Abstract
Successful use of woody species in reducing climatic and environmental risks of energy shortage and spreading pollution requires deeper understanding of the physiological functions controlling biomass productivity and phytoremediation efficiency. Targets in the breeding of energy willow include the size and the functionality of the root system. For the combination of polyploidy and heterosis, we have generated triploid hybrids (THs) of energy willow by crossing autotetraploid willow plants with leading cultivars (Tordis and Inger). These novel Salix genotypes (TH3/12, TH17/17, TH21/2) have provided a unique experimental material for characterization of Mid-Parent Heterosis (MPH) in various root traits. Using a root phenotyping platform, we detected heterosis (TH3/12: MPH 43.99%; TH21/2: MPH 26.93%) in the size of the root system in soil. Triploid heterosis was also recorded in the fresh root weights, but it was less pronounced (MPH%: 9.63–19.31). In agreement with root growth characteristics in soil, the TH3/12 hybrids showed considerable heterosis (MPH: 70.08%) under in vitro conditions. Confocal microscopy-based imaging and quantitative analysis of root parenchyma cells at the division–elongation transition zone showed increased average cell diameter as a sign of cellular heterosis in plants from TH17/17 and TH21/2 triploid lines. Analysis of the hormonal background revealed that the auxin level was seven times higher than the total cytokinin contents in root tips of parental Tordis plants. In triploid hybrids, the auxin–cytokinin ratios were considerably reduced in TH3/12 and TH17/17 roots. In particular, the contents of cytokinin precursor, such as isopentenyl adenosine monophosphate, were elevated in all three triploid hybrids. Heterosis was also recorded in the amounts of active gibberellin precursor, GA19, in roots of TH3/12 plants. The presented experimental findings highlight the physiological basics of triploid heterosis in energy willow roots.
1. Introduction
Short rotation plantations of the fast-growing trees, such as energy willow (Salix spp.), can play a key role in combating climate change and improving energy security. Maximizing the biomass yield requires integrated contributions from willow breeding and agrotechnology, which are both supported by plant research. Shrub willows grown as a woody crop have outstanding potential to serve as an optimal feedstock to produce bioenergy, biofuels, and bioproducts with environmental and rural development benefits [1]. As wood pellets, it can be used in gasifier to produce syngas [2,3]; alternatively, it can serve as raw material for cellulosic ethanol production [4,5]. In addition, the energy-willow-based pyrolysis system for biomethane production led to increased energy performance and negative global warming potential [6]. Recent studies by Kakuk et al. [7] demonstrated that green willow biomass harvested as less than one-year-old shrubs could be efficient biogas substrate compared to woody tissues.
Out of serious global ecological problems, the pollution of the soil with toxic heavy metals or with organic pollutants urges application of phytoremediation as a cheap and environmentally sustainable method for land decontamination, preferentially based on woody species [8,9,10]. Use of willow genotypes capable of heavy metal and organic phytoremediation associated with biomass production gains an increasing interest world-wide [11,12]. Willow cultivation on saline-alkali soil lands is dependent on the salt tolerance of Salix species [13].
The environmental and economical sustainability of the above-listed multipurpose applications exploits some key properties of energy willow genotypes that can be generated by improved breeding technologies (see the review by Hanley and Karp, [14]). The increase in the biomass yield and energy production is a central goal [15]. Willow breeding can rely on the extended biodiversity of the genus Salix, which includes 330–500 species and more than 200 hybrids [16]. In the genus Salix, the formation of new species involved hybridization and subsequent polyploidization [17]. Crossing between Salix miyabeana as a tetraploid species native to Japan, Korea, and China and diploid S. purpurea or its hybrids resulted in triploid genotypes with higher green biomass yield than their diploid or tetraploid parental species [18]. In these studies, the highest-yielding genotype was a triploid hybrid (S. koriyanagi × S. purpurea) × S. miyabeana. These interspecific triploid hybrids displayed significant heterosis for the harvestable biomass and biomass-related growth traits in the greenhouse and in the field [19]. Cheng et al. [20] reported a large number of differentially expressed genes by comparing synthetic Populus allotriploid (2n = 3x = 57) and their diploid parents (2n = 2x = 38). These allotriploid plants exhibited higher heights.
Considering the above outlined extensive applications of energy willow plants grown as short rotation coppice for biomass production or phytoremediation, it is evident that the root system parameters can play a key role in the efficiency of these activities. Improvement of the root characteristics is a special breeding target that can be supported by phenotyping technologies (reviewed by Tracy et al. [21]). In only a few cases, heterosis was recorded in root growth characteristics. Hund et al. [22] reported the expression of marginal overdominance with respect to different traits of root system growth in maize. The relative Mid-Parent Heterosis for maize root traits was higher under stress generated by low phosphorus availability [23]. Natural or artificial multiplication of the plant genome size (that is, polyploidization) can also cause essential alterations in different traits of the plant anatomy and functions or even in the stress responses (see review by Ruiz et al. [24]). The interplay between ploidy and heterosis was concluded from the studies on triploid maize hybrids. Yao et al. [25] showed that the heterosis is subject to dosage effects. In contrast, the paternal genome dosage in F1 triploid hybrids of sugar beet does not enhance heterosis effects beyond what can be achieved in F1 diploid hybrids [26]. The progenies from the crossings between the diploid Camellia sinensis and tetraploid variants showed heterosis for caffeine and epigallocatechin-3-gallate contents [27].
Previously, we reported the production and the detailed characterization of a set of autotetraploid S. viminalis var. Energo genotypes (polyploid Energo [PP-E]; (2n = 4x = 76, [28]). Comparison of diploid and tetraploid variants revealed complex, multiple changes at the anatomical and morphological levels and in growth parameters of energy willow organs. Duplication of the genome size resulted in significant stimulation of root development. In parallel, we showed an improved salt tolerance of tetraploid energy willow genotypes [29]. As an extension of our energy willow breeding program, we crossed leading Swedish diploid cultivars (Tordis and Inger) with our autotetraploid genotypes. The detailed characterization of the aboveground traits clearly indicated the expression of Mid-Parent Heterosis (MPH%) in the shoot growth rate or in the CO2 assimilation rate [30]. The present paper is focused on the characterization of triploid heterosis controlling the biomass, growth rate, cellular parameters, and hormonal status of roots. The presented data provide deeper insight into how root traits can be improved through the combination of hybrid vigor with polyploidization.
2. Materials and Methods
2.1. Crossing Diploid Tordis or Inger Female Plants with Autotetraploid Male Plants and Propagation of Triploid Energy Willow Genotypes
Previously, we produced autotetraploid (2n = 4x = 76) energy willow plants and described in detail the main characteristics generated by genome duplication [28]. Parental stem cuttings were collected in wintertime and dormancy was released in water under greenhouse conditions. Male catkins from the tetraploid PP-E7 and PP-E15 plants were removed at anthesis for the collection of pollen to accomplish artificial pollination; pollen was applied on the receptive stigma of Tordis and Inger cultivars. After the isolation of 10- and 14-day-old hybrid embryos, they were cultured on the half MS hormone-free medium. The in vitro grown plants were rooted and transferred to soil and propagated by stem cuttings to be used in phenotyping studies.
2.2. Flow Cytometry
Determination of ploidy levels of obtained hybrids was conducted by flow cytometry. Root tips (approximately 5–7 mm) were excised from the plants grown in Knop’s solution: (Ca(NO3)2: 0.55 mM; KNO3: 1.90 mM; NaCl: 0.10 mM; KH2PO4: 0.40 mM; MgSO4: 0.50 mM; and FeCl3: 3.8 µM, pH of 5.5). Determination of ploidy levels was conducted by flow cytometry (BD FACSCalibur) equipped with a 532 nm green solid-state laser operating at 30 mW, as we published before [28]. The suspension of released nuclei was stained with 1 μg mL−1 propidium iodide (Sigma, Livonia, MI, USA) for 10 min. The cell cycle phase distribution was analyzed using ModFit LT™ version 3.0 (Verity Software House, Toshan, ME, USA).
2.3. Phenotyping of Triploid Heterosis in Root Growth in Soil
Single dormant stem cuttings were planted into radio-tagged Plexiglas columns (100 mm in diameter and 400 mm deep) with a mixture of 80% Terra peat soil and 20% sandy soil. Each genotype was represented by 10 replications. The Plexiglas columns were photographed from four different side positions and from the bottom [28]. The root-related white pixels were identified by subtracting the black soil background from the images. To characterize the root surface area (cm2) appearing at the surface of the chamber, the metric values of the areas of the four side view projections (90° rotation) were summarized, and the metric value of the area of the bottom view was added.
After completion of the 8-week phenotyping experiment, the rooted stems were removed from the soil, and the roots were separated from the soil. The root weights were measured immediately after removal from the soil (fresh weight).
Mid-Parent Heterosis (MPH) values were calculated as heterosis over mid-parent (MPH%) = [(F1 − MP)/MP × 100], where F1 is the numerical value trait measurement in the hybrid and MP values are the mean values of the parents (P1 + P2)/2.
2.4. Quantification of Triploid Heterosis by Analyses of Root Growth Intensity Parameters in Knop’s Solution
Stem cuttings (10 cm) of triploid hybrid and parental genotypes were placed into holes of plastic foam floating in a pool filled with Knop’s solution. The length of the main roots was measured in the most developed four root systems from each genotype represented by three cuttings. For determination of root growth intensity, we measured the length of the same main roots twice in 48 h intervals. We also estimated heterosis over the cultivar parent (CPH%) = [(F1 − CP)/BP × 100].
2.5. Root Sections, Microscopy, and Measurement of Cell Size
Stem cuttings were rooted in Knop’s-solution-saturated zeolite-containing jars. The roots were harvested when they reached more than 5 cm in length. Slices of disks were cut manually under a stereozoom microscope at the beginning of vasculature at the root tip (between the end of the division zone and the beginning of the elongation zone). The disks were mounted with 10 × 10 mm square coverslips using phosphate-buffered saline (PBS) onto standard glass microscope slides. Cell wall autofluorescence was captured using 405 nm laser excitation and a wide spectral emission detection between 415 nm and 602 nm detection of Leica SP5 AOBS confocal laser scanning microscope (Leica Microsystems GmbH, Wetzlar, Germany). A 40× dry objective (HCX PL Fluotar, N.A 0.75, Leica Microsystems CMS GmbH, Wetzlar, Germany) was used with pinhole (set to 1 Airy unit (113 µm). Bright field images were captured with a transmitted detector using the same laser line. The diameters of the root parenchyma cells were measured using FIJI software (v1.53c). Two concentric circles (0.33r and 0.83r, r = radius of the disc section) were overlaid onto the root disk image to exclude heterogeneous population of cells in the central vascular area and epidermal region. Diameters of ~50 parenchyma cells (per disk) were recorded within the area delineated by the two overlaid circles using a macro in FIJI [31]. The macro written for the FIJI program (v1.53c) was used to establish the initial settings of the image imported in Leica LIF file format. The macro commands are the following. 1. Select fluorescence channel, 2. Revert grayscale image to cyan LUT, and 3. Set the line tool for the measurement. The physical lengths of the selected lines were determined based on the scale imported from the microscope from the LIF file. The macro allowed streamlining of the measurement protocol. We measured the diameters of a total of 1649 root cells from 6 different lines. Charts were prepared after exporting of the measurements to Microsoft Excel 2010 (Microsoft, Redmond, WA, USA). Composite images were prepared using CorelDraw Graphics Suite X7 (Corel Corporation, Ottawa, ON, Canada)
2.6. Analysis of Hormone Contents in Main Root Tips
Root tissue samples (30–80 mg fresh weight) were purified and analyzed according to Dobrev and Kamínek [32] and Dobrev and Vankova [33], as described in our previous publication [28]. The root tip tissues were collected in two replications, and the mean values are provided. Briefly, samples were, after homogenization, extracted with cold (−20 °C) methanol:water:formic acid (15:4:1, v/v/v). The labelled internal standards (10 pmol per sample) were added. The phytohormones were separated with a mixed-mode reverse phase cation-exchange SPE column (Oasis-MCX; Waters, Milford, MA, USA) into the acid fraction by elution with methanol (auxins, ABA, SA, JA, GA) and into the basic fraction by elution with 0.35 M NH4OH in 60% methanol (CKs). Hormone metabolites were analyzed using HPLC (Ultimate 3000; Dionex, Sunnyvale, CA, USA) coupled to a hybrid triple quadrupole/linear ion-trap mass spectrometer (3200 Q TRAP; Applied Biosystems, Foster City, CA, USA).
2.7. Statistical Analyses
In the statistical analyses, the significance of differences among genotypes was evaluated using Student’s t-test for comparisons between the traits of diploid parents and triploid hybrids. Statistically significant events in the traits compared with the triploid hybrids’ mean values are indicated with mean labels as p ≤ 0.01 ***, p ≤ 0.05 **, and p ≤ 0.1 *.
3. Results
3.1. Generation of Triploid Hybrids by Crossing Autotetraploid and Diploid Energy Willow Plants
Cuttings of woody stems of willow genotypes were rooted in water and used for nuclei isolation from root tips. DNA ploidy levels were screened by flow cytometry for identification of the triploid lines. Typical output histograms of the flow cytometry for selected hybrids and their parents studied in the present work are shown in Figure 1. Based on these DNA content indicators, we have established a collection of triploid hybrid (TH) lines for studies on the expression of hybrid vigor in root traits.
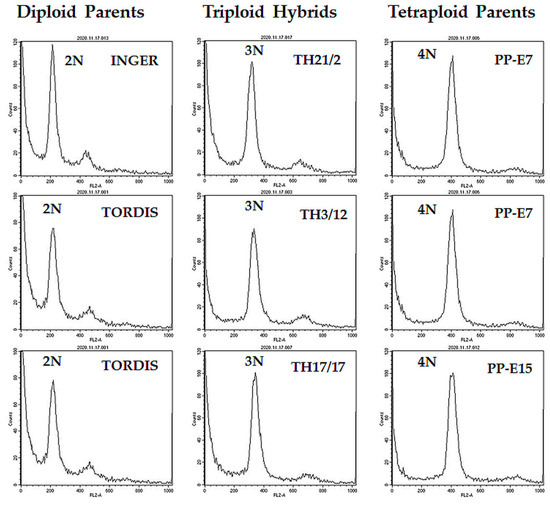
Figure 1.
Identification of triploid willow hybrids by flow cytometry analysis of relative DNA content in root tip cells (using propidium iodide staining).
3.2. Expression of Triploid Heterosis in Root Biomass as Shown by Root Phenotyping of Willow Plants Grown in Soil
Previously, we reported the use of a root phenotyping platform for characterization of the willow root system [28]. As shown in Figure 2, significant differences in root formation can be recorded between the parental plants and their triploid hybrids. The two triploid hybrids (TH3/12 and TH21/2) developed an enlarged root system in comparison to their parental plants. Differences in root formation are more recognizable on pictures from the bottom side.
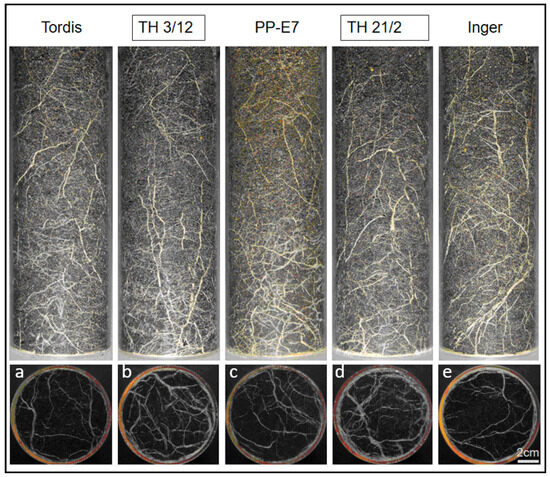
Figure 2.
Root growth comparison of hybrid and parental plants. (a–e) Side and bottom views of roots represent root systems of willow plants grown in soil in transparent wall Plexiglas columns. (b,d) The triploid hybrids (TH3/12, TH21/2) produced an enlarged root system with thicker roots relative to both of the diploid cultivars (Tordis, Inger in panels (a,e)) and the autotetraploid male parents (c). Digital images were taken after eight weeks of cultivation. Bar is 2 cm for all images.
To quantify genotype-dependent root biomass production, Table 1 presents the total surface area values (in cm2) occupied by white pixels and root wet weights. In each crossing combination, we have calculated Mid-Parent Heterosis (MPH) % values for comparison of heterosis levels.

Table 1.
Triploid hybrid willow plants show heterosis in soil-grown root biomass produced in relation to parental combinations.
In the present experimental design, we could compare heterosis effects in hybrids with different parental combinations. In the root surface area parameter, we recognized significantly different responses after crossing the Swedish Tordis male cultivar with the PP-E7 or the PP-E15 tetraploid plants. High triploid heterosis was expressed in the TH3/12 plants (female parent PP-E7), while the TH17/17 plants (female parent PP-E15) failed to exhibit a positive heterosis response. The PP-E7 plants as crossing partners could also ensure heterosis in other crossing combinations, as with the Inger plants. In the case of TH21/2 plants, the MPH% was lower, which is possibly due to the larger surface area of the root system of Inger parental plants compared to Tordis plants. After eight weeks of the growing period, the roots were separated from the soil, and the root biomasses were measured as fresh weights (Table 1). This parameter has indicated MPH heterosis in all three hybrid combinations, but with lower values as detected by the root surface area data shown by phenotyping, even in the case of the TH17/17 roots. This difference in recording by phenotyping and root weight measurement of the root biomass may originate from the limitation of digital imaging of roots grown in Plexiglas columns. Fresh weight values for the TH plants were close to tetraploid parental roots and exceeded the root formation of diploid parents. Subsequently, the MPH% values were found to be low in this trait. However, the fresh root weight of TH21/2 hybrid plants exceeded the root weights of both parental genotypes. The average increase reached 10% even when compared to the PP-E7 tetraploid plants. This difference was found to be significant.
To gain more insight into the dependence of root development on the genotype of willow plants, we have recorded the root surface area values during eight weeks of the growing period (Figure 3). The sum of the side and bottom images of the Plexiglas columns indicated root biomass differences after four weeks.
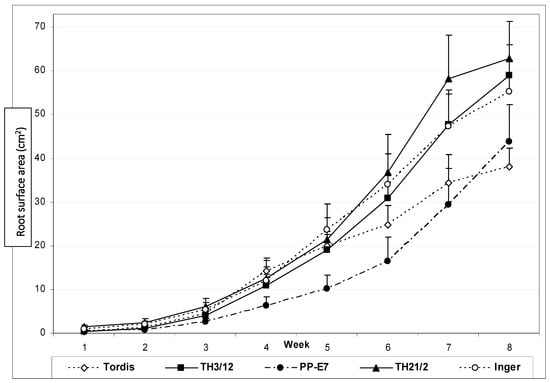
Figure 3.
Differential expression of genotype-dependent root formation potential during the growth period of eight weeks in soil. Comparison of white pixel-based root surface area monitored by digital photography to record the root biomass growth of energy willow triploid hybrid (TH) plants and tetraploid parental plants (PP-E7) in comparison with roots of the diploid commercial cultivars (Inger and Tordis).
The tetraploid PP-E7 plants showed lower root surface area during almost the whole growing period. After four weeks, the diploid plants of the Tordis cultivar also formed less roots in comparison to the triploid hybrids. The TH21/2 triploids produced the highest root surface area values, especially during the last two weeks of culture. Out of parental lines, Inger plants have good root formation potential.
3.3. Deeper Analysis of the Triploid Hybrid Vigor in Root Growth Rate In Vitro
In attempt to gain more information about the expression of triploid heterosis in root characteristics, willow stem cuttings were rooted in Knop’s solution in a thermostat room. As shown in Table 2, the rooting parameters varied significantly during the early phase of root formation. After 16 days of the growing period, the root length of parental diploid Tordis plants was longer than that of Inger plants.

Table 2.
Monitoring the expression of triploid heterosis in root length, growth intensity, and frequency of G2/M cells in Knop’s solution.
Under these experimental conditions, the tetraploid stem cuttings produced the longest primary roots, especially in the case of the PP-E7 plants. Characterization of this trait indicated an advantage of hybridity expressed in root growth of the TH21/2 plants. The Mid-Parent Heterosis (30.67%) was detectable in this triploid hybrid combination. As a common trend, we recorded the fastest growth rate in the roots of tetraploid parental plants. This feature could contribute to the low or the zero MPH% values recorded in these crossing combinations. Because in the use of the triploid hybrids for bioenergy production or phytoremediation any hybrid will be compared to commercial cultivars, we present the heterosis values relative to the parent cultivars (CPH%). Taking this into consideration, we can postulate that all of the triploid hybrids, especially the TH3/12 plants, showed significant hybrid vigor.
To better understand the physiological backgrounds of triploid heterosis, we have determined frequencies of root cells in various cell cycle phases. The data generated by flow cytometry failed to show a general tendency, except in the ratio of the G2/M phase cells (Table 2). These cell cycle phases were represented with the lowest frequency in the tetraploid root cells. In the triploid hybrid roots, the occurrence of G2/M cells showed intermediate frequency values between the parental genotypes with zero MPH%.
3.4. Comparison of Cellular Structures in Roots of Triploid Hybrids and Their Parents
To assess the influence of triploid hybrid vigor at the cellular and tissue levels, cell wall autofluorescence was imaged by laser scanning confocal microscopy on hand-sectioned, unfixed root tips (Figure 4).
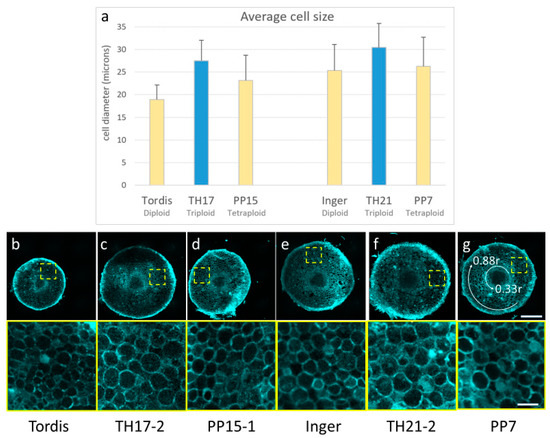
Figure 4.
Differences in cell size of root parenchyma cells of triploid hybrid plants (TH) compared to diploid (Tordis, Inger) or tetraploid (PP-E15) plants. (a) Average cell diameter of root parenchyma cells measured on hand-sections prepared from root tips of indicated plants. (b–g) Representative confocal microscopy images of root disks displaying intrinsic cell wall fluorescence upon violet laser excitation (top panel). Bottom panel shows close-up images of the indicated rectangular ROIs. The last panel shows the position of 0.33r and 0.88r circles between which the parenchyma cell measurements were taken. Scale bars are 250 µm (top) and 50 µm (bottom).
In general, the root tissue architecture of the analyzed triploid (TH17/17, TH21/2) and tetraploid (PP7, PP15) plants was comparable to that of diploid Inger and Tordis lines, with a well-defined stele, cortex (parenchyma), and epidermis without notable abnormalities (Figure 4b–g). On the other hand, a marked increase in cell size was discernable in the root cortex of triploid lines during microscopy observations. The root cortex contains multiple files of parenchyma cells, which play an important role in determining the root thickness. Cell size measurements showed that roots of the TH17 and the TH21 triploid plants contain larger average parenchyma cell diameter compared to their diploid and tetraploid parents, indicating a clear heterosis effect at the cellular level (Figure 4).
3.5. Hormonal Background of Growth Responses in Roots of Triploid Hybrids and Their Parents in Knop’s Solution
It is well established that plant hormones are involved in the regulation of root development. Therefore, determination of the actual concentration of these compounds may shed light on the molecular factors controlling triploid heterosis in energy willow. In the present study, the rooting of stem cuttings from the hybrids and their parents offered a unique experimental system to screen concentration differences in key growth regulators as factors with potential involvement in the generation of hybrid vigor. Quantification of individual plant hormones resulted in a complex picture with differences between various hybrid combinations (Table 3).

Table 3.
Hormonal status of root tip regions/energy willow root tip regions with different genetic origins.
The interpretation of these data should be linked to growth intensity data presented in Table 2 showing a fast-growing rate for the tetraploid parents and the triploid plants with Mid-Parent Heterosis (TH3/12:70.08%; TH21/2 10.17%). The TH3/12 plants presented significant MPH% in levels of CK precursors, especially isopentenyl adenosine monophosphate and gibberellin precursor GA19. Furthermore, accumulation of auxin and ABA deactivation metabolites, such as IAA-glutamate, or phaseic acid were characteristic of roots of the TH3/12 plants with only moderate MPH% in amounts of the trans-zeatin riboside monophosphate and of ABA catabolite dihydrophaseic acid. Reduced concentration of indole-3-acetic acid, salicylic acid, as well as CK deactivation metabolites CK O-glucosides was recorded in TH3/12 hybrids relative to their parents. We could identify TH3/12 plants with intermediate values in contents of phenylacetic acid, benzoic acid, and jasmonic acid.
The TH21/2 triploid hybrid roots with low MPH% value in growth rate showed high MPH% in the concentration of IAA-glutamate and in total cytokinin content. In the case of dihydrophaseic acid and phaseic acid, the MPH% values were low. In these roots, reduced concentrations of salicylic acid, jasmonic acid, abscisic acid, and CK O-glucosides were detected in comparison to the parental roots. A considerable fraction of hormones represented intermediate amounts relative to the parental roots. The TH17/17 triploid hybrids from crossing between Tordis and PP-E15 plants failed to express MPH% in root growth intensity. Similarly, several hormones exhibited low or intermediate amounts, except CK ribosides and CK phosphates (i.e., trans-zeatin riboside monophosphate, isopentenyl adenosine monophosphate, and cis-zeatin riboside monophosphate) with MPH varied between 2.26% and 21.33% (see Table 3).
4. Discussion
It is evident that plant root systems exhibit a multitude of important roles both in the control of aboveground biomass production and in the efficiency of phytoremediation. Therefore, enhancing the capacity of root functions determined by several physiological parameters is also a key breeding target. The utilization of natural and artificial genetic variability for root trait improvement is more efficient when performed through the application of root trait phenotyping (see review by Tracy et al. [21]). Out of several breeding techniques, we previously reported that the autotetraploidization of energy willow plants from the Hungarian cultivar, Energo, significantly stimulated root development, with a simultaneous increase in the amounts of indole acetic acid, cytokinins, gibberellin, and salicylic acid in the root tips of these plants [28]. This artificially generated genetic stock opened an avenue for further breeding of triploid genotypes by crossing between these tetraploid willow plants with leading diploid Swedish cultivars [30]. Previously, Serapiglia et al. [18] published that triploid species hybrids (S. koriyanagi × S. purpurea) × S. miyabeana produced the highest biomass yield under field conditions. Later, these high-yielding genotypes were selected for stable performance [19]. An artificial triploid Populus family was published after hybridization between Populus simonii × (P. nigra var. Italica) × (P. popularis) species. Zhang et al. [34] reported a “Gigas” effect on leaf size. Populus allotriploid plants (2n = 3n = 57) were generated from the hybridization of induced 2n female gametes of Populus pseudosimonii × P. nigra and P. × beijingensis [35]. These poplar plants showed more vigorous growth and more efficient photosynthesis, carbon fixation, sucrose, and starch synthesis. None of these cited research reports provided information about the expression of heterosis in root traits, particularly in woody plants. In the present program, the triploid heterosis was analyzed in hybrid plants from commercial cultivars with a focus on the root system.
4.1. Control of the Size of the Root System
Cultivation of energy willow plants in the Plexiglas columns allowed for the production of two types of parameters for the quantification of root biomass. The root surface area data were generated by a phenotyping platform determining root-related pixels. Table 1 presents these values collected at the end of a growing period of eight weeks in comparison to root fresh weight values measured after separation of root tissues from the soil. Both approaches have a margin of error because digital images did not record roots deep inside the columns; therefore, we also determined the root surface areas at the bottom surfaces. We also have to consider that the separation of roots from the soil cannot be complete. Despite these limitations, the comparison of genotypes clearly indicated Mid-Parent Heterosis to a varying degree. According to the root surface area values, hybrids (TH3/12 and TH21/2) from crossings between PP-E7 tetraploid plants and Tordis and Inger showed clear hybrid vigor (MPH 43.99% and MPH 26.93%) compared to the average of both parents. In contrast, progeny plants of PP-E15 tetraploid parent (TH17/17) failed to produce larger root systems than their parental genotypes. The ranking of genotypes according to the root surface area or the fresh weight values showed similar trends (Table 1). The fresh weight data also showed heterosis, but with lower MPH% values. According to this parameter, the TH17/17 hybrid plants also express low hybrid vigor. Differences in the level of combining ability between crossing partners can have a direct effect on heterosis [36]. In this respect, we may suggest that the tetraploid PP-E7 plants are more optimal partners than plants of PP-E15 tetraploid genotypes in the generation of triploid heterosis.
The root initiation of stem cuttings and the early root development showed significant differences between various willow genotypes, as indicated by the root surface area data monitored by phenotyping. Figure 3 presents root growth curves for two hybrids expressing heterosis (TH3/12 and TH21/2) in addition to their parental plants. Differences between genotypes are increased after six weeks, except for the tetraploid PP-E7 parental plants, which showed delayed rooting. However, these plants present larger root surface area values than plants of the Tordis cultivar at the termination of the experiments. The rooting vigor is significantly higher using Inger plants as crossing partners than the hybridization carried out with plants of the Tordis cultivar. This feature is expected to contribute to the hybrid vigor of TH21/2 hybrids showing the highest root biomass (Table 1). In contrast, the MPH% value indicates enhanced triploid hybrid heterosis for the TH3/12 plants, which are the progenies of the Tordis cultivar with a moderate root system. Because both hybrids are progenies of the tetraploid PP-E7 plants, we can propose that the differences might be due to the combination of these two diploid cultivars.
4.2. Primary Factor in Generation of Triploid Heterosis in Root Systems
To obtain deeper insight into the characteristics of root system, the use of in vitro cultures can be a prerequisite [22,37]. In the present study, willow cuttings were rooted in Knop’s solution for quantification of early growth intensity for the triploid hybrids and their parents. Table 2 provides root length data as a reference for the developmental stage during the last two days of measurement. In all three crossing combinations, the tetraploid parents developed the longest roots. Under the culture conditions used, the stem cuttings of TH3/12 showed delayed rooting and reduced early growth. The Mid-Parent Heterosis of 28.46% could be recorded in the root system of TH21/2 hybrid plants. Root growth rates showed opposite trends when we compared plants from the commercial cultivars and the tetraploid parents. For two days, roots of the PP-E7and the PP-E15 genotypes produced 35–38 mm growth, while roots of Inger and Tordis plants grew only 13–18 mm. The MPH value (70.08%) reflected hybrid vigor only in the TH3/12 plants. This response is in good agreement with the highest heterosis in the root surface area value of the TH3/12 plants in soil (Table 1). The interpretation of the data of Mid-Parent Heterosis in the triploid willow hybrids needs to consider the high growth intensity of the tetraploid parents. Even though the Mid-Parent Heterosis % is the most used parameter for the presentation of hybrid vigor, we also used growth intensity data of commercial cultivars as a reference. As shown in Table 2, significant hybrid vigor can be seen if the Cultivar-Parent Heterosis (CPH%) is analyzed. In this regard, all triploid willow hybrids showed heterosis, with the highest root growth rate in the case of TH3/12 plants.
Increase in the root length during growth is dependent on the cell division rate in the meristematic region and elongation in the differentiation zone (reviewed by Perilli et al. [38]). In attempt to discover the cellular basis of different root growth rates, we also used flow cytometry to identify frequencies of root tip cells at various cell cycle phases. Table 2 presents only the ratio of G2/M cells, because we could detect a general trend only in this parameter during comparison between triploid hybrids and their parents. A high number of G2/M cells was characteristic for roots of the diploid parental cultivars with low root growth intensity, while the tetraploid roots contained a low number of G2/M cells during the fast growth. In maize roots, the frequency of cells in the G2 phase significantly increased in the elongation zone [37]. It is possible that the high growth rate in tetraploids may originate from the more extended elongation zones. In the hybrid roots, the numbers of G2/M cells are intermediate between the parental values, with an intermediate growth intensity rate, except for TH3/12 roots. The triploid hybrid vigor could be seen in the size of the parenchyma cells in the root cortex. This result is in accordance with earlier observations that in maize hybrid leaves, overdominance increased the cell length and width [39].
4.3. Dependence of Triploid Heterosis on a Shift in the Auxin–Cytokinin Ratio in Willow Roots
Assessing a possible link between root growth and plant hormones can highlight molecular regulatory elements in triploid heterosis observed in the energy willow root system. As reviewed by Saini et al. [40], the crosstalk between auxin and other plant hormones is a central mechanism in the regulation of root behavior. Considering the level of triploid heterosis in the root surface area (cm2), the TH3/12 and TH21/2 plants showed increases relative to both parental genotypes. In the growth intensity, heterosis was detectable compared to the diploid parental cultivars. In the present study, 5–7 mm sections of root tips representing meristematic and short elongation zones were collected for hormone analyses. The strongest heterosis and most significant increase in hybrid vigor were found in the roots of TH3/12 plants. The contents of the indole-3-acetic acid and the phenylacetic acid were lower in these F1 primary roots than in the roots of parents (Table 3). In addition, significant heterosis was realized in the elevation of the IAA-glutamate contents, which may indicate enhanced IAA turn-over. Auxin conjugation to glutamate produces indole-3-acetyl glutamic acid (IAAGlu) as an inactive product (see review by Korasick et al. [41]). In 10-day-old seedlings of Theobroma cacao (cacao), a sharp reduction in IAA concentration and an increase in IAA-Glu content was reported, especially in meristematic and elongation zones [42].
The low auxin levels in the primary roots of triploid hybrids TH3/12 and TH17/17 can alter the auxin–cytokinin ratio, given that high Mid-Parent Heterosis values were recorded for cytokinin precursors (namely, cytokinin phosphates and their ribosides) (see Table 3). In root tips from Tordis plants, the auxin content (IAA + phenylacetic acid) was seven times higher than the sum of the cytokinin metabolites. This ratio was essentially lowered in the PP-E7 (1.90) and the PP-E15 (4.84) tetraploids roots. In triploid hybrid progenies, even more cytokinins were detected relative to the auxin contents. The auxin–cytokinin ratios were 1.68 for the TH3/12 root and 3.31 for the TH17/17 root. In root tips from diploid plants of the Inger cultivar and its TH21/2 triploid hybrids, the auxin–cytokinin ratios were 3.21 and 2.89, which also indicate the dominant role of cytokinins. Antagonistic auxin and cytokinin interaction plays a critical role in the regulation of root meristem development (see reviews by Su et al. [43]). Auxin is required for meristem cell division, while cytokinin antagonizing auxin signaling in the transition zone affects cell differentiation and elongation [44]. Stimulation of cytokinin signaling via receptor AHK3 and type-B response regulator represses auxin signaling. Positive cytokinin-response regulators ARR1 and ARR12 promote the expression of the SHORT HYPOCOTYL 2 (SHY2/IAA3) gene in the vascular tissues of roots in the transient zone. The SHY2 protein acts as a repressor of auxin signaling through negative regulation of the PINs as the auxin transporters. As such, cytokinins cause auxin redistribution, prompting cell differentiation [45]. In the present study, the lower auxin–cytokinin ratios could be responsible for the larger cell size in roots of triploid hybrids with heterotic responses, which may explain the observation of considerably thicker roots in triploid hybrids (Figure 2 and Figure 4c–f).
Elevated contents of the isopentenyl adenosine monophosphate (iPMP) indicated heterosis in all three triploid hybrids, with the highest MPH % in roots of TH3/12 hybrids (Table 3). The iPMP is a cytokinin precursor, which can be hydroxylated to trans-zeatin riboside-5-monophosphate. Both precursors (isopentenyl adenosine monophosphate and trans-zeatin riboside monophosphate) provide active cytokinin bases either directly through LOG enzyme catalysis or via riboside formation [46]. Accumulation of the cis-zeatin riboside monophosphate in the roots of triploid hybrids also indicates heterosis (Table 3). In the roots of TH3/12, considerable heterosis was recorded in the amount of active gibberellin precursor GA19 (Table 3), which can further support the central role of increased cell size in root heterosis. In Arabidopsis roots, the gibberellin-deficient mutants showed shorter root length, reduced meristem size, and smaller cell length in comparison to the wild-type roots [47]. In root tips of the two triploid hybrids (TH3/12 and TH21/2), contents of abscisic acid (ABA) and its metabolites (dihydrophaseic acid, phaseic acid) reflect the variable degree of heterosis (Table 3). Especially high MPH % was correlated with the amount of phaseic acid, the primary ABA catabolite. In Arabidopsis, the biphasic effect of ABA included the stimulatory impact. Exogenously applied 0.1 μM ABA caused a 30% faster root growth rate [48].
The triploid heterosis detected in root growth parameters can be a consequence of a reduction in the amount of growth-inhibitory hormones salicylic acid and jasmonic acid in comparison to the parental roots. The benzoic acid levels showed intermediates in the triploid hybrid roots (Table 3). The salicylic acid can influence root meristem patterning in a concentration-dependent manner [49]. Inhibition of root elongation by jasmonic acid via a reduction in the cell number and size was reported (see review by Xu et al. [50]).
5. Conclusions
In the present study, we integrated polyploidy and heterosis effects through the production of triploid hybrids for detailed analysis of consequences in the root development of energy willow plants. Because root functions play a pivotal role in biomass productivity and in phytoremediation, the presented data can support an extended use of this energy plant in a short rotation cultivation system. The use of a phenotyping platform allowed us to characterize the root parameters of the triploid hybrids and their parents, while the in vitro culture in Knop’s solution provided root samples for monitoring the growth rate, cell cycle events, and differences in hormonal status of roots. The Mid-Parent Heterosis (MPH%) in the root biomass yield showed variation between crossing combinations, with an essential contribution from the tetraploid parents having enlarged root system. In particular, differences in the growth rate were significant between roots from the diploid cultivars and the tetraploid genotypes. Therefore, we provide Cultivar-Parent Heterosis (CPH%) values in all three crossing combinations. The search for the hormonal background of triploid heterosis highlighted the role of several plant hormones and their ratios.
The reduction of the auxin–cytokinin ratios in the roots of triploid hybrids indicated a dominant role for cytokinins. The hormone analysis revealed heterosis in additional hormones. These findings emphasize a complex regulation in the expression of the triploid heterosis during the formation of the willow root system. The hormonal influences in MPH% can be responsible for alterations at the cellular level. The enlarged root system in defined crossing combinations (TH3/12 and TH21/2 triploid hybrids) can have a significant role in biomass yield, drought tolerance, or the efficiency of phytoremediation. Therefore, these genotypes can be integrated in energy willow breeding programs.
Author Contributions
D.D. conceived the project, carried out the crossings, and wrote the article with contributions from all of the authors; A.C. and Z.Z. performed phenotyping; K.T. performed the in vitro experiments; L.S. and G.S. performed bioinformatics and data analyses; R.V. and P.I.D. performed hormone analysis; G.F. performed results interpretation; F.A. and I.K.-V. performed microscopy, statistical analyses, and coordination of experiments. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by grant no. GINOP-2.2.1-15-2017-00081 and 2020-1.1.2-PIACI-KFI-2020-00117 from the Hungarian government. F.A. received support from the European Union’s Horizon 2020 research and innovation program with grant number 739593.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Edit Kotogány for flow cytometric analysis. The authors express their great appreciation to Feríz Rádi (Kiskunkutatóközpont Kft. Kiskunhalas, Hungary) and Antal Beck for providing the experimental field.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Clifton-Brown, J.; Harfouche, A.; Casler, M.D.; Dylan Jones, H.; Macalpine, W.J.; Murphy-Bokern, D.; Smart, L.B.; Adler, A.; Ashman, C.; Awty-Carroll, D.; et al. Breeding Progress and Preparedness for Mass-Scale Deployment of Perennial Lignocellulosic Biomass Crops Switchgrass, Miscanthus, Willow and Poplar. GCB Bioenergy 2019, 11, 118–151. [Google Scholar] [CrossRef]
- De Kam, M.J.; Vance Morey, R.; Tiffany, D.G. Biomass Integrated Gasification Combined Cycle for Heat and Power at Ethanol Plants. Energy Convers. Manag. 2009, 50, 1682–1690. [Google Scholar] [CrossRef]
- Dias, G.M.; Ayer, N.W.; Kariyapperuma, K.; Thevathasan, N.; Gordon, A.; Sidders, D.; Johannesson, G.H. Life Cycle Assessment of Thermal Energy Production from Short-Rotation Willow Biomass in Southern Ontario, Canada. Appl. Energy 2017, 204, 343–352. [Google Scholar] [CrossRef]
- Dey, P.; Pal, P.; Kevin, J.D.; Das, D.B. Lignocellulosic Bioethanol Production: Prospects of Emerging Membrane Technologies to Improve the Process—A Critical Review. Rev. Chem. Eng. 2020, 36, 333–367. [Google Scholar] [CrossRef]
- Gaykawad, S.S.; Zha, Y.; Punt, P.J.; van Groenestijn, J.W.; van der Wielen, L.A.M.; Straathof, A.J.J. Pervaporation of Ethanol from Lignocellulosic Fermentation Broth. Bioresour. Technol. 2013, 129, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi Moghaddam, E.; Ericsson, N.; Hansson, P.A.; Nordberg, Å. Exploring the Potential for Biomethane Production by Willow Pyrolysis Using Life Cycle Assessment Methodology. Energy Sustain. Soc. 2019, 9, 6. [Google Scholar] [CrossRef]
- Kakuk, B.; Bagi, Z.; Rákhely, G.; Maróti, G.; Dudits, D.; Kovács, K.L. Methane Production from Green and Woody Biomass Using Short Rotation Willow Genotypes for Bioenergy Generation. Bioresour. Technol. 2021, 333, 125223. [Google Scholar] [CrossRef] [PubMed]
- Tangahu, B.V.; Sheikh Abdullah, S.R.; Basri, H.; Idris, M.; Anuar, N.; Mukhlisin, M. A Review on Heavy Metals (As, Pb, and Hg) Uptake by Plants through Phytoremediation. Int. J. Chem. Eng. 2011, 2011, 939161. [Google Scholar] [CrossRef]
- Capuana, M. A Review of the Performance of Woody and Herbaceous Ornamental Plants for Phytoremediation in Urban Areas. Iforest-Biogeosci. For. 2020, 13, 139–151. [Google Scholar] [CrossRef]
- Hauptvogl, M.; Kotrla, M.; Prčík, M.; Pauková, Ž.; Kováčik, M.; Lošák, T. Phytoremediation Potential of Fast-Growing Energy Plants: Challenges and Perspectives—A Review. Pol. J. Environ. Stud. 2020, 29, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Marmiroli, M.; Pietrini, F.; Maestri, E.; Zacchini, M.; Marmiroli, N.; Massacci, A. Growth, Physiological and Molecular Traits in Salicaceae Trees Investigated for Phytoremediation of Heavy Metals and Organics. Tree Physiol. 2011, 31, 1319–1334. [Google Scholar] [CrossRef] [PubMed]
- Wani, K.A.; Sofi, Z.M.; Malik, J.A.; Wani, J.A. Phytoremediation of Heavy Metals Using Salix (Willows). In Bioremediation and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; Volume 2, pp. 161–174. [Google Scholar]
- Feng, X.; Xu, S.; Li, J.; Yang, Y.; Chen, Q.; Lyu, H.; Zhong, C.; He, Z.; Shi, S. Molecular Adaptation to Salinity Fluctuation in Tropical Intertidal Environments of a Mangrove Tree Sonneratia Alba. BMC Plant Biol. 2020, 20, 178. [Google Scholar] [CrossRef]
- Hanley, S.J.; Karp, A. Genetic Strategies for Dissecting Complex Traits in Biomass Willows (Salix spp.). Tree Physiol. 2014, 34, 1167–1180. [Google Scholar] [CrossRef]
- Kulig, B.; Gacek, E.; Wojciechowski, R.; Oleksy, A.; Kołodziejczyk, M.; Szewczyk, W.; Klimek-Kopyra, A. Biomass Yield and Energy Efficiency of Willow Depending on Cultivar, Harvesting Frequency and Planting Density. Plant Soil Environ. 2019, 65, 377–386. [Google Scholar] [CrossRef]
- Isebrands, J.; Richardson, J. Poplars and Willows: Trees for Society and the Environment; CABI: Wallingford, UK, 2014. [Google Scholar]
- Suda, Y.; Argus, G.W. Chromosome Numbers of Some North American Salix. Brittonia 1968, 20, 191–197. [Google Scholar] [CrossRef]
- Serapiglia, M.J.; Gouker, F.E.; Smart, L.B. Early Selection of Novel Triploid Hybrids of Shrub Willow with Improved Biomass Yield Relative to Diploids. BMC Plant Biol. 2014, 14, 74. [Google Scholar] [CrossRef][Green Version]
- Carlson, C.H.; Smart, L.B. Heterosis for Biomass-Related Traits in Interspecific Triploid Hybrids of Willow (Salix spp.). Bioenergy Res. 2021, 15, 1042–1056. [Google Scholar] [CrossRef]
- Cheng, S.; Zhu, X.; Liao, T.; Li, Y.; Yao, P.; Suo, Y.; Zhang, P.; Wang, J.; Kang, X. Gene Expression Differences between High-Growth Populus Allotriploids and Their Diploid Parents. Forests 2015, 6, 839. [Google Scholar] [CrossRef]
- Tracy, S.R.; Nagel, K.A.; Postma, J.A.; Fassbender, H.; Wasson, A.; Watt, M. Crop Improvement from Phenotyping Roots: Highlights Reveal Expanding Opportunities. Trends Plant Sci. 2020, 25, 105–118. [Google Scholar] [CrossRef]
- Hund, A.; Reimer, R.; Stamp, P.; Walter, A. Can We Improve Heterosis for Root Growth of Maize by Selecting Parental Inbred Lines with Different Temperature Behaviour? Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1580–1588. [Google Scholar] [CrossRef][Green Version]
- Liu, Z.; Liu, X.; Craft, E.J.; Yuan, L.; Cheng, L.; Mi, G.; Chen, F. Physiological and Genetic Analysis for Maize Root Characters and Yield in Response to Low Phosphorus Stress. Breed Sci. 2018, 68, 268–277. [Google Scholar] [CrossRef]
- Ruiz, M.; Oustric, J.; Santini, J.; Morillon, R. Synthetic Polyploidy in Grafted Crops. Front. Plant Sci. 2020, 11, 540894. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; Gray, A.D.; Auger, D.L.; Birchler, J.A. Genomic Dosage Effects on Heterosis in Triploid Maize. Proc. Natl. Acad. Sci. USA 2013, 110, 2665–2669. [Google Scholar] [CrossRef]
- Hallahan, B.F.; Fernandez-Tendero, E.; Fort, A.; Ryder, P.; Dupouy, G.; Deletre, M.; Curley, E.; Brychkova, G.; Schulz, B.; Spillane, C. Hybridity Has a Greater Effect than Paternal Genome Dosage on Heterosis in Sugar Beet (β vulgaris). BMC Plant Biol. 2018, 18, 120. [Google Scholar] [CrossRef] [PubMed]
- Das, S.K.; Sabhapondit, S.; Ahmed, G.; Das, S. Biochemical Evaluation of Triploid Progenies of Diploid × Tetraploid Breeding Populations of Camellia for Genotypes Rich in Catechin and Caffeine. Biochem. Genet. 2013, 51, 358–376. [Google Scholar] [CrossRef]
- Dudits, D.; Török, K.; Cseri, A.; Paul, K.; Nagy, A.V.; Nagy, B.; Sass, L.; Ferenc, G.; Vankova, R.; Dobrev, P.; et al. Response of Organ Structure and Physiology to Autotetraploidization in Early Development of Energy Willow Salix viminalis. Plant Physiol. 2016, 170, 1504–1523. [Google Scholar] [CrossRef]
- Cseri, A.; Borbély, P.; Poór, P.; Fehér, A.; Sass, L.; Jancsó, M.; Penczi, A.; Rádi, F.; Gyuricza, C.; Digruber, T.; et al. Increased Adaptation of an Energy Willow Cultivar to Soil Salinity by Duplication of Its Genome Size. Biomass Bioenergy 2020, 140, 105655. [Google Scholar] [CrossRef]
- Dudits, D.; Cseri, A.; Török, K.; Sass, L.; Zombori, Z.; Ferenc, G.; Poór, P.; Borbély, P.; Czékus, Z.; Vankova, R.; et al. Triploid Hybrid Vigor in Above-Ground Growth and Methane Fermentation Efficiency of Energy Willow. Front. Plant Sci. 2022, 13, 770284. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Ivanov Dobrev, P.; Kamínek, M. Fast and Efficient Separation of Cytokinins from Auxin and Abscisic Acid and Their Purification Using Mixed-Mode Solid-Phase Extraction. J. Chromatogr. A 2002, 950, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, P.I.; Vankova, R. Quantification of Abscisic Acid, Cytokinin, and Auxin Content in Salt-Stressed Plant Tissues. In Plant Salt Tolerance: Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 913. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, B.; Qi, S.; Dong, M.; Wang, Z.; Li, Y.; Chen, S.; Li, B.; Zhang, J. Ploidy and Hybridity Effects on Leaf Size, Cell Size and Related Genes Expression in Triploids, Diploids and Their Parents in Populus. Planta 2019, 249, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Liao, T.; Ren, Y.; Geng, X.; Kang, X. Molecular Mechanism of Vegetative Growth Advantage in Allotriploid Populus. Int. J. Mol. Sci. 2020, 21, 441. [Google Scholar] [CrossRef]
- Liu, Z.; Jiang, J.; Ren, A.; Xu, X.; Zhang, H.; Zhao, T.; Jiang, X.; Sun, Y.; Li, J.; Yang, H. Heterosis and Combining Ability Analysis of Fruit Yield, Early Maturity, and Quality in Tomato. Agronomy 2021, 11, 807. [Google Scholar] [CrossRef]
- Alarcon, M.V.; Salguero, J. Transition Zone Cells Reach G2 Phase before Initiating Elongation in Maize Root Apex. Biol. Open 2017, 6, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Perilli, S.; Di Mambro, R.; Sabatini, S. Growth and Development of the Root Apical Meristem. Curr. Opin. Plant Biol. 2012, 15, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Pavlikova, E.; Rood, S.B. Cellular basis of heterosis for leaf area in maize. Can. J. Plant Sci. 1987, 67, 99–104. [Google Scholar] [CrossRef]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin: A Master Regulator in Plant Root Development. Plant Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin Biosynthesis and Storage Forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef]
- Noah, A.M.; Casanova-Sáez, R.; Ango, R.E.M.; Antoniadi, I.; Karady, M.; Novák, O.; Niemenak, N.; Ljung, K. Dynamics of Auxin and Cytokinin Metabolism during Early Root and Hypocotyl Growth in Theobroma Cacao. Plants 2021, 10, 967. [Google Scholar] [CrossRef]
- Su, Y.H.; Liu, Y.B.; Zhang, X.S. Auxin-Cytokinin Interaction Regulates Meristem Development. Mol. Plant 2011, 4, 616–625. [Google Scholar] [CrossRef]
- Chapman, E.J.; Estelle, M. Mechanism of Auxin-Regulated Gene Expression in Plants. Annu. Rev. Genet. 2009, 43, 265–285. [Google Scholar] [CrossRef] [PubMed]
- Dello Ioio, R.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A Genetic Framework for the Control of Cell Division and Differentiation in the Root Meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Takei, K.; Sakakibara, H.; Taniguchi, M.; Sugiyama, T. Nitrogen-Dependent Accumulation of Cytokinins in Root and the Translocation to Leaf: Implication of Cytokinin Species That Induces Gene Expression of Maize Response Regulator. Plant Cell Physiol. 2001, 42, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Barker, R.; Fernandez Garcia, M.N.; Powers, S.J.; Vaughan, S.; Bennett, M.J.; Phillips, A.L.; Thomas, S.G.; Hedden, P. Mapping Sites of Gibberellin Biosynthesis in the Arabidopsis Root Tip. New Phytol. 2021, 229, 1521–1534. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, L.; Forde, B.G.; Davies, W.J. The Biphasic Root Growth Response to Abscisic Acid in Arabidopsis Involves Interaction with Ethylene and Auxin Signalling Pathways. Front. Plant Sci. 2017, 8, 1493. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, T.; Groot, E.P.; Kazantsev, F.V.; Teale, W.; Omelyanchuk, N.; Kovrizhnykh, V.; Palme, K.; Mironova, V.V. Salicylic Acid Affects Root Meristem Patterning via Auxin Distribution in a Concentration-Dependent Manner. Plant Physiol. 2019, 180, 1725–1739. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Zhao, P.X.; Cai, X.T.; Mao, J.L.; Miao, Z.Q.; Xiang, C. Bin Integration of Jasmonic Acid and Ethylene into Auxin Signaling in Root Development. Front. Plant Sci. 2020, 11, 271. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).