Exploring the Genetic Causality of Discordant Phenotypes in Familial Apparently Balanced Translocation Cases Using Whole Exome Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Whole Exome Sequencing
2.3. WES Data Processing and Variant Annotation
2.4. Candidate Variant Validation
2.5. In Silico Analysis of WES Variants
2.6. RNA Extraction
2.7. Reverse-Transcription PCR
2.8. Gene and Variant Nomenclature
3. Results
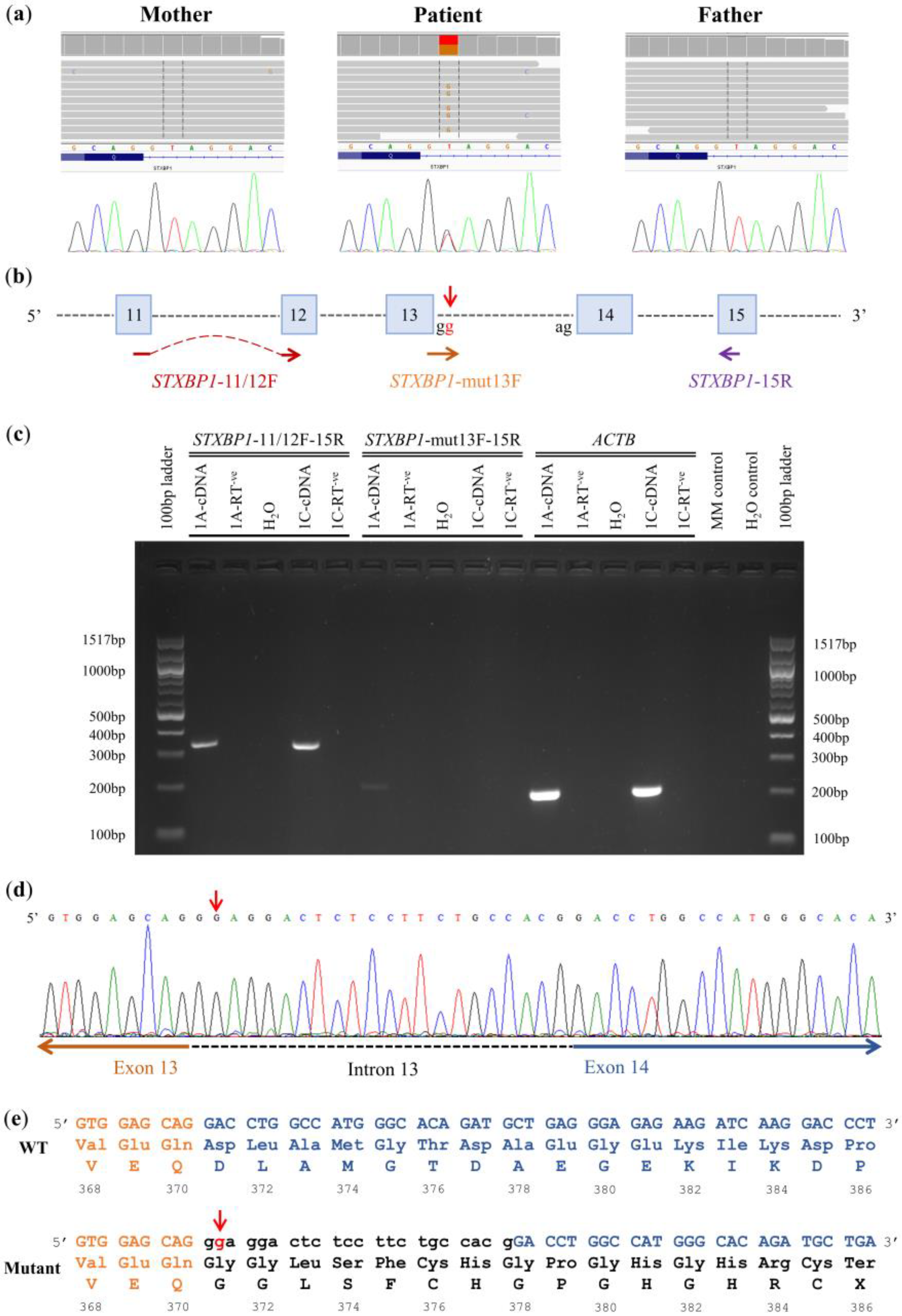
3.1. Family 1
3.2. Family 2
3.3. Family 3
3.4. Family 4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scriven, P.N.; Handyside, A.H.; Ogilvie, C.M. Chromosome translocations: Segregation modes and strategies for preimplantation genetic diagnosis. Prenat. Diagn. 1998, 18, 1437–1449. [Google Scholar] [CrossRef]
- Madan, K. Balanced complex chromosome rearrangements: Reproductive aspects. A review. Am. J. Med. Genet. A. 2012, 158A, 947–963. [Google Scholar] [CrossRef] [PubMed]
- Warburton, D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: Clinical significance and distribution of breakpoints. Am. J. Hum. Genet. 1991, 49, 995–1013. [Google Scholar] [PubMed]
- Halgren, C.; Nielsen, N.M.; Nazaryan-Petersen, L.; Silahtaroglu, A.; Collins, R.L.; Lowther, C.; Kjaergaard, S.; Frisch, M.; Kirchhoff, M.; Brøndum-Nielsen, K.; et al. Risks and Recommendations in Prenatally Detected De Novo Balanced Chromosomal Rearrangements from Assessment of Long-Term Outcomes. Am. J. Hum. Genet. 2018, 102, 1090–1103. [Google Scholar] [CrossRef] [PubMed]
- Young, I.D. Introduction to Risk Calculation in Genetic Counseling; Oxford University Press: Oxford, UK, 2007. [Google Scholar]
- Stengel-Rutkowski, S.; Stene, J.; Gallano, P. Risk Estimates in Balanced Parental Reciprocal Translocations: Analysis of 1120 Pedigrees; Expansion Scientifique Francaise: Paris, France, 1988. [Google Scholar]
- Gorski, J.L.; Kistenmacher, M.L.; Punnett, H.H.; Zackai, E.H.; Emanuel, B.S.; Optiz, J.M.; Reynolds, J.F. Reproductive risks for carriers of complex chromosome rearrangements: Analysis of 25 families. Am. J. Med. Genet. 1988, 29, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Neri, G.; Serra, A.; Campana, M.; Tedeschi, B. Reproductive risks for translocation carriers: Cytogenetic study and analysis of pregnancy outcome in 58 families. Am. J. Med. Genet. 1983, 16, 535–561. [Google Scholar] [CrossRef]
- Stene, J.; Stengel-Rutowski, S. Genetic risks for familial reciprocal translocations with special emphasis on those leading to 9p, 10p and 12p trisomies. Ann. Hum. Genet. 1982, 46, 41–74. [Google Scholar] [CrossRef]
- Midro, A.T.; Stengel-Rutkowski, S.; Stene, J. Experiences with risk estimates for carriers of chromosomal reciprocal translocations. Clin. Genet. 1992, 41, 113–122. [Google Scholar] [CrossRef]
- Sismani, C.; Kitsiou-Tzeli, S.; Ioannides, M.; Christodoulou, C.; Anastasiadou, V.; Stylianidou, G.; Papadopoulou, E.; Kanavakis, E.; Kosmaidou-Aravidou, Z.; Patsalis, P.C. Cryptic genomic imbalances in patients with de novo or familial apparently balanced translocations and abnormal phenotype. Mol. Cytogenet. 2008, 1, 15. [Google Scholar] [CrossRef]
- Aristidou, C.; Koufaris, C.; Theodosiou, A.; Bak, M.; Mehrjouy, M.M.; Behjati, F.; Tanteles, G.; Christophidou-Anastasiadou, V.; Tommerup, N.; Sismani, C. Accurate breakpoint mapping in apparently balanced translocation families with discordant phenotypes using whole genome mate-pair sequencing. PLoS ONE 2017, 12, e0169935. [Google Scholar] [CrossRef]
- Wenger, S.L.; Steele, M.W.; Boone, L.Y.; Lenkey, S.G.; Cummins, J.H.; Chen, X.Q. “Balanced” karyotypes in six abnormal offspring of balanced reciprocal translocation normal carrier parents. Am. J. Med. Genet. 1995, 55, 47–52. [Google Scholar] [CrossRef]
- Schluth-Bolard, C.; Delobel, B.; Sanlaville, D.; Boute, O.; Cuisset, J.-M.M.; Sukno, S.; Labalme, A.; Duban-Bedu, B.n.d.B.; Plessis, G.; Jaillard, S.; et al. Cryptic genomic imbalances in de novo and inherited apparently balanced chromosomal rearrangements: Array CGH study of 47 unrelated cases. Eur. J. Med. Genet. 2009, 52, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Kanakavalli, M.; Padmalatha, V.; Nallari, P.; Singh, L. Paternally derived translocation t(8;18)(q22.1;q22)pat associated in a patient with developmental delay: Case report and review. J. Pediatr. Neurosci. 2010, 5, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Utami, K.H.; Hillmer, A.M.; Aksoy, I.; Chew, E.G.; Teo, A.S.; Zhang, Z.; Lee, C.W.; Chen, P.J.; Seng, C.C.; Ariyaratne, P.N.; et al. Detection of chromosomal breakpoints in patients with developmental delay and speech disorders. PLoS ONE 2014, 9, e90852. [Google Scholar] [CrossRef] [PubMed]
- Aristidou, C.; Theodosiou, A.; Bak, M.; Mehrjouy, M.M.; Constantinou, E.; Alexandrou, A.; Papaevripidou, I.; Christophidou-Anastasiadou, V.; Skordis, N.; Kitsiou-Tzeli, S.; et al. Position effect, cryptic complexity, and direct gene disruption as disease mechanisms in de novo apparently balanced translocation cases. PLoS ONE 2018, 13, e0205298. [Google Scholar] [CrossRef] [PubMed]
- Redin, C.; Brand, H.; Collins, R.L.; Kammin, T.; Mitchell, E.; Hodge, J.C.; Hanscom, C.; Pillalamarri, V.; Seabra, C.M.; Abbott, M.-A.; et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat. Genet. 2017, 49, 36–45. [Google Scholar] [CrossRef]
- Schluth-Bolard, C.; Diguet, F.; Chatron, N.; Rollat-Farnier, P.A.; Bardel, C.; Afenjar, A.; Amblard, F.; Amiel, J.; Blesson, S.; Callier, P.; et al. Whole genome paired-end sequencing elucidates functional and phenotypic consequences of balanced chromosomal rearrangement in patients with developmental disorders. J. Med. Genet. 2019, 56, 526–535. [Google Scholar] [CrossRef]
- Bugge, M.; Bruun-Petersen, G.; Brøndum-Nielsen, K.; Friedrich, U.; Hansen, J.; Jensen, G.; Jensen, P.K.; Kristoffersson, U.; Lundsteen, C.; Niebuhr, E.; et al. Disease associated balanced chromosome rearrangements: A resource for large scale genotype-phenotype delineation in man. J. Med. Genet. 2000, 37, 858–865. [Google Scholar] [CrossRef]
- Schluth-Bolard, C.; Labalme, A.; Cordier, M.-P.P.; Till, M.; Nadeau, G.; Tevissen, H.; Lesca, G.; Boutry-Kryza, N.; Rossignol, S.; Rocas, D.; et al. Breakpoint mapping by next generation sequencing reveals causative gene disruption in patients carrying apparently balanced chromosome rearrangements with intellectual deficiency and/or congenital malformations. J. Med. Genet. 2013, 50, 144–150. [Google Scholar] [CrossRef]
- Nilsson, D.; Pettersson, M.; Gustavsson, P.; Förster, A.; Hofmeister, W.; Wincent, J.; Zachariadis, V.; Anderlid, B.-M.; Nordgren, A.; Mäkitie, O.; et al. Whole-Genome Sequencing of Cytogenetically Balanced Chromosome Translocations Identifies Potentially Pathological Gene Disruptions and Highlights the Importance of Microhomology in the Mechanism of Formation. Hum. Mutat. 2017, 38, 180–192. [Google Scholar] [CrossRef]
- Patsalis, P.C.; Evangelidou, P.; Charalambous, S.; Sismani, C. Fluorescence in situ hybridization characterization of apparently balanced translocation reveals cryptic complex chromosomal rearrangements with unexpected level of complexity. Eur. J. Hum. Genet. 2004, 12, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Gribble, S.M.; Prigmore, E.; Burford, D.C.; Porter, K.M.; Ng, B.L.; Douglas, E.J.; Fiegler, H.; Carr, P.; Kalaitzopoulos, D.; Clegg, S.; et al. The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J. Med. Genet. 2005, 42, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Kleinjan, D.-J.; van Heyningen, V. Position Effect in Human Genetic Disease. Hum. Mol. Genet. 1998, 7, 1611–1618. [Google Scholar] [CrossRef] [PubMed]
- Velagaleti, G.V.N.; Bien-Willner, G.A.; Northup, J.K.; Lockhart, L.H.; Hawkins, J.C.; Jalal, S.M.; Withers, M.; Lupski, J.R.; Stankiewicz, P. Position Effects Due to Chromosome Breakpoints that Map ~900 Kb Upstream and ~1.3 Mb Downstream of SOX9 in Two Patients with Campomelic Dysplasia. Am. J. Hum. Genet. 2005, 76, 652–662. [Google Scholar] [CrossRef]
- Finelli, P.; Sirchia, S.M.; Masciadri, M.; Crippa, M.; Recalcati, M.P.; Rusconi, D.; Giardino, D.; Monti, L.; Cogliati, F.; Faravelli, F.; et al. Juxtaposition of heterochromatic and euchromatic regions by chromosomal translocation mediates a heterochromatic long-range position effect associated with a severe neurological phenotype. Mol. Cytogenet. 2012, 5, 16. [Google Scholar] [CrossRef][Green Version]
- Choi, M.; Scholl, U.I.; Ji, W.; Liu, T.; Tikhonova, I.R.; Zumbo, P.; Nayir, A.; Bakkaloǧlu, A.; Özen, S.; Sanjad, S.; et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl. Acad. Sci. USA 2009, 106, 19096. [Google Scholar] [CrossRef]
- Ng, S.B.; Buckingham, K.J.; Lee, C.; Bigham, A.W.; Tabor, H.K.; Dent, K.M.; Huff, C.D.; Shannon, P.T.; Jabs, E.W.; Nickerson, D.A.; et al. Exome sequencing identifies the cause of a Mendelian disorder. Nat. Genet. 2010, 42, 30. [Google Scholar] [CrossRef]
- Goh, G.; Choi, M. Application of Whole Exome Sequencing to Identify Disease-Causing Variants in Inherited Human Diseases. Genomics Inform. 2012, 10, 214. [Google Scholar] [CrossRef]
- Robinson, P.N.; Köhler, S.; Bauer, S.; Seelow, D.; Horn, D.; Mundlos, S. The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease. Am. J. Hum. Genet. 2008, 83, 610. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 11, 11.10.1–11.10.33. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Paila, U.; Chapman, B.A.; Kirchner, R.; Quinlan, A.R. GEMINI: Integrative Exploration of Genetic Variation and Genome Annotations. PLoS Comput. Biol. 2013, 9, e1003153. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBNet J. 2011 2011, 17, 3. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pedersen, B.S.; Quinlan, A.R. Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am. J. Hum. Genet. 2017, 100, 406–413. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Bëroud, G.; Claustres, M.; Béroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, 1–14. [Google Scholar] [CrossRef]
- Vissers, L.E.L.M.; Gilissen, C.; Veltman, J.A. Genetic studies in intellectual disability and related disorders. Nat. Rev. Genet. 2015, 17, 9–18. [Google Scholar] [CrossRef]
- Plagnol, V.; Curtis, J.; Epstein, M.; Mok, K.Y.; Stebbings, E.; Grigoriadou, S.; Wood, N.W.; Hambleton, S.; Burns, S.O.; Thrasher, A.J.; et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics 2012, 28, 2747–2754. [Google Scholar] [CrossRef] [PubMed]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Albarca Aguilera, M.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Richards, C.S.; Bale, S.; Bellissimo, D.B.; Das, S.; Grody, W.W.; Hegde, M.R.; Lyon, E.; Ward, B.E. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008, 10, 294–300. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405. [Google Scholar] [CrossRef] [PubMed]
- Ellard, S.; Baple, E.L.; Callaway, A.; Berry, I.; Forrester, N.; Turnbull, C.; Owens, M.; Eccles, D.M.; Abbs, S.; Scott, R.; et al. ACGS Best Practice Guidelines for Variant Classification in Rare Disease 2020. Available online: https://www.acgs.uk.com/media/11631/uk-practice-guidelines-for-variant-classification-v4-01-2020.pdf (accessed on 25 October 2022).
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef]
- Ittisoponpisan, S.; Islam, S.A.; Khanna, T.; Alhuzimi, E.; David, A.; Sternberg, M.J.E. Can Predicted Protein 3D Structures Provide Reliable Insights into whether Missense Variants Are Disease Associated? J. Mol. Biol. 2019, 431, 2197–2212. [Google Scholar] [CrossRef]
- Venselaar, H.; Te Beek, T.A.; Kuipers, R.K.; Hekkelman, M.L.; Vriend, G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010, 11, 548. [Google Scholar] [CrossRef]
- Tweedie, S.; Braschi, B.; Gray, K.; Jones, T.E.M.; Seal, R.L.; Yates, B.; Bruford, E.A. Genenames.org: The HGNC and VGNC resources in 2021. Nucleic Acids Res. 2021, 49, D939. [Google Scholar] [CrossRef]
- den Dunnen, J.T.; Dalgleish, R.; Maglott, D.R.; Hart, R.K.; Greenblatt, M.S.; McGowan-Jordan, J.; Roux, A.F.; Smith, T.; Antonarakis, S.E.; Taschner, P.E.M. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 2016, 37, 564–569. [Google Scholar] [CrossRef]
- Burkhardt, P.; Hattendorf, D.A.; Weis, W.I.; Fasshauer, D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008, 27, 923–933. [Google Scholar] [CrossRef]
- Han, G.A.; Malintan, N.T.; Collins, B.M.; Meunier, F.A.; Sugita, S. Munc18-1 as a key regulator of neurosecretion. J. Neurochem. 2010, 115, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Saitsu, H.; Kato, M.; Mizuguchi, T.; Hamada, K.; Osaka, H.; Tohyama, J.; Uruno, K.; Kumada, S.; Nishiyama, K.; Nishimura, A.; et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat. Genet. 2008, 40, 782–788. [Google Scholar] [CrossRef] [PubMed]
- Saitsu, H.; Kato, M.; Okada, I.; Orii, K.E.; Higuchi, T.; Hoshino, H.; Kubota, M.; Arai, H.; Tagawa, T.; Kimura, S.; et al. STXBP1 mutations in early infantile epileptic encephalopathy with suppression-burst pattern. Epilepsia 2010, 51, 2397–2405. [Google Scholar] [CrossRef] [PubMed]
- Deprez, L.; Weckhuysen, S.; Holmgren, P.; Suls, A.; Van Dyck, T.; Goossens, D.; Del-Favero, J.; Jansen, A.; Verhaert, K.; Lagae, L.; et al. Clinical spectrum of early-onset epileptic encephalopathies associated with STXBP1 mutations. Neurology 2010, 75, 1159–1165. [Google Scholar] [CrossRef]
- Barcia, G.; Chemaly, N.; Gobin, S.; Milh, M.; Van Bogaert, P.; Barnerias, C.; Kaminska, A.; Dulac, O.; Desguerre, I.; Cormier, V.; et al. Early epileptic encephalopathies associated with STXBP1 mutations: Could we better delineate the phenotype? Eur. J. Med. Genet. 2014, 57, 15–20. [Google Scholar] [CrossRef]
- Saitsu, H.; Kato, M.; Shimono, M.; Senju, A.; Tanabe, S.; Kimura, T.; Nishiyama, K.; Yoneda, Y.; Kondo, Y.; Tsurusaki, Y.; et al. Association of genomic deletions in the STXBP1 gene with Ohtahara syndrome. Clin. Genet. 2012, 81, 399–402. [Google Scholar] [CrossRef]
- Hamdan, F.F.; Piton, A.; Gauthier, J.; Lortie, A.; Dubeau, F.; Dobrzeniecka, S.; Spiegelman, D.; Noreau, A.; Pellerin, S.; Côté, M.; et al. De novo STXBP1 mutations in mental retardation and nonsyndromic epilepsy. Ann. Neurol. 2009, 65, 748–753. [Google Scholar] [CrossRef]
- Saitsu, H.; Hoshino, H.; Kato, M.; Nishiyama, K.; Okada, I.; Yoneda, Y.; Tsurusaki, Y.; Doi, H.; Miyake, N.; Kubota, M.; et al. Paternal mosaicism of an STXBP1 mutation in OS. Clin. Genet. 2011, 80, 484–488. [Google Scholar] [CrossRef]
- Ortega-Moreno, L.; Giráldez, B.G.; Verdú, A.; García-Campos, O.; Sánchez-Martín, G.; Serratosa, J.M.; Guerrero-López, R. Novel mutation in STXBP1 gene in a patient with non-lesional Ohtahara syndrome. Neurologia 2015, 31, 523–527. [Google Scholar] [CrossRef]
- Yamamoto, T.; Shimojima, K.; Yano, T.; Ueda, Y.; Takayama, R.; Ikeda, H.; Imai, K. Loss-of-function mutations of STXBP1 in patients with epileptic encephalopathy. Brain Dev. 2016, 38, 280–284. [Google Scholar] [CrossRef]
- Cogliati, F.; Giorgini, V.; Masciadri, M.; Bonati, M.T.; Marchi, M.; Cracco, I.; Gentilini, D.; Peron, A.; Savini, M.N.; Spaccini, L.; et al. Pathogenic Variants in STXBP1 and in Genes for GABAa Receptor Subunities Cause Atypical Rett/Rett-like Phenotypes. Int. J. Mol. Sci. 2019, 20, 3621. [Google Scholar] [CrossRef] [PubMed]
- Lykke-Andersen, S.; Jensen, T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015, 16, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Misura, K.M.; Scheller, R.H.; Weis, W.I. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 2000, 404, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Aiken, J.; Buscaglia, G.; Bates, E.A.; Moore, J.K. The α-Tubulin gene TUBA1A in Brain Development: A Key Ingredient in the Neuronal Isotype Blend. J. Dev. Biol. 2017, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Keays, D.A.; Tian, G.; Poirier, K.; Huang, G.J.; Siebold, C.; Cleak, J.; Oliver, P.L.; Fray, M.; Harvey, R.J.; Molnár, Z.; et al. Mutations in α-Tubulin Cause Abnormal Neuronal Migration in Mice and Lissencephaly in Humans. Cell 2007, 128, 45–57. [Google Scholar] [CrossRef]
- Poirier, K.; Keays, D.A.; Francis, F.; Saillour, Y.; Bahi, N.; Manouvrier, S.; Fallet-Bianco, C.; Pasquier, L.; Toutain, A.; Tuy, F.P.D.; et al. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1A (TUBA1A). Hum. Mutat. 2007, 28, 1055–1064. [Google Scholar] [CrossRef]
- Kumar, R.A.; Pilz, D.T.; Babatz, T.D.; Cushion, T.D.; Harvey, K.; Topf, M.; Yates, L.; Robb, S.; Uyanik, G.; Mancini, G.M.S.; et al. TUBA1A mutations cause wide spectrum lissencephaly (smooth brain) and suggest that multiple neuronal migration pathways converge on alpha tubulins. Hum. Mol. Genet. 2010, 19, 2817–2827. [Google Scholar] [CrossRef]
- Hebebrand, M.; Hüffmeier, U.; Trollmann, R.; Hehr, U.; Uebe, S.; Ekici, A.B.; Kraus, C.; Krumbiegel, M.; Reis, A.; Thiel, C.T.; et al. The mutational and phenotypic spectrum of TUBA1A-associated tubulinopathy. Orphanet J. Rare Dis. 2019, 14, 38. [Google Scholar] [CrossRef]
- Aiken, J.; Buscaglia, G.; Aiken, A.S.; Moore, J.K.; Bates, E.A. Tubulin mutations in brain development disorders: Why haploinsufficiency does not explain TUBA1A tubulinopathies. Cytoskeleton 2020, 77, 40–54. [Google Scholar] [CrossRef]
- Catterall, W.A. From ionic currents to molecular mechanisms: The structure and function of voltage-gated sodium channels. Neuron 2000, 26, 13–25. [Google Scholar] [CrossRef]
- Mulley, J.C.; Scheffer, I.E.; Petrou, S.; Dibbens, L.M.; Berkovic, S.F.; Harkin, L.A. SCN1A mutations and epilepsy. Hum. Mutat. 2005, 25, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Claes, L.R.F.; Deprez, L.; Suls, A.; Baets, J.; Smets, K.; Van Dyck, T.; Deconinck, T.; Jordanova, A.; De Jonghe, P. The SCN1A variant database: A novel research and diagnostic tool. Hum. Mutat. 2009, 30-10, E904–E920. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Xu, H.Q.; Yu, L.; Lin, G.W.; He, N.; Su, T.; Shi, Y.W.; Li, B.; Wang, J.; Liu, X.R.; et al. The SCN1A Mutation Database: Updating Information and Analysis of the Relationships among Genotype, Functional Alteration, and Phenotype. Hum. Mutat. 2015, 36, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Escayg, A.; MacDonald, B.T.; Meisler, M.H.; Baulac, S.; Huberfeld, G.; An-Gourfinkel, I.; Brice, A.; LeGuern, E.; Moulard, B.; Chaigne, D.; et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat. Genet. 2000, 24, 343–345. [Google Scholar] [CrossRef]
- Claes, L.; Del-Favero, J.; Ceulemans, B.; Lagae, L.; Van Broeckhoven, C.; De Jonghe, P. De Novo Mutations in the Sodium-Channel Gene SCN1A Cause Severe Myoclonic Epilepsy of Infancy. Am. J. Hum. Genet. 2001, 68, 1327–1332. [Google Scholar] [CrossRef]
- Kanai, K.; Hirose, S.; Oguni, H.; Fukuma, G.; Shirasaka, Y.; Miyajima, T.; Wada, K.; Iwasa, H.; Yasumoto, S.; Matsuo, M.; et al. Effect of localization of missense mutations in SCN1A on epilepsy phenotype severity. Neurology 2004, 63, 329–334. [Google Scholar] [CrossRef]
- Ceulemans, B.P.G.M.; Claes, L.R.F.; Lagae, L.G. Clinical correlations of mutations in the SCN1A gene: From febrile seizures to severe myoclonic epilepsy in infancy. Pediatr. Neurol. 2004, 30, 236–243. [Google Scholar] [CrossRef]
- de Lange, I.M.; Koudijs, M.J.; van ’t Slot, R.; Gunning, B.; Sonsma, A.C.M.; van Gemert, L.J.J.M.; Mulder, F.; Carbo, E.C.; van Kempen, M.J.A.; Verbeek, N.E.; et al. Mosaicism of de novo pathogenic SCN1A variants in epilepsy is a frequent phenomenon that correlates with variable phenotypes. Epilepsia 2018, 59, 690–703. [Google Scholar] [CrossRef]
- Escayg, A.; Goldin, A.L. Sodium channel SCN1A and epilepsy: Mutations and mechanisms. Epilepsia 2010, 51, 1650–1658. [Google Scholar] [CrossRef]
- Ohmori, I.; Ouchida, M.; Ohtsuka, Y.; Oka, E.; Shimizu, K. Significant correlation of the SCN1A mutations and severe myoclonic epilepsy in infancy. Biochem. Biophys. Res. Commun. 2002, 295, 17–23. [Google Scholar] [CrossRef]
- Gurrieri, F.; Franco, B.; Toriello, H.; Neri, G. Oral-facial-digital syndromes: Review and diagnostic guidelines. Am J Med Genet A 2007, 143A, 3314–3323. [Google Scholar] [CrossRef] [PubMed]
- Franco, B.; Thauvin-Robinet, C. Update on oral-facial-digital syndromes (OFDS). Cilia 2016, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Bruel, A.L.; Franco, B.; Duffourd, Y.; Thevenon, J.; Jego, L.; Lopez, E.; Deleuze, J.F.; Doummar, D.; Giles, R.H.; Johnson, C.A.; et al. Fifteen years of research on oral-facial-digital syndromes: From 1 to 16 causal genes. J. Med. Genet. 2017, 54, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Strong, A.; Simone, L.; Krentz, A.; Vaccaro, C.; Watson, D.; Ron, H.; Kalish, J.M.; Pedro, H.F.; Zackai, E.H.; Hakonarson, H. Expanding the genetic landscape of oral-facial-digital syndrome with two novel genes. Am. J. Med. Genet. A 2021, 185, 2409–2416. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Deignan, J.L.; Dorrani, N.; Strom, S.P.; Kantarci, S.; Quintero-Rivera, F.; Das, K.; Toy, T.; Harry, B.; Yourshaw, M.; et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA 2014, 312, 1880–1887. [Google Scholar] [CrossRef] [PubMed]
- Wright, C.F.; Fitzgerald, T.W.; Jones, W.D.; Clayton, S.; McRae, J.F.; Van Kogelenberg, M.; King, D.A.; Ambridge, K.; Barrett, D.M.; Bayzetinova, T.; et al. Genetic diagnosis of developmental disorders in the DDD study: A scalable analysis of genome-wide research data. Lancet 2015, 385, 1305. [Google Scholar] [CrossRef]
- Burdick, K.J.; Cogan, J.D.; Rives, L.C.; Robertson, A.K.; Koziura, M.E.; Brokamp, E.; Duncan, L.; Hannig, V.; Pfotenhauer, J.; Vanzo, R.; et al. Limitations of exome sequencing in detecting rare and undiagnosed diseases. Am. J. Med. Genet. A. 2020, 182, 1400–1406. [Google Scholar] [CrossRef]
- Belkadi, A.; Bolze, A.; Itan, Y.; Cobat, A.; Vincent, Q.B.; Antipenko, A.; Shang, L.; Boisson, B.; Casanova, J.L.; Abel, L. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc. Natl. Acad. Sci. USA 2015, 112, 5473–5478. [Google Scholar] [CrossRef] [PubMed]
- Meienberg, J.; Bruggmann, R.; Oexle, K.; Matyas, G. Clinical sequencing: Is WGS the better WES? Hum. Genet. 2016, 135, 359–362. [Google Scholar] [CrossRef]
- Ebbert, M.T.W.; Jensen, T.D.; Jansen-West, K.; Sens, J.P.; Reddy, J.S.; Ridge, P.G.; Kauwe, J.S.K.; Belzil, V.; Pregent, L.; Carrasquillo, M.M.; et al. Systematic analysis of dark and camouflaged genes reveals disease-relevant genes hiding in plain sight. Genome Biol. 2019, 20, 97. [Google Scholar] [CrossRef]
- Mantere, T.; Kersten, S.; Hoischen, A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019, 10, 426. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.E.; Sulovari, A.; Wang, T.; Loucks, H.; Hoekzema, K.; Munson, K.M.; Lewis, A.P.; Fuerte, E.P.A.; Paschal, C.R.; Walsh, T.; et al. Targeted long-read sequencing identifies missing disease-causing variation. Am. J. Hum. Genet. 2021, 108, 1436–1449. [Google Scholar] [CrossRef] [PubMed]
- Mantere, T.; Neveling, K.; Pebrel-Richard, C.; Benoist, M.; van der Zande, G.; Kater-Baats, E.; Baatout, I.; van Beek, R.; Yammine, T.; Oorsprong, M.; et al. Optical genome mapping enables constitutional chromosomal aberration detection. Am. J. Hum. Genet. 2021, 108, 1409–1422. [Google Scholar] [CrossRef] [PubMed]
| Family Number | Sample Name | Reason for Referral | Phenotypes (HPO) | Karyotype |
|---|---|---|---|---|
| 1 | 1A | Affected individual with intellectual disability, psychomotor delay, epilepsy and multifocal epileptiform discharges | HP:0001249 HP:0001263 HP:0001250 HP:0010841 | 46,XY,t(1;7)(p36.1;q22)mat |
| 1 | 1B | Mother of sample 1A | Non-affected | 46,XX,t(1;7)(p36.1;q22) |
| 1 | 1C | Father of sample 1A | Non-affected | 46,XY |
| 2 | 2A | Affected individual with severe intellectual disability and microcephaly | HP:0010864 HP:0000252 | 46,XX,t(7;8)(q32;q24.13) |
| 2 | 2B | Sibling of sample 2A | Non-affected | 46,XX,t(7;8)(q32;q24.13) |
| 3 | 3A | Affected individual with mild intellectual disability, short stature, low set ears, flat mid face, long neck, high arched palate, and simian palmar crease | HP:0001256 HP:0004322 HP:0000369 HP:0011800 HP:0000472 HP:0000218 HP:0000954 | 46,XX,t(4;10)(q35;q11.2)mat |
| 3 | 3B | Mother of sample 3A | Non-affected | 46,XX,t(4;10)(q35;q11.2) |
| 3 | 3C | Sibling of sample 3A | Non-affected | 46,XX,t(4;10)(q35;q11.2)mat |
| 4 | 4A | Affected individual with polysyndactyly and oral anomalies | HP:0001159 HP:0010442 HP:0000153 | 46,XY,t(1;20)(p35.3;q13.3)mat |
| 4 | 4B | Mother of sample 4A | Non-affected | 46,XX,t(1;20)(p35.3;q13.3) |
| 4 | 4C | Father of sample 4A | Non-affected | 46,XY |
| Candidate Variant | Candidate Gene | Classification | MOI 1 | Sample Name 3 | Zygosity | Allelic Balance | Coverage |
|---|---|---|---|---|---|---|---|
| Family 1: NM_003165.6:c.1110+2T>G NP_003156.1:p.(Asp371GlyfsTer16) | STXBP1 | Pathogenic PVS1_very strong PS2_moderate PM2_suporting | AD 2 | 1A | heterozygous | 0.52 | 181 |
| 1B | homozygous reference | 0.0122 | 164 | ||||
| 1C | homozygous reference | 0 | 140 | ||||
| Family 2: NM_006009.4:c.875C>T NP_006000.2:p.(Thr292Ile) | TUBA1A | Likely Pathogenic PM1_supporting PM2_supporting PP3_strong | AD 2 | 2A | heterozygous | 0.48 | 211 |
| 2B | homozygous reference | 0 | 225 | ||||
| Family 3: NM_006920.6:c.5060A>G NP_008851.3:p.(Glu1687Gly) | SCN1A | VUS PM1_moderate PM2_supporting PP3_moderate | AD 2 | 3A | heterozygous | 0.46 | 194 |
| 3B | homozygous reference | 0 | 231 | ||||
| 3C | homozygous reference | 0 | 205 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aristidou, C.; Theodosiou, A.; Alexandrou, A.; Papaevripidou, I.; Evangelidou, P.; Kosmaidou-Aravidou, Z.; Behjati, F.; Christophidou-Anastasiadou, V.; Tanteles, G.A.; Sismani, C. Exploring the Genetic Causality of Discordant Phenotypes in Familial Apparently Balanced Translocation Cases Using Whole Exome Sequencing. Genes 2023, 14, 82. https://doi.org/10.3390/genes14010082
Aristidou C, Theodosiou A, Alexandrou A, Papaevripidou I, Evangelidou P, Kosmaidou-Aravidou Z, Behjati F, Christophidou-Anastasiadou V, Tanteles GA, Sismani C. Exploring the Genetic Causality of Discordant Phenotypes in Familial Apparently Balanced Translocation Cases Using Whole Exome Sequencing. Genes. 2023; 14(1):82. https://doi.org/10.3390/genes14010082
Chicago/Turabian StyleAristidou, Constantia, Athina Theodosiou, Angelos Alexandrou, Ioannis Papaevripidou, Paola Evangelidou, Zoe Kosmaidou-Aravidou, Farkhondeh Behjati, Violetta Christophidou-Anastasiadou, George A. Tanteles, and Carolina Sismani. 2023. "Exploring the Genetic Causality of Discordant Phenotypes in Familial Apparently Balanced Translocation Cases Using Whole Exome Sequencing" Genes 14, no. 1: 82. https://doi.org/10.3390/genes14010082
APA StyleAristidou, C., Theodosiou, A., Alexandrou, A., Papaevripidou, I., Evangelidou, P., Kosmaidou-Aravidou, Z., Behjati, F., Christophidou-Anastasiadou, V., Tanteles, G. A., & Sismani, C. (2023). Exploring the Genetic Causality of Discordant Phenotypes in Familial Apparently Balanced Translocation Cases Using Whole Exome Sequencing. Genes, 14(1), 82. https://doi.org/10.3390/genes14010082






