to perform over a longer period of time, plays a major role here. Therefore, this criterion is not only of interest for endurance rides, but for almost all types of use of the horse. However, there is only insufficient knowledge about the biochemical–physiological basis of this characteristic. In addition, it is still unclear how it can be quantified at all, since it is extremely complex and involves the interaction of numerous factors. Studies in this area have shown repeatedly how closely intertwined the various metabolic pathways are, on the one hand, and how environmental factors can have a modeling effect on expression, on the other hand. Bou et al. [39] showed that endurance training leads to alternative splicing and exon-skipping events as well as differentially expressed noncoding RNAs. The modulation of noncoding RNAs, which ultimately causes altered post-transcriptional regulation of gene expression and, thus, metabolic as well as immune adaptation, in response to exercise were shown in various studies [25]. As circulating miRNAs, they correlate with different reactions to endurance stress and are therefore considered to be more accurate biomarkers than serum markers [26,40].
A particularly important characteristic for good endurance horses is the moderate handling of stress. Stress can have an impact on different levels. Important types of stress include oxidative, heat, hypoxic, hormonal, and glucose stress [41]. In addition to adrenal gland hormones, immunological markers react to stress, meaning that their level can be seen as a marker for stress. This has been demonstrated, for example, for various interleukins and TNF-α both at the mRNA expression level [42,43] and at the protein level (serum concentration) [44,45,46]. These studies are important because precise knowledge of stress indicators, such as cytokines, offers an opportunity to correctly assess the training level of horses and thus help not to overload horses in sporting competitions. This addresses welfare issues that play a major role in dealing with horses today.
In our study, RNA sequencing data from seven Kabardian horses, generated before and after runs over two different distances (70 km and 15 km), are used to identify genes involved in endurance and gain an insight into the underlying physiological processes of sustained work. We pursued two goals: (i) Search for genes associated with stress-coping in order to find indicators to monitor health and the level of effort; (ii) Identification of candidate genes that are directly related to performance and can therefore be of interest for breeding and selection of endurance horses.
2. Material and Methods
2.1. Animals, Endurance Ride Conditions, and Ethics Statement
We studied seven adult Kabardian horses (Equus caballus) in endurance riding at two different locations. Three animals participated in an endurance ride over a 70 km distance in the Caucasus (Russia). After 35 km, there was a one-hour break and a vet check (long-distance group). The other four animals took part in a ride in the Bavarian Forest (Germany) covering a 15 km distance without break (short-distance group). The terms long- and short-distance do not refer to the categories used in endurance races, where only runs of 100 km or more are considered long-distance runs and runs of 15 km are not considered endurance rides. We use them to make a brief and clear distinction between our two distance groups. In the Caucasus, the route was in the first stage at a height of 630 to 750 m above sea level with an average speed of 14 km/h and in the second stage at 630 m to 1200 m above sea level with an average of 10 km/h. In the Bavarian Forest, the height was between 450 m and 600 m above sea level with an average speed of 12 km/h. Temperature and humidity were moderate. The endurance rides were practice rides. The field and forest paths were easy to ride on. All horses were well trained and had already completed such distance rides. All experimental procedures used in this study were performed in accordance with the animal welfare laws of both countries and were approved by the respective animal welfare organizations (Russia: No. 1571, Germany: 55.2-1-54-2532-128-1514). The horses were monitored by veterinarians during the rides.
2.2. Sample Collection and RNA Sequencing
A total of 10 mL of EDTA blood was taken from the jugular vein of each horse shortly before, T1 (15 min), and immediately after the end, T2, of the endurance ride. The total RNA from this blood was immediately secured using the LeukoLOOK-KIT Total RNA System from Ambion by Life Technologies (Carlsbad, CA, USA). With this kit, cellular fractionation of the whole blood was carried out immediately after blood collection. The leukocyte population obtained was transported in a stabilizing RNAlater for further processing in the laboratory. After extraction of total RNA according to the manufacturer’s instructions, which also significantly reduced the proportion of globin mRNA (negative effect on downstream expression analyses), the RNA quality was checked using a BioAnalyzer 2100 (Agilent, Santa Clara, CA, USA) with an RNA chip, and storage took place at −80 degrees Celsius until sequencing.
The total RNA sequencing, including the library preparation, was carried out by LGC Genomics (Berlin, Germany) following Illumina’s specifications. The NuGEN Ovation RNA-seq library preparation system (Illumina Inc., San Diego, CA, USA) was used to create the mRNA libraries. The 75 bp paired-end reads were generated with the Illumina NextSeq 500 V2 system (Illumina Inc., San Diego, CA, USA) for all seven animals. Altogether, 28 libraries (7 animals × 2 time points (before and after the run) × 2 directions (forward and reverse)) of RNA sequences comprising 582 million reads were generated. The number of reads in a library ranged from 11.4 million to 28.8 million, with a mean of 18.8 million reads.
2.3. Data Processing
We downloaded the horse reference genome Equus caballus version EquCab3.0 (file: Equus_caballus.EquCab3.0.DNA.toplevel.fa.gz) and the gene annotation file (Equus_caballus.EquCab3.0.108.gtf.gz) from the Ensembl ftp-server https://www.ensembl.org/info/data/ftp/ndex.html (accessed on 7 November 2022). The adapter clipped short reads were mapped to the reference genome using STAR 2.7.4a [47], which yielded alignment files (so-called bam files). From these alignments, we extracted the gene counts using htseq-count [48]. The R programming environment was used for general data handling and standard analysis. The Venn diagram was produced with the online tool available under https://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 14 November 2022).
2.4. Analysis of Differentially Expressed Genes
For the identification of differentially expressed genes, the two groups of animals were considered separately: the three animals (n = 3) that completed the long distance (70 km) and the four animals (n = 4) that completed the short distance (15 km). We term these groups as the following: “long-distance” and “short-distance” groups.
Differentially expressed genes (DEGs) were identified with the R-package DESeq2 [49] based upon unnormalized count data. The function DESeq of that package is used to calculate the dual logarithm of the fold changes (log2FC). Adjusted p-values and false discovery rates (FDR) were calculated with the R-function p.adjust. Genes with an absolute value of the log fold change > 0.58 (i.e., a fold change of over 1.5 or under 0.67), and an adjusted p-value < 0.05 were considered as differentially expressed, as was performed in other studies [16,50,51]. Hence, we speak of (statistically significant) upregulation of a gene if its expression was, on average, at least 50% increased after the run (T2, also termed “post-run”) with respect to its expression before the run (T1, also termed “pre-run”). Likewise, we speak of downregulation of a gene if its expression was, on average, at least 33% decreased after the run with respect to its expression before the run. To easily distinguish up- and downregulated genes, we use, in all tables, a minus sign to indicate downregulated genes while upregulated genes are shown as a positive number, which is in line with the nomenclature used in [52,53].
2.5. Functional Enrichment Analysis of Differentially Expressed Genes
Functional enrichment analysis of the identified DEGs was performed with g:Profiler https://biit.cs.ut.ee/gprofiler (accessed 30 August 2023) [54], using the GOSt functional profiling to obtain the gene ontology (GO) terms for biological processes (BPs) version e109_eg56_p17_1d3191d (accessed on 30 August 2023) and for cellular component (CC) and molecular function (MF), as well as KEGG version e110_eg57_p18_4b54a898 (accessed on 28 September 2023). We use a g:SCS threshold of 0.05.
3. Results and Discussion
3.1. Differentially Expressed Genes (DEGs)
In the long-distance group, we found 2175 DEGs (1484 upregulated and 691 downregulated). In the short-distance group, there were only 21 DEGs (13 upregulated and 8 downregulated). The complete lists of all found DEGs in both long- and short-distance runs with gene names, fold change (FC), and false discovery rates (FDR) are given in Tables S1 and S2. The number of up- and downregulated genes that overlap between both the long- and short-distance groups can be seen in the Venn diagram (Figure 1; see also Table 1). In particular, 16 of the 21 DEGs of the short-distance group overlap with the DEGs of the long-distance group, while five apply only to the short-distance DEGs (three upregulated, two downregulated).
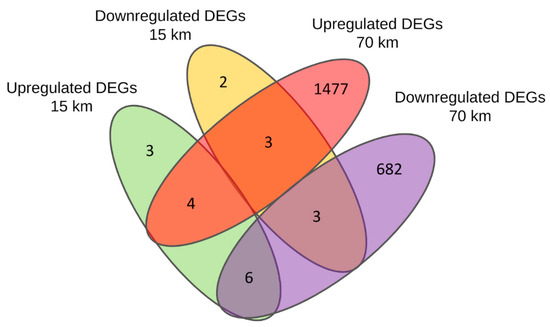
Figure 1.
Venn diagram of the number of differentially expressed genes (DEGs) obtained for the horses pre-run and post-run that covered distances of 70 km (long-distance) and 15 km (short-distance), respectively. The diagram shows the number of upregulated and downregulated DEGs which are common or specific to the two distance groups. The 16 DEGs overlapping between long- and short-distance groups are listed in Table 1.

Table 1.
DEGs which were found in both long- and short-distance runs.
Since there is still a lot of nonannotated (NA) sequence information in horses, which cannot be used for the evaluation of the activated metabolic pathways, the further analyses were only carried out with the annotated genes. This means that 1275 upregulated and 580 downregulated DEGs are available for the long-distance as well as 13 up- and 5 downregulated DEGs for the short-distance group.
The large difference in the numbers of DEGs in the two distance groups is striking. The remarkably smaller number of DEGs in the short-distance group may be due to the fact that the horses were well trained and generally not ridden at maximum speed. This possibly means that the differences in gene expression between pre-run and post-run are only relatively small, because the metabolic processes have not changed much and, consequently, the individual variation between the few animals in a distance group is too large to find more significant changes. Despite the clear methodological differences in the various studies, a certain trend can be observed overall. There are usually more upregulated than downregulated genes, while the total number of DEGs is greater after heavy physical activity than after light physical activity [51]. For example, Mach et al. [25] found 2453 DEGs after a 160 km endurance ride, while our own results show 2175 DEGs after 70 km and 21 after 15 km. In contrast, after a 30 min trot, only 142 DEGs were detected in blood [17].
3.2. Gene Ontology Enrichment and Pathway Analysis of DEGs
The GO analysis revealed several biological processes that are associated with the endurance of the horses in long- and short-distance runs. The varying distance covered by the horses in the two runs, and thus the different endurance requirements, can give first insight into the active biological processes.
Long-distance group: Functional enrichment analysis of the 1275 upregulated DEGs representing known genes in the long-distance group shows that they are involved in 343 biological processes. In order to have a better chance of finding really important genes and metabolic processes, we focused on the processes in which the genes found account for at least 10% of the total number of genes (intersection percentage) and in which no more than 250 genes are involved in total. This constraint reduced the number of biological processes to 109, involving 457 upregulated genes (Table S3). A reduction from 43 to 15 was achieved in the cellular components (CCs) and from 40 to 10 in the molecular functions (MFs). Figure 2 show the top-10 GO terms (sorted after intersection percentage) of BPs, CCs, and MFs, respectively.
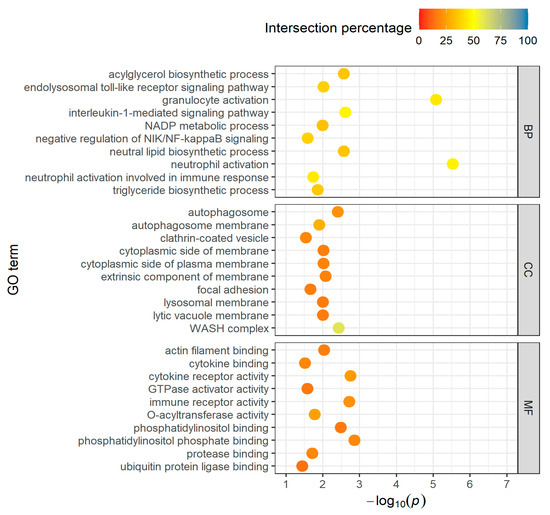
Figure 2.
Top-10 GO terms of BPs, CCs and MFs for upregulated DEGs in the long-distance group, taking into account the proportion of found genes which are overlapping with the total number of genes in the GO term (intersection percentage) and the negative log10 of the adjusted p-value.
Among the top-10 biological processes (BPs), the focus is on fatty acid metabolism (acylglycerol, neutral lipid, and triglyceride biosynthetic process) and immune response (neutrophil and granulocyte activation, toll-like receptor signaling pathway). The cellular components (CCs) primarily list reactions that influence membranes, and the molecular functions (MFs) refer to enzyme reactions that are particularly related to the biological processes (O-acyltransferase and cytokine activity) mentioned but also to cell proliferation and cell membrane actions (phosphatidylinositol binding).
In the downregulated DEGs of the long-distance group, the number of biological processes decreased from 209 to 104, of cellular component terms from 16 to 6, as well as of the molecular functions from 8 to 6. That results in a decrease of involved genes from 580 to 111 (Table S4). The top-10 GO BPs terms of this group are shown in Figure 3. Only six terms are available for CCs and MFs.
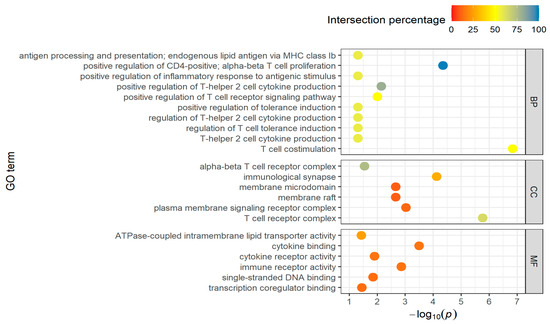
Figure 3.
Top-10 GO terms of BPs, CCs, and MFs for downregulated DEGs in the long-distance group, taking into account the proportion of found genes that are overlapping with the total number of genes in the GO term (intersection percentage) and the negative log10 of the adjusted p-value.
The identified processes are often involved in regulatory tasks and immune and stress response (T cell activation, interleukin production, leukocyte proliferation), as well as in metabolic processes (carbohydrate catabolic process, lipid biosynthesis, NADP metabolic process). The T cell receptor complex is also strongly represented in the GO:CC terms and the cytokine activity can be found under the molecular functions.
Among the top-10 upregulated genes (sorted according to FC, Table 2) involved in the important biological processes and found to be differentially expressed in our study, six genes are active in the inflammatory and the immune system (IL22RA2, IL20RA, IL1R2, LFT, SPI2) as well as in apoptotic processes (CLU). Four genes are involved in energy supply: the STBD1 protein, which is important as a cargo receptor for the transport of glycogen to the lysosomes; the GYG1 gene, which encodes a glycosyltransferase involved in the glycogen metabolism in muscle; MOGAT1, which catalyzes the diacylglycerol synthesis; and VDR, which regulates a variety of metabolic pathways. The downregulated genes, often transcription factors, are mainly involved in immune processes. In addition to the interleukin IL5RA, the system of T and B cell differentiation and proliferation is particularly represented (CD40LG, EPHB6, UBASH3A, HES1, FCER2, TRAT1, FOXP3).

Table 2.
Top-10 up- as well as downregulated DEGs (sorted according to FC) that are involved in the selected GO terms (after filtering according to intersection percentage).
In numerous studies, an increased expression of various interleukins as well as an activation of T-cell-associated processes could be observed [55,56,57]. While IL1R2 and IL22RA2 were differentially expressed in accordance with other studies [53,58], IL6 and IL8 were not identified as DEGs in our study [53,58,59,60]. The strong activation of regulatory processes and immune response-related genes seems to play crucial roles in the endurance of horses [51,58,61].
The metabolic active gene GYG1 was also found in earlier studies [16,17]. In addition to MOGAT1, some more genes from the lipid metabolism, such as PLIN5 (FC = 5.66) or SIK1 (FC = 3.5), were identified as DEGs, albeit not in the top-20 group.
Short-distance group: In the short-distance group, only few DEGs could be found (see Table S2). The 13 upregulated DEGs take part in such different processes that no significant ontology enrichment analysis was possible. Some genes could only be assigned to interesting biological processes. For example, FOS, FOSB, and PRF1 play a role in “cellular response to calcium ion” (GO:0071277). CCL5, FASLG, and TRAT1 are involved in the “positive regulation of phosphatidylserine exposure on apoptotic cell surface” (GO:1905782). IGF2 acts in the “glucose metabolic process” (GO:0006006) while PRF1 is involved in the “Type I diabetes mellitus signaling” pathway.
The five annotated downregulated genes are integrated in 32 biological processes, no cellular component, and one molecular function term (Table S5). After filtering using the intersection percentage, eleven biological processes and one molecular function GO term with four genes each appear to be particularly relevant (Figure 4).
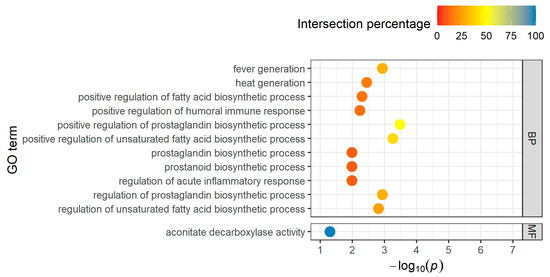
Figure 4.
Top-11 BP terms and one MF term for downregulated DEGs in the short-distance group, taking into account the proportion of found genes that overlapped with the total number of genes in the GO term (intersection percentage) and the negative log10 of the adjusted p-value.
The two main genes, IL1B and PTGS2, are important for the “regulation of prostaglandin biosynthetic process” (GO:0031392, GO:0031394) and the “regulation of fatty acid biosynthetic process” (GO:0045723, GO:2001279, GO:2001280). For the immune and inflammatory response, which also plays a role in this group, ACOD1 and NLRP3 are also involved. ACOD1 is the only gene which acts in the molecular term “aconitate decarboxylase activity”.
Despite the considerably lower number of DEGs compared to the long-distance group, regulatory processes come first. The strong involvement of the response to calcium ion is interesting and can be a reaction to oxidative stress within the blood during the endurance run. When examining Standardbred racehorses with recurrent exertional rhabdomyolysis, calcium ion regulation was significantly downregulated [37]. The activation of the Ca2+-sensitive K+ channel increases the K+ efflux and results in shrinkage of the cell and breakdown of phosphatidylserine asymmetry. It also increases the flow of Ca2+ within the cells and further makes phosphatidylserine translocate to the membrane of erythrocytes, thus triggering the death of erythrocytes [62].
This assumption is supported by the upregulation of FOS in muscle found in a test leading to maximum heartrate immediately [16]. Interestingly, the GO category “regulation of prostaglandin biosynthetic process” is relatively well represented here. Genes in this group are involved in blood flow, anti-inflammatory effect, and pain perception [63,64]. The importance of prostaglandin and proinflammatory cytokines (e.g., IL8B) in connection with endurance was shown in a stress test of Thoroughbred horses, where, among other things, a significant increase in prostaglandin plasma concentration was found [60].
Pathway analysis: Using KEGG analysis, we obtained 81 significantly overrepresented pathways in the long-distance group. Of these, 48 pathways are of particular interest because 8 are primarily associated with cancer and 25 with infectious diseases (Table S6). The top-20 (sorted according to intersection percentage) are shown in Figure 5.
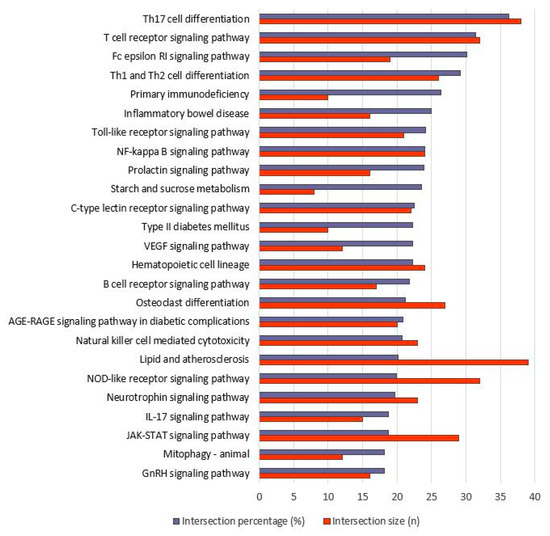
Figure 5.
Top-20 KEGG pathways of up- as well as downregulated genes after long-distance run (sorted after intersection percentage). KEGG categories are shown along the Y-axis, while both intersection percentage (proportion of intersection size to term size in %) and intersection size (number of genes that overlapped between KEGG category genes and found DEGs) are shown along the X-axis.
Upon closer inspection, it can be seen that pathways that are integrated into both the immune response and the anti-inflammatory system are significantly overrepresented. This group includes T, T helper, and B cell cascades which are playing an important role in the adaptive immune system. A total of 38 of the genes we found are integrated into the “Th17 cell differentiation” cascade and 32 genes are involved in the “T cell receptor signaling pathway”, each of which accounts for around a third of all genes contained in this pathway. Another pathway is the “Fc epsilon RI signaling” cascade, which regulates the release of inflammatory mediators such as cytokines, interleukins, and prostaglandins. The importance of the genes in this complex in coping with stress when faced with high demands has already been demonstrated in various studies both at the molecular and protein levels. For example, well-trained horses showed an anti-inflammatory reaction after an 800 m race, while untrained horses showed a proinflammatory reaction, which is considered a training effect [42,65]. After an appropriate period of rest, these changes appear to return to normal, at least partially [44]. IL1RA, which correlates with cortisol concentration, appears to be of particular importance in restoring the homeostatic equilibrium [45]. In our studies, we also found some of these genes, for example, CD4 (FC = −1.74) or FOXP3 (FC = −2.44), to be differentially expressed as well as the overrepresented “IL-17 pathway”.
The top-25 pathways also include the “toll-like receptor (TLR) signaling” and the “NF-kappa B signaling” cascade. TLRs induce kinases and pathways including IL6 (chemokine), which results in the expression of inflammation- and immune-response-related genes [58]. In our study, the MYD88 gene was significantly upregulated (FC = 2.15). This gene encodes an adaptor protein which acts as a mediator of the NF-kappa B signaling starting from toll-like receptors until the nucleus, finally resulting in the expression of interferon, and inflammation-related cytokine genes [66]. As a consequence, inflammation occurs as a stress response, which in turn could be activating pathways for maintaining homeostasis. This could be the major reason for the overrepresentation of several immune-response-related pathways, especially in the long-distance group. Platelet activation and aggregation could be occurring as a result of the activation of inflammation-related pathways, particularly for tissue repair following physiological adaptation and homeostasis [67]. The “hematopoietic cell lineage pathway”, in which 24 of our DEGs are involved, also works in this direction. “TLR signaling” and “NF-kappa B signaling” as well as “NOD-like receptor signaling” (cooperating with TLR in inflammatory and apoptotic response) and “JAK-STAT signaling” (playing a crucial role in myogenesis) pathways were found as overrepresented and to be involved in stress response after exercises in various studies [68,69,70].
For the downregulated genes of the short-distance group, only five pathways are overrepresented. Three are connected with “C-type lectin receptor signaling”, “IL-17 signaling”, and “cytosolic DNA-sensing” pathways, while the first two are also found in the top-25 pathways of the long-distance group. Two further pathways are strongly associated with infectious diseases (Table S7).
The significantly changed expression in several genes and the reflection in the pathway activation reflects the metabolic changes of an organism as a reaction to the demands of an endurance run. Some genes are characterized by a high reproducibility of both their induced expression and changed serum level of the proteins. This group includes the interleukins IL1B, IL1R2, and IL22RA2, whose genes were also expressed differentially in our study, as well as IL6, IL8, IL13, and IL17, which were found in other expression or enzyme studies [42,43,44,45,46,55,56,60,65,71]. Various studies on other serum values are interesting in this context. While, previously, the lactate concentration [72,73] was primarily used as a parameter for the degree of exhaustion, today, hormones such as testosterone and cortisol are also used for evaluation in addition to cytokines. An anabolic index determined from the ratio of these hormones is a possible parameter but is more suitable for racing horses than for endurance horses [74]. Differences in expression of these genes could therefore possibly indicate individually varying adaptability and could thus be markers for the monitoring to avoid training overload as well as for identifying horses with higher stress resistance [68]. Although there are already numerous studies in this area that determine gene expression or enzyme serum levels, there is still a lack of data and reference values, especially in the combination of such datasets. In addition to the individual variation in these values, this is primarily due to the extremely different test conditions (trained vs. untrained horses, training effect over a certain period of time, short races vs. long and very long endurance rides, different breeds, and sick vs. healthy horses [21,40,43,46,75]. In the future, determining the expression of these genes or the concentration of their proteins in the serum could provide information about the adaptation of individuals to the requirements of performance challenges and thus be markers for stress management. This would enable the training and sporting use of horses to be optimized as well as monitored and overtraining syndrome (OTS) to be avoided [40,51,57,76,77]. This would be a big step towards improving welfare. Whether the horse can be a model for humans in the field of endurance [46,65] should be discussed, as the horse’s physiology shows considerable differences in how it copes with heavy muscle strain and oxygen supply.
3.3. Search for Candidate Genes for Endurance Performance
The analyses of our data so far show that genes and biological processes that are associated with cellular regulation, apoptosis, or immune, as well as inflammatory, response are subject to strong activation, regardless of the specific exercise type, playing an important role in coping with stress. In addition to this group, genes are also of interest, which can be candidates for performance selection for endurance based on their function in the metabolism. Therefore, we focused our attention on 16 DEGs which are represented in both distance groups (see Table 1). Two of these (upregulated in long-distance and downregulated in short-distance runs), which have no annotation will not be considered in detail.
An example of the changes in expression of three genes in terms of transcripts per million (TPM) before and after the run is given in Figure 6. The expression changes in all genes in both groups (long- and short-distance) are given in Figure S1.
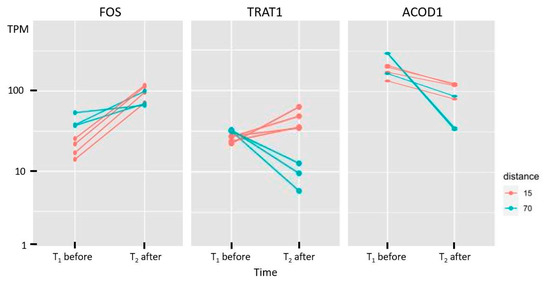
Figure 6.
Comparison of the expression level (in transcripts per million, TPM) before and after the run in three DEGs that are congruent (FOS: up–up, ACOD1: down–down) or opposite (TRAT1: down–up) in both distance groups (long: 70 km, short: 15 km).
However, a look at the function of these genes shows that most of them are also associated with inflammatory processes and stress response. The already-mentioned genes FOS and FOSB, upregulated for long- and short-distance runs, are AP-1 transcription factor subunits which play a major role in regulating the osteoblast differentiation at the early stages and are known for their role in phospholipid synthesis [78]. An upregulation of FOS was also found in previous analyses in both muscle and blood [16,17]. This could be the reason for the crucial role of FOS in adaptation of the animal after an endurance run. Both genes are also involved in processes of lipodystrophy. But in context with endurance, their function in cell death activation of T cells by binding on the FASLG promotor, IL-17 pathway, and apoptosis cannot be underestimated and may be the main reason for their differential expression. LMNA, which is extensively involved in the maintenance of skeletal muscle strength and osteoblastogenesis, as well as in apoptosis, and NLRP12, acting as regulator of inflammatory response, are also upregulated genes for both distances. An interesting group includes genes with opposite changes in the two distance groups. NLRP3, a paralog of NLRP12, is one of the genes for which expression levels increased in animals that performed the long-distance run, whereas they decreased in animals during a short-distance run. It plays a major role in the formation of the cytosolic multiprotein complex called NLRP3 inflammasome, and is therefore involved in the NF-kappa B signaling and the regulation of innate immune responses, cell death, and inflammation [71,79,80]. This group also comprises the FASLG gene, a member of the tumor necrosis factor superfamily which is important for the regulation of the immune system [81], TRAT1, part of the T cell receptor complex, and the chemokine gene CCL5, which has been reported as a candidate gene in previous studies [51,53]. The opposite direction of regulation, also seen in ADGRG1 (inhibitor of tumor progression) and RETREG1 (acting in neuro survival), can point to a lack of available energy in the long-distance run when all nonessential biological processes are shut down as much as possible. It is remarkable to note the downregulation in the long-distance group of PSD3, which reduces the intracellular lipid content of the liver [82], and which is therefore integrated into the process of energy supply. The genes ACOD1, IFIT2, and PTGS2, which are downregulated in both distances, are involved in processes of apoptosis and in the inflammatory system.
In the search for candidate genes, Park et al. [17] compared their expression profiles with known racing-performance-related genes [83] and found, among other things, HIF1A to be upregulated. This transcription factor subunit is involved in energy metabolism, which is increasing oxygen delivery and metabolic adaptation to hypoxia. In our analysis, HIF1A is not in the top 10, but is also significantly upregulated (FC = 2.33). This is also in agreement with the results of a study in Thoroughbred horses [84]. Furthermore, HIF1A can stimulate promotor elements of PFKFB3 [85], a regulatory molecule that controls glycolysis, which is upregulated (FC = 9.14) in our study but also in other studies [16]. Changes in the glucose as well as the insulin pathway were repeatedly pointed out [26,29], which we can confirm based on specific DEGs (MOGAT1: FC = 16.01; GYG1: FC = 12.45) and pathways (“Type II diabetes mellitus”) that we have identified.
The DEG PPARD involved in the “PPAR signaling pathway” and found as a significant DEG in many studies [76,86,87] did not show significantly changed expression in our analysis. Only PPARG was differentially expressed. Of the other genes influencing potential racing performance [83], GYG1 and VDR, which is a transcription factor and mainly involved in mineral metabolism, were upregulated. In blood, the transcription factor GATA2, which is important for development and proliferation of hematopoietic stem cells, was downregulated (FC = −5.49), which was also reported in previous studies [17,53]. Above all, the provision of energy is important for a horse’s performance. Of course, it must be taken into account that when switching from short-term exercise to endurance exercise, the horse also switches from the anaerobic to the aerobic metabolism [34]. For humans, there is already a review that includes DNA polymorphisms associated with athletic performance. There are already 41 endurance-related, 45 power-related, and 42 strength-related genetic biomarkers [88]. Such a gene and SNP collection could be a model for research on horses.
In order to achieve an even better understanding about the importance of the various DEGs for endurance performance and to foster a more targeted selection of the genes to be examined in more detail, the results of expression studies should be combined with GWAS analyses [89], investigations of transcription regulation (e.g., miRNAs or methylation analysis [25,29,40]), and SNP identification. Last, but not least, these findings should be compared with the response in the blood and muscles [90].
4. Conclusions
In our study, mainly DEGs from the area of general regulation and both the immune and stress response were found after endurance runs in horses. These genes show a fast response in blood and could therefore be candidates for quick and easy monitoring of stress of horses in training as well as in competition, improving animal welfare and avoiding the overtraining syndrome and overload in runs. On the other hand, there is interest in genes that could be responsible for individual endurance disposition. It would be promising to look for mutations in the genes themselves as well as in their regulatory regions. These specific candidate genes might serve as potential markers for selection in future breeding programs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14111982/s1, Figure S1: Gene expression in terms of transcripts per million (TPM) of genes which are significantly differentially expressed in both short- and long-distance runs; Table S1: List of all DEGs for the long-distance run with FC ≥ 1.5 as well as FC ≤ −1.5; Table S2: List of all DEGs for the short-distance run with FC ≥ 1.5 as well as FC ≤ −1.5; Table S3: Gene ontology (GO) enrichment analysis for upregulated genes involved in the long-distance run; Table S4: Gene ontology (GO) enrichment analysis for downregulated genes involved in the long-distance run; Table S5: Gene ontology (GO) enrichment analysis for downregulated genes involved in the short-distance run; Table S6: KEGG pathway analysis for up- and downregulated genes found in the long-distance run; Table S7: KEGG pathway analysis for downregulated genes found in the short-distance run.
Author Contributions
M.R. and Z.A.K. planned the project and organized the endurance runs; M.R. performed the laboratory work; A.O.S. designed and supervised the bioinformatics analyses; A.R. participated in the design of the study and conducted analyses together with A.O.S.; A.R. and M.R. performed the literature survey and interpreted the results; A.R., M.R. and A.O.S. wrote the final version of the manuscript; M.R. and A.O.S. conceived and managed the project. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Volkswagen Stiftung, Germany (Az.: 85 858).
Institutional Review Board Statement
All experimental procedures used in this study were carried out in accordance with the Animal Protection Laws in Russia and Germany, respectively, and the animal study protocol was approved by the respective animal welfare organizations (Russia: No. 1571, Germany: 55.2-1-54-2532-128-1514).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data will be made available after acceptance of the publication.
Acknowledgments
We acknowledge support by the Open Access Publication Funds of the Göttingen University. We also thank all Kabardian horse breeders for their support, especially the heads of the breeding associations in Russia and Germany, Khazhismel K. Amshokov and Tobias Knoll, respectively. The authors would also like to thank Paula Korkuc for her support in developing the figures.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Metallinos, D.L.; Bowling, A.T.; Rine, J. A missense mutation in the endothelin-B receptor gene is associated with Lethal White Foal Syndrome: An equine version of Hirschsprung disease. Mamm. Genome 1998, 9, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Marklund, L.; Moller, M.J.; Sandberg, K.; Andersson, L. A missense mutation in the gene for melanocyte-stimulating hormone receptor (MC1R) is associated with the chestnut coat color in horses. Mamm. Genome 1996, 7, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Morgenthaler, C.; Diribarne, M.; Capitan, A.; Legendre, R.; Saintilan, R.; Gilles, M.; Esquerre, D.; Juras, R.; Khanshour, A.; Schibler, L.; et al. A missense variant in the coil1A domain of the keratin 25 gene is associated with the dominant curly hair coat trait (Crd) in horse. Genet. Sel. Evol. 2017, 49, 85. [Google Scholar] [CrossRef]
- Littiere, T.O.; Castro, G.H.F.; Rodriguez, M.; Bonafe, C.M.; Magalhaes, A.F.B.; Faleiros, R.R.; Vieira, J.I.G.; Santos, C.G.; Verardo, L.L. Identification and Functional Annotation of Genes Related to Horses’ Performance: From GWAS to Post-GWAS. Animals 2020, 10, 1173. [Google Scholar] [CrossRef]
- Petersen, J.L.; Mickelson, J.R.; Rendahl, A.K.; Valberg, S.J.; Andersson, L.S.; Axelsson, J.; Bailey, E.; Bannasch, D.; Binns, M.M.; Borges, A.S.; et al. Genome-wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genet. 2013, 9, e1003211. [Google Scholar] [CrossRef]
- Staiger, E.A.; Albright, J.D.; Brooks, S.A. Genome-wide association mapping of heritable temperament variation in the Tennessee Walking Horse. Genes. Brain Behav. 2016, 15, 514–526. [Google Scholar] [CrossRef] [PubMed]
- Hill, E.W.; McGivney, B.A.; Gu, J.; Whiston, R.; Machugh, D.E. A genome-wide SNP-association study confirms a sequence variant (g.66493737C>T) in the equine myostatin (MSTN) gene as the most powerful predictor of optimum racing distance for Thoroughbred racehorses. BMC Genom. 2010, 11, 552. [Google Scholar] [CrossRef] [PubMed]
- Tozaki, T.; Miyake, T.; Kakoi, H.; Gawahara, H.; Sugita, S.; Hasegawa, T.; Ishida, N.; Hirota, K.; Nakano, Y. A genome-wide association study for racing performances in Thoroughbreds clarifies a candidate region near the MSTN gene. Anim. Genet. 2010, 41 (Suppl. S2), 28–35. [Google Scholar] [CrossRef]
- Rooney, M.F.; Porter, R.K.; Katz, L.M.; Hill, E.W. Skeletal muscle mitochondrial bioenergetics and associations with myostatin genotypes in the Thoroughbred horse. PLoS ONE 2017, 12, e0186247. [Google Scholar] [CrossRef]
- Farries, G.; McGettigan, P.A.; Gough, K.F.; McGivney, B.A.; MacHugh, D.E.; Katz, L.M.; Hill, E.W. Genetic contributions to precocity traits in racing Thoroughbreds. Anim. Genet. 2018, 49, 193–204. [Google Scholar] [CrossRef] [PubMed]
- Tozaki, T.; Sato, F.; Hill, E.W.; Miyake, T.; Endo, Y.; Kakoi, H.; Gawahara, H.; Hirota, K.; Nakano, Y.; Nambo, Y.; et al. Sequence variants at the myostatin gene locus influence the body composition of Thoroughbred horses. J. Vet. Med. Sci. 2011, 73, 1617–1624. [Google Scholar] [CrossRef]
- Fontanel, M.; Todd, E.; Drabbe, A.; Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Mycka, G.; Velie, B.D. Variation in the SLC16A1 and the ACOX1 Genes Is Associated with Gallop Racing Performance in Arabian Horses. J. Equine Vet. Sci. 2020, 93, 103202. [Google Scholar] [CrossRef]
- Pereira, G.L.; Malheiros, J.M.; Ospina, A.M.T.; Chardulo, L.A.L.; Curi, R.A. Exome sequencing in genomic regions related to racing performance of Quarter Horses. J. Appl. Genet. 2019, 60, 79–86. [Google Scholar] [CrossRef]
- Brard, S.; Ricard, A. Genome-wide association study for jumping performances in French sport horses. Anim. Genet. 2015, 46, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Andersson, L.S.; Larhammar, M.; Memic, F.; Wootz, H.; Schwochow, D.; Rubin, C.J.; Patra, K.; Arnason, T.; Wellbring, L.; Hjalm, G.; et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 2012, 488, 642–646. [Google Scholar] [CrossRef] [PubMed]
- McGivney, B.A.; Eivers, S.S.; MacHugh, D.E.; MacLeod, J.N.; O’Gorman, G.M.; Park, S.D.; Katz, L.M.; Hill, E.W. Transcriptional adaptations following exercise in thoroughbred horse skeletal muscle highlights molecular mechanisms that lead to muscle hypertrophy. BMC Genom. 2009, 10, 638. [Google Scholar] [CrossRef] [PubMed]
- Park, K.D.; Park, J.; Ko, J.; Kim, B.C.; Kim, H.S.; Ahn, K.; Do, K.T.; Choi, H.; Kim, H.M.; Song, S.; et al. Whole transcriptome analyses of six thoroughbred horses before and after exercise using RNA-Seq. BMC Genom. 2012, 13, 473. [Google Scholar] [CrossRef]
- Bao, T.; Han, H.; Li, B.; Zhao, Y.; Bou, G.; Zhang, X.; Du, M.; Zhao, R.; Mongke, T.; Laxima; et al. The distinct transcriptomes of fast-twitch and slow-twitch muscles in Mongolian horses. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 33, 100649. [Google Scholar] [CrossRef]
- Lee, H.Y.; Kim, J.Y.; Kim, K.H.; Jeong, S.; Cho, Y.; Kim, N. Gene Expression Profile in Similar Tissues Using Transcriptome Sequencing Data of Whole-Body Horse Skeletal Muscle. Genes 2020, 11, 1359. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk-Szmukier, M.; Szmatola, T.; Latka, J.; Dlugosz, B.; Ropka-Molik, K. The Blood and Muscle Expression Pattern of the Equine TCAP Gene during the Race Track Training of Arabian Horses. Animals 2019, 9, 574. [Google Scholar] [CrossRef]
- Khummuang, S.; Lee, H.G.; Joo, S.S.; Park, J.W.; Choi, J.Y.; Oh, J.H.; Kim, K.H.; Youn, H.H.; Kim, M.; Cho, B.W. Comparison for immunophysiological responses of Jeju and Thoroughbred horses after exercise. Asian-Australas J. Anim. Sci. 2020, 33, 424–435. [Google Scholar] [CrossRef]
- Srikanth, K.; Kim, N.Y.; Park, W.; Kim, J.M.; Kim, K.D.; Lee, K.T.; Son, J.H.; Chai, H.H.; Choi, J.W.; Jang, G.W.; et al. Comprehensive genome and transcriptome analyses reveal genetic relationship, selection signature, and transcriptome landscape of small-sized Korean native Jeju horse. Sci. Rep. 2019, 9, 16672. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Nanaei, H.A.; Wei, B.; Wang, Y.; Wang, X.; Li, Z.; Dai, X.; Wang, Z.; Jiang, Y.; Shao, J. Comparative Transcriptome Profiling Analysis Uncovers Novel Heterosis-Related Candidate Genes Associated with Muscular Endurance in Mules. Animals 2020, 10, 980. [Google Scholar] [CrossRef]
- Farries, G.; Bryan, K.; McGivney, C.L.; McGettigan, P.A.; Gough, K.F.; Browne, J.A.; MacHugh, D.E.; Katz, L.M.; Hill, E.W. Expression Quantitative Trait Loci in Equine Skeletal Muscle Reveals Heritable Variation in Metabolism and the Training Responsive Transcriptome. Front. Genet. 2019, 10, 1215. [Google Scholar] [CrossRef] [PubMed]
- Mach, N.; Plancade, S.; Pacholewska, A.; Lecardonnel, J.; Riviere, J.; Moroldo, M.; Vaiman, A.; Morgenthaler, C.; Beinat, M.; Nevot, A.; et al. Integrated mRNA and miRNA expression profiling in blood reveals candidate biomarkers associated with endurance exercise in the horse. Sci. Rep. 2016, 6, 22932. [Google Scholar] [CrossRef]
- Cappelli, K.; Capomaccio, S.; Viglino, A.; Silvestrelli, M.; Beccati, F.; Moscati, L.; Chiaradia, E. Circulating miRNAs as Putative Biomarkers of Exercise Adaptation in Endurance Horses. Front. Physiol. 2018, 9, 429. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S.; Haghani, A.; Peng, S.; Hales, E.N.; Zoller, J.A.; Raj, K.; Larison, B.; Robeck, T.R.; Petersen, J.L.; Bellone, R.R.; et al. DNA methylation aging and transcriptomic studies in horses. Nat. Commun. 2022, 13, 40. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, Y.; Ma, H.; Cao, H.; Liu, W. Integrating genome-wide methylation and transcriptome-wide analyses to reveal the genetic mechanism of milk traits in Kazakh horses. Gene 2023, 856, 147143. [Google Scholar] [CrossRef]
- Gim, J.A.; Hong, C.P.; Kim, D.S.; Moon, J.W.; Choi, Y.; Eo, J.; Kwon, Y.J.; Lee, J.R.; Jung, Y.D.; Bae, J.H.; et al. Genome-wide analysis of DNA methylation before-and after exercise in the thoroughbred horse with MeDIP-Seq. Mol. Cells 2015, 38, 210–220. [Google Scholar] [CrossRef] [PubMed]
- McQueen, C.M.; Whitfield-Cargile, C.M.; Konganti, K.; Blodgett, G.P.; Dindot, S.V.; Cohen, N.D. TRPM2 SNP genotype previously associated with susceptibility to Rhodococcus equi pneumonia in Quarter Horse foals displays differential gene expression identified using RNA-Seq. BMC Genom. 2016, 17, 993. [Google Scholar] [CrossRef] [PubMed]
- Miyata, H.; Itoh, R.; Sato, F.; Takebe, N.; Hada, T.; Tozaki, T. Effect of Myostatin SNP on muscle fiber properties in male Thoroughbred horses during training period. J. Physiol. Sci. 2018, 68, 639–646. [Google Scholar] [CrossRef]
- McGivney, B.A.; Browne, J.A.; Fonseca, R.G.; Katz, L.M.; Machugh, D.E.; Whiston, R.; Hill, E.W. MSTN genotypes in Thoroughbred horses influence skeletal muscle gene expression and racetrack performance. Anim. Genet. 2012, 43, 810–812. [Google Scholar] [CrossRef]
- Mach, N.; Ramayo-Caldas, Y.; Clark, A.; Moroldo, M.; Robert, C.; Barrey, E.; Lopez, J.M.; Le Moyec, L. Understanding the response to endurance exercise using a systems biology approach: Combining blood metabolomics, transcriptomics and miRNomics in horses. BMC Genom. 2017, 18, 187. [Google Scholar] [CrossRef] [PubMed]
- Klein, D.J.; Anthony, T.G.; McKeever, K.H. Metabolomics in equine sport and exercise. J. Anim. Physiol. Anim. Nutr. 2021, 105, 140–148. [Google Scholar] [CrossRef]
- Karagianni, A.E.; Kurian, D.; Cillan-Garcia, E.; Eaton, S.L.; Wishart, T.M.; Pirie, R.S. Training associated alterations in equine respiratory immunity using a multiomics comparative approach. Sci. Rep. 2022, 12, 427. [Google Scholar] [CrossRef] [PubMed]
- Williams, Z.J.; Velez-Irizarry, D.; Gardner, K.; Valberg, S.J. Integrated proteomic and transcriptomic profiling identifies aberrant gene and protein expression in the sarcomere, mitochondrial complex I, and the extracellular matrix in Warmblood horses with myofibrillar myopathy. BMC Genom. 2021, 22, 438. [Google Scholar] [CrossRef] [PubMed]
- Valberg, S.J.; Velez-Irizarry, D.; Williams, Z.J.; Henry, M.L.; Iglewski, H.; Herrick, K.; Fenger, C. Enriched Pathways of Calcium Regulation, Cellular/Oxidative Stress, Inflammation, and Cell Proliferation Characterize Gluteal Muscle of Standardbred Horses between Episodes of Recurrent Exertional Rhabdomyolysis. Genes 2022, 13, 1853. [Google Scholar] [CrossRef]
- Foury, A.; Mach, N.; Ruet, A.; Lansade, L.; Moisan, M.P. Transcriptomic signature related to poor welfare of sport horses. Compr. Psychoneuroendocrinol. 2023, 16, 100201. [Google Scholar] [CrossRef]
- Bou, T.; Ding, W.; Ren, X.; Liu, H.; Gong, W.; Jia, Z.; Zhang, X.; Dugarjaviin, M.; Bai, D. Muscle Fibre Transition and Transcriptional Changes of Horse Skeletal Muscles during Traditional Mongolian Endurance Training. Equine Vet. J. 2023; Early View. [Google Scholar] [CrossRef]
- Cappelli, K.; Mecocci, S.; Capomaccio, S.; Beccati, F.; Palumbo, A.R.; Tognoloni, A.; Pepe, M.; Chiaradia, E. Circulating Transcriptional Profile Modulation in Response to Metabolic Unbalance Due to Long-Term Exercise in Equine Athletes: A Pilot Study. Genes 2021, 12, 1965. [Google Scholar] [CrossRef]
- Sp, N.; Kang, D.Y.; Kim, D.H.; Lee, H.G.; Park, Y.M.; Kim, I.H.; Lee, H.K.; Cho, B.W.; Jang, K.J.; Yang, Y.M. Methylsulfonylmethane inhibits cortisol-induced stress through p53-mediated SDHA/HPRT1 expression in racehorse skeletal muscle cells: A primary step against exercise stress. Exp. Ther. Med. 2020, 19, 214–222. [Google Scholar] [CrossRef]
- Witkowska-Pilaszewicz, O.; Baska, P.; Czopowicz, M.; Zmigrodzka, M.; Szarska, E.; Szczepaniak, J.; Nowak, Z.; Winnicka, A.; Cywinska, A. Anti-Inflammatory State in Arabian Horses Introduced to the Endurance Training. Animals 2019, 9, 616. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.D.; Ivester, K.M.; Stewart, C.; Page, A.E.; Horohov, D.W.; Couetil, L.L. The effect of lower airway inflammation on inflammatory cytokine gene expression in bronchoalveolar lavage fluid and whole blood in racing Thoroughbreds. Vet. Immunol. Immunopathol. 2021, 237, 110266. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.N.; Hughes, K.J.; Hall, S.; Labens, R. The effect of exercise on cytokine concentration in equine autologous conditioned serum. Equine Vet. J. 2023, 55, 551–556. [Google Scholar] [CrossRef]
- Arfuso, F.; Giudice, E.; Panzera, M.; Rizzo, M.; Fazio, F.; Piccione, G.; Giannetto, C. Interleukin-1Ra (Il-1Ra) and serum cortisol level relationship in horse as dynamic adaptive response during physical exercise. Vet. Immunol. Immunopathol. 2022, 243, 110368. [Google Scholar] [CrossRef] [PubMed]
- Plisak, U.; Szczepaniak, J.; Zmigrodzka, M.; Giercuszkiewicz-Hecold, B.; Witkowska-Pilaszewicz, O. Changes in novel anti-infalmmatory cytokine concetration in the bood of endurance and race horses at different levels of training. Comput. Struct. Biotechnol. J. 2023, 21, 418–424. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Black, M.B.; Andersen, M.E.; Pendse, S.N.; Borghoff, S.J.; Streicker, M.; McMullen, P.D. RNA-sequencing (transcriptomic) data collected in liver and lung of male and female B6C3F1 mice exposed to various dose levels of 4-methylimidazole for 2, 5, or 28 days. Data Brief. 2021, 38, 107420. [Google Scholar] [CrossRef]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Zukowski, K.; Piorkowska, K.; Gurgul, A.; Bugno-Poniewierska, M. Transcriptome profiling of Arabian horse blood during training regimens. BMC Genet. 2017, 18, 31. [Google Scholar] [CrossRef][Green Version]
- Kumar, S.; Jung, J.K.; Kim, Y. Characterization of joining sites of a viral histone H4 on host insect chromosomes. PLoS ONE 2017, 12, e0177066. [Google Scholar] [CrossRef]
- Capomaccio, S.; Vitulo, N.; Verini-Supplizi, A.; Barcaccia, G.; Albiero, A.; D’Angelo, M.; Campagna, D.; Valle, G.; Felicetti, M.; Silvestrelli, M.; et al. RNA sequencing of the exercise transcriptome in equine athletes. PLoS ONE 2013, 8, e83504. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Liburt, N.R.; Adams, A.A.; Betancourt, A.; Horohov, D.W.; McKeever, K.H. Exercise-induced increases in inflammatory cytokines in muscle and blood of horses. Equine Vet. J. Suppl. 2010, 42, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Page, A.E.; Stewart, J.C.; Fielding, C.L.; Horohov, D.W. The Effect of a 160-Kilometer Competitive Endurance Ride on Inflammatory Marker mRNA Expression in Horses. J. Equine Vet. Sci. 2019, 79, 45–49. [Google Scholar] [CrossRef]
- Wilson, J.; De Donato, M.; Appelbaum, B.; Garcia, C.T.; Peters, S. Differential Expression of Innate and Adaptive Immune Genes during Acute Physical Exercise in American Quarter Horses. Animals 2023, 13, 308. [Google Scholar] [CrossRef]
- Cappelli, K.; Felicetti, M.; Capomaccio, S.; Nocelli, C.; Silvestrelli, M.; Verini-Supplizi, A. Effect of training status on immune defence related gene expression in Thoroughbred: Are genes ready for the sprint? Vet. J. 2013, 195, 373–376. [Google Scholar] [CrossRef]
- Lee, H.G.; Choi, J.Y.; Park, J.W.; Park, T.S.; Song, K.D.; Shin, D.; Cho, B.W. Effects of exercise on myokine gene expression in horse skeletal muscles. Asian-Australas. J. Anim. Sci. 2019, 32, 350–356. [Google Scholar] [CrossRef]
- Donovan, D.C.; Jackson, C.A.; Colahan, P.T.; Norton, N.; Hurley, D.J. Exercise-induced alterations in pro-inflammatory cytokines and prostaglandin F2alpha in horses. Vet. Immunol. Immunopathol. 2007, 118, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Donovan, D.C.; Jackson, C.A.; Colahan, P.T.; Norton, N.N.; Clapper, J.L.; Moore, J.N.; Hurley, D.J. Assessment of exercise-induced alterations in neutrophil function in horses. Am. J. Vet. Res. 2007, 68, 1198–1204. [Google Scholar] [CrossRef]
- Mao, T.Y.; Fu, L.L.; Wang, J.S. Hypoxic exercise training causes erythrocyte senescence and rheological dysfunction by depressed Gardos channel activity. J. Appl. Physiol. 2011, 111, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Simon, L.S. Role and regulation of cyclooxygenase-2 during inflammation. Am. J. Med. 1999, 106, 37S–42S. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.; Xu, W. The function of the selective inhibitors of cyclooxygenase 2. Mini Rev. Med. Chem. 2006, 6, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Witkowska-Pilaszewicz, O.; Pingwara, R.; Winnicka, A. The Effect of Physical Training on Peripheral Blood Mononuclear Cell Ex Vivo Proliferation, Differentiation, Activity, and Reactive Oxygen Species Production in Racehorses. Antioxidants 2020, 9, 1155. [Google Scholar] [CrossRef]
- Mottahedin, A.; Paidikondala, M.; Cholleti, H.; Baule, C. NF-kappaB activation by equine arteritis virus is MyD88 dependent and promotes viral replication. Arch. Virol. 2013, 158, 701–705. [Google Scholar] [CrossRef] [PubMed]
- Aldemir, H.; Kilic, N. The effect of time of day and exercise on platelet functions and platelet-neutrophil aggregates in healthy male subjects. Mol. Cell Biochem. 2005, 280, 119–124. [Google Scholar] [CrossRef]
- Kim, D.H.; Lee, H.G.; Sp, N.; Kang, D.Y.; Jang, K.J.; Lee, H.K.; Cho, B.W.; Yang, Y.M. Validation of exercise-response genes in skeletal muscle cells of Thoroughbred racing horses. Asian-Australas. J. Anim. Sci. 2021, 34, 134–142. [Google Scholar] [CrossRef]
- Kim, H.; Lee, T.; Park, W.; Lee, J.W.; Kim, J.; Lee, B.Y.; Ahn, H.; Moon, S.; Cho, S.; Do, K.T.; et al. Peeling back the evolutionary layers of molecular mechanisms responsive to exercise-stress in the skeletal muscle of the racing horse. DNA Res. 2013, 20, 287–298. [Google Scholar] [CrossRef][Green Version]
- Park, J.W.; Kim, K.H.; Choi, J.K.; Park, T.S.; Song, K.D.; Cho, B.W. Regulation of toll-like receptors expression in muscle cells by exercise-induced stress. Anim. Biosci. 2021, 34, 1590–1599. [Google Scholar] [CrossRef]
- Ahn, H.; Kim, J.; Lee, H.; Lee, E.; Lee, G.S. Characterization of equine inflammasomes and their regulation. Vet. Res. Commun. 2020, 44, 51–59. [Google Scholar] [CrossRef]
- King, C.M.; Evans, D.L.; Rose, R.J. Significance for exercise capacity of some electrocardiographic findings in racehorses. Aust. Vet. J. 1994, 71, 200–202. [Google Scholar] [CrossRef] [PubMed]
- Rainger, J.E.; Evans, D.L.; Hodgson, D.R.; Rose, R.J. Distribution of lactate in plasma and erythrocytes during and after exercise in horses. Br. Vet. J. 1995, 151, 299–310. [Google Scholar] [CrossRef]
- Grzedzicka, J.; Dabrowska, I.; Malin, K.; Witkowska-Pilaszewicz, O. Exercise-related changes in the anabolic index (testosterone to cortisol ratio) and serum amyloid A concentration in endurance and racehorses at different fitness levels. Front. Vet. Sci. 2023, 10, 1148990. [Google Scholar] [CrossRef] [PubMed]
- Barrey, E.; Mucher, E.; Robert, C.; Amiot, F.; Gidrol, X. Gene expression profiling in blood cells of endurance horses completing competition or disqualified due to metabolic disorder. Equine Vet. J. Suppl. 2006, 38, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Ropka-Molik, K.; Stefaniuk-Szmukier, M.; Z˙ukowski, K.; Piórkowska, K.; Bugno-Poniewierska, M. Exercise-induced modification of the skeletal muscle transcriptome in Arabian horses. Physiol. Genom. 2017, 49, 318–326. [Google Scholar] [CrossRef]
- Miglio, A.; Falcinelli, E.; Mezzasoma, A.M.; Cappelli, K.; Mecocci, S.; Gresele, P.; Antognoni, M.T. Effect of First Long-Term Training on Whole Blood Count and Blood Clotting Parameters in Thoroughbreds. Animals 2021, 11, 447. [Google Scholar] [CrossRef]
- Bussolino, D.F.; Guido, M.E.; Gil, G.A.; Borioli, G.A.; Renner, M.L.; Grabois, V.R.; Conde, C.B.; Caputto, B.L. c-Fos associates with the endoplasmic reticulum and activates phospholipid metabolism. FASEB J. 2001, 15, 556–558. [Google Scholar] [CrossRef]
- Papin, S.; Cuenin, S.; Agostini, L.; Martinon, F.; Werner, S.; Beer, H.D.; Grutter, C.; Grutter, M.; Tschopp, J. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1beta processing. Cell Death Differ. 2007, 14, 1457–1466. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Jain, A.; Choi, S.W.; Mandell, M.A.; Schroder, K.; Johansen, T.; Deretic, V. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J. Cell Biol. 2015, 210, 973–989. [Google Scholar] [CrossRef]
- Liu, W.; Ramagopal, U.; Cheng, H.; Bonanno, J.B.; Toro, R.; Bhosle, R.; Zhan, C.; Almo, S.C. Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3. Structure 2016, 24, 2016–2023. [Google Scholar] [CrossRef]
- Mancina, R.M.; Sasidharan, K.; Lindblom, A.; Wei, Y.; Ciociola, E.; Jamialahmadi, O.; Pingitore, P.; Andreasson, A.C.; Pellegrini, G.; Baselli, G.; et al. PSD3 downregulation confers protection against fatty liver disease. Nat. Metab. 2022, 4, 60–75. [Google Scholar] [CrossRef]
- Schroder, W.; Klostermann, A.; Distl, O. Candidate genes for physical performance in the horse. Vet. J. 2011, 190, 39–48. [Google Scholar] [CrossRef]
- Ghosh, M.; Cho, H.W.; Park, J.W.; Choi, J.Y.; Chung, Y.H.; Sharma, N.; Singh, A.K.; Kim, N.E.; Mongre, R.K.; Huynh, D.; et al. Comparative Transcriptomic Analyses by RNA-seq to Elucidate Differentially Expressed Genes in the Muscle of Korean Thoroughbred Horses. Appl. Biochem. Biotechnol. 2016, 180, 588–608. [Google Scholar] [CrossRef]
- Obach, M.; Navarro-Sabate, A.; Caro, J.; Kong, X.; Duran, J.; Gomez, M.; Perales, J.C.; Ventura, F.; Rosa, J.L.; Bartrons, R. 6-Phosphofructo-2-kinase (pfkfb3) gene promoter contains hypoxia-inducible factor-1 binding sites necessary for transactivation in response to hypoxia. J. Biol. Chem. 2004, 279, 53562–53570. [Google Scholar] [CrossRef]
- Eivers, S.S.; McGivney, B.A.; Fonseca, R.G.; MacHugh, D.E.; Menson, K.; Park, S.D.; Rivero, J.L.; Taylor, C.T.; Katz, L.M.; Hill, E.W. Alterations in oxidative gene expression in equine skeletal muscle following exercise and training. Physiol. Genom. 2010, 40, 83–93. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Eivers, S.S.; McGivney, B.A.; Gu, J.; MacHugh, D.E.; Katz, L.M.; Hill, E.W. PGC-1alpha encoded by the PPARGC1A gene regulates oxidative energy metabolism in equine skeletal muscle during exercise. Anim. Genet. 2012, 43, 153–162. [Google Scholar] [CrossRef]
- Semenova, E.A.; Hall, E.C.R.; Ahmetov, I.I. Genes and Athletic Performance: The 2023 Update. Genes 2023, 14, 1235. [Google Scholar] [CrossRef] [PubMed]
- Ricard, A.; Robert, C.; Blouin, C.; Baste, F.; Torquet, G.; Morgenthaler, C.; Riviere, J.; Mach, N.; Mata, X.; Schibler, L.; et al. Endurance Exercise Ability in the Horse: A Trait with Complex Polygenic Determinism. Front. Genet. 2017, 8, 89. [Google Scholar] [CrossRef] [PubMed]
- Le Moyec, L.; Robert, C.; Triba, M.N.; Billat, V.L.; Mata, X.; Schibler, L.; Barrey, E. Protein catabolism and high lipid metabolism associated with long-distance exercise are revealed by plasma NMR metabolomics in endurance horses. PLoS ONE 2014, 9, e90730. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).