Genome-Wide Identification of DUF668 Gene Family and Expression Analysis under Drought and Salt Stresses in Sweet Potato [Ipomoea batatas (L.) Lam]
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Identification and Bioinformatics Analysis of the IbDUF668 Gene
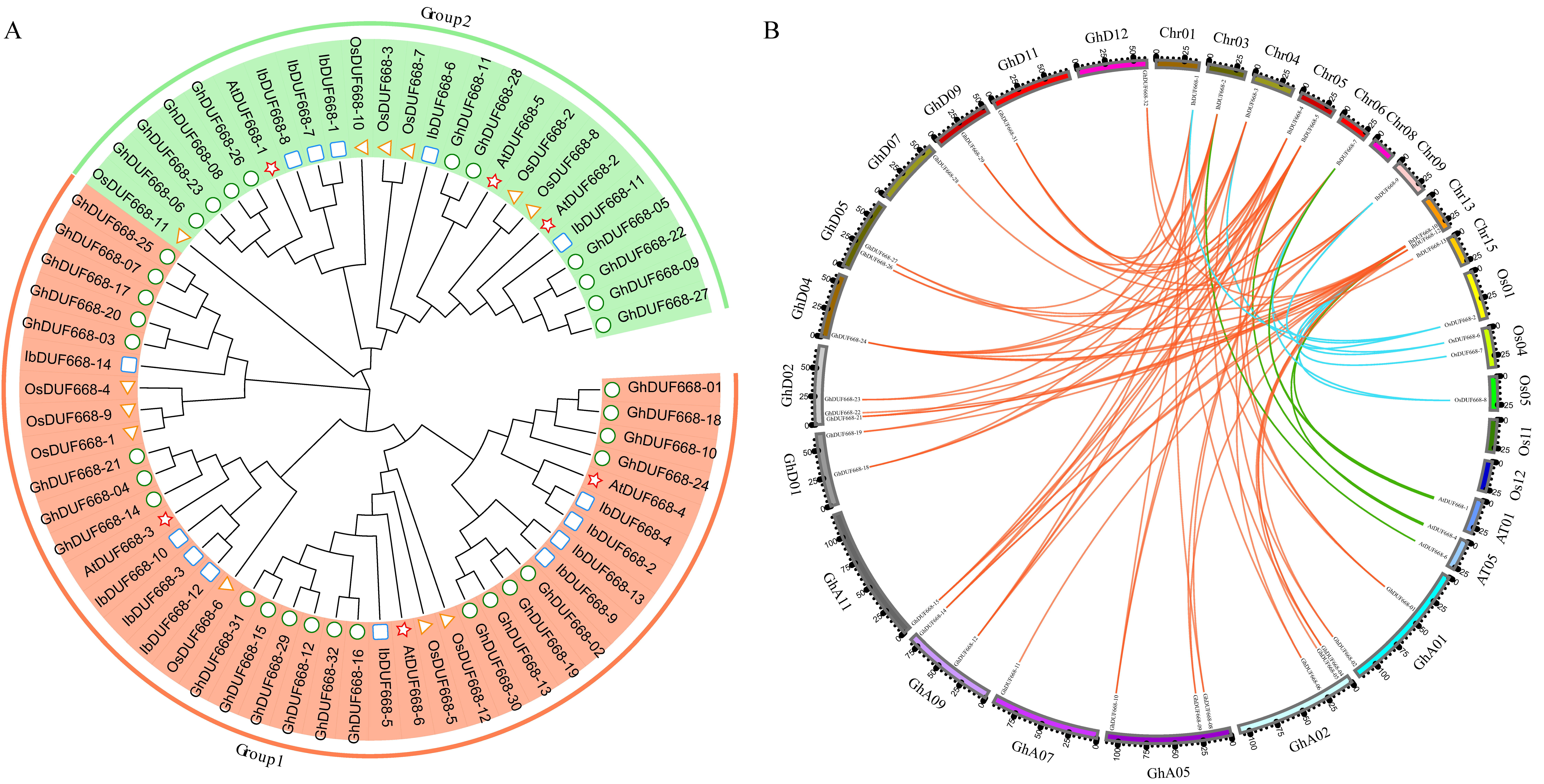
2.3. Collinear Analysis and Phylogenetic of the IbDUF668 Gene Family
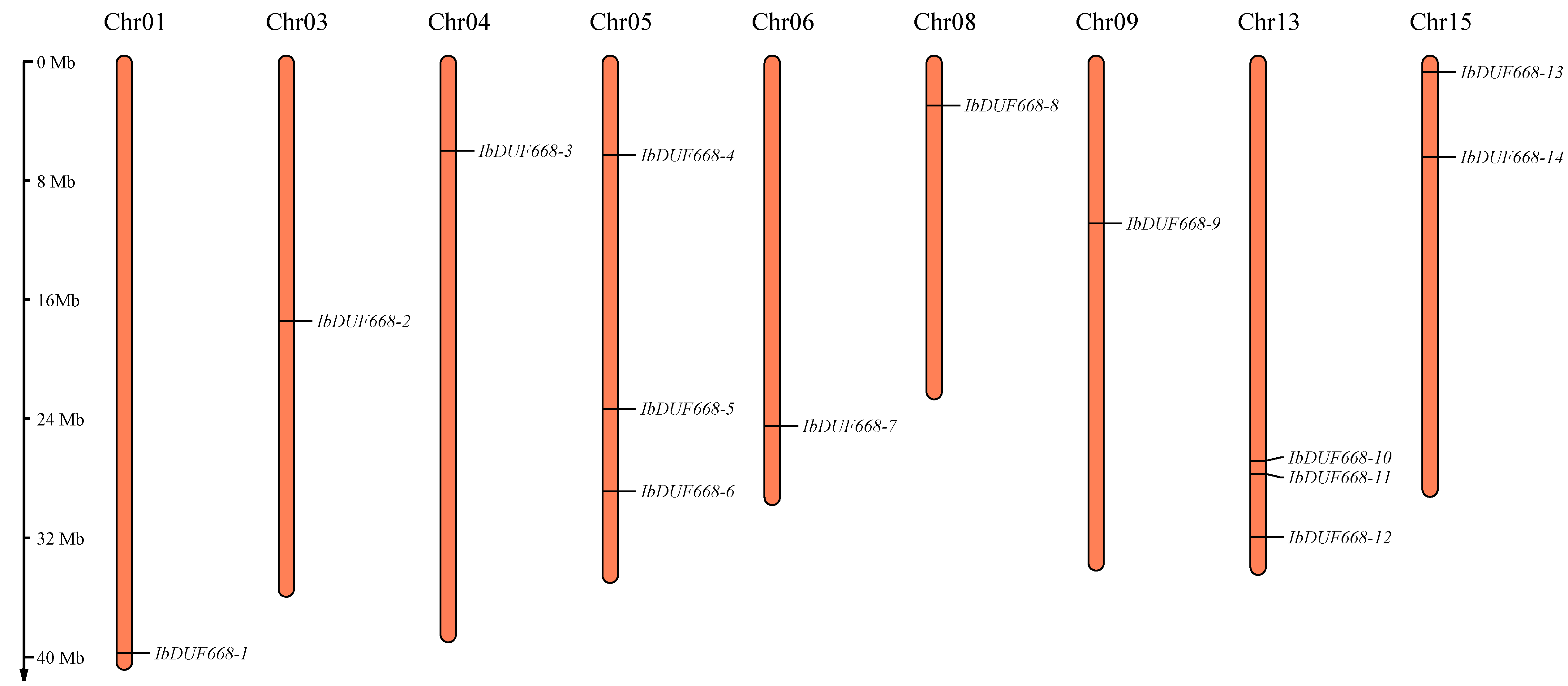
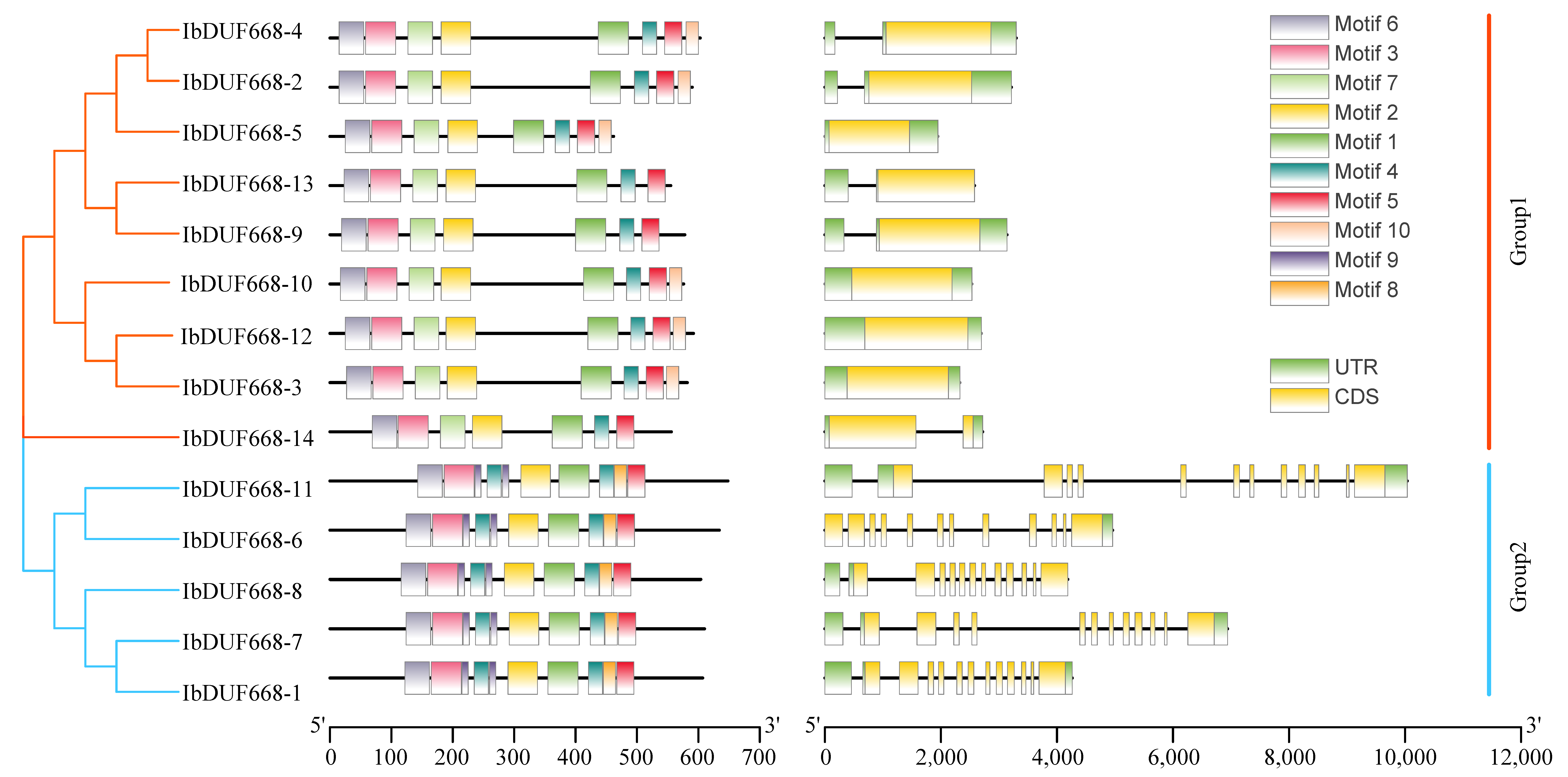
2.4. Chromosomal Location, Gene Structure and Motif Analysis of the IbDUF668 Gene Family
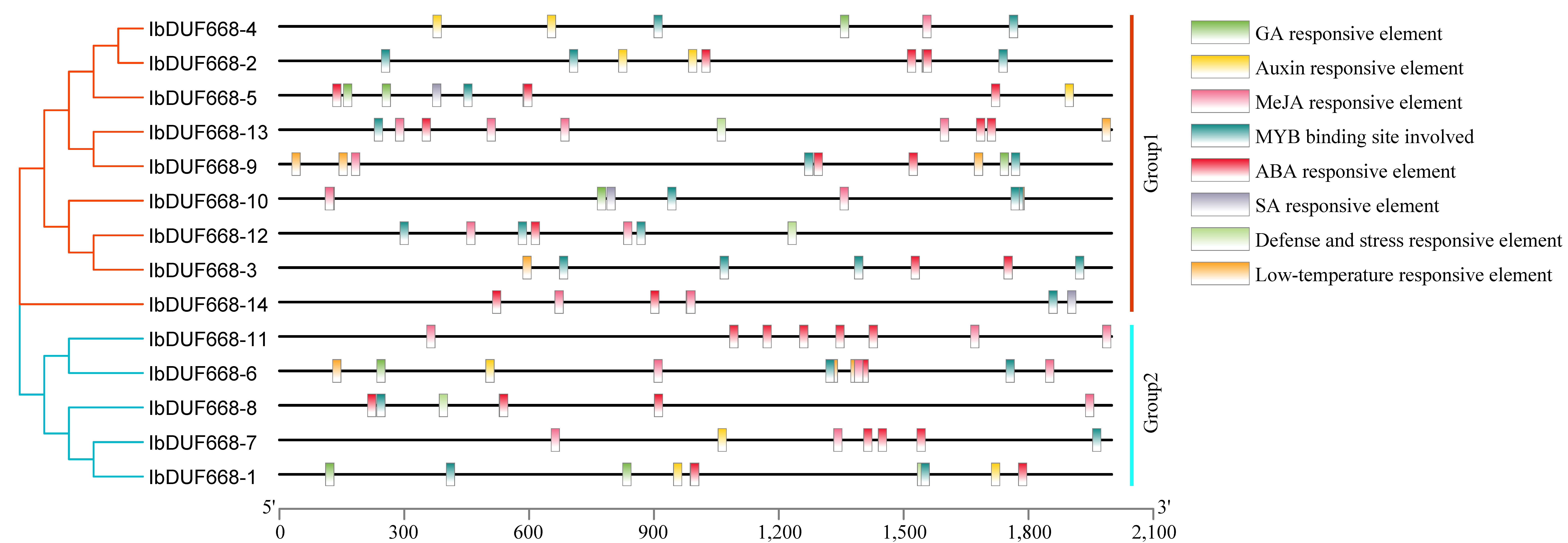
2.5. Analysis of Upstream Cis-Acting Elements of the IbDUF668 Gene
2.6. RNA-Seq Analysis
2.7. Total RNA Extraction, cDNA Synthesis, and qPCR
3. Result
3.1. Identification of DUF668 Gene Family in Sweet Potato
3.2. Evolutionary Analysis of the Sweet Potato DUF668 Gene
3.3. Evolutionary Tree, Gene Structure and Motif Analysis of Sweet Potato DUF668 Gene
3.4. Analysis of Promoter Cis-Acting Elements of DUF668 Gene in Sweet Potato
3.5. RNA-Seq Analysis of the Sweet Potato DUF668 Gene
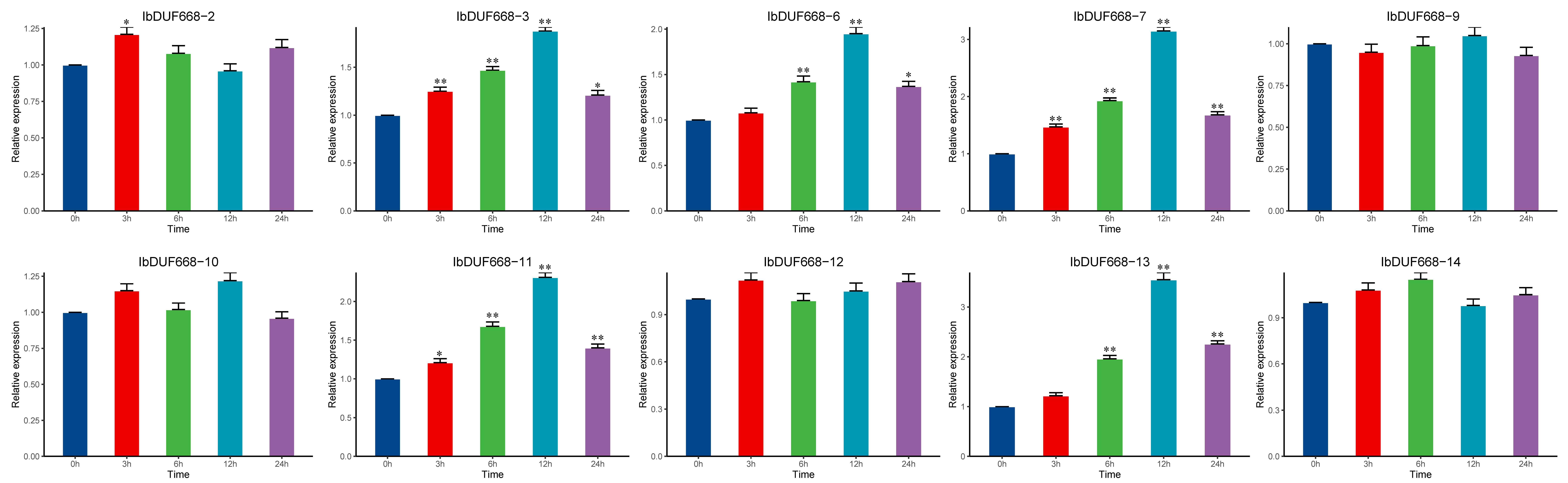
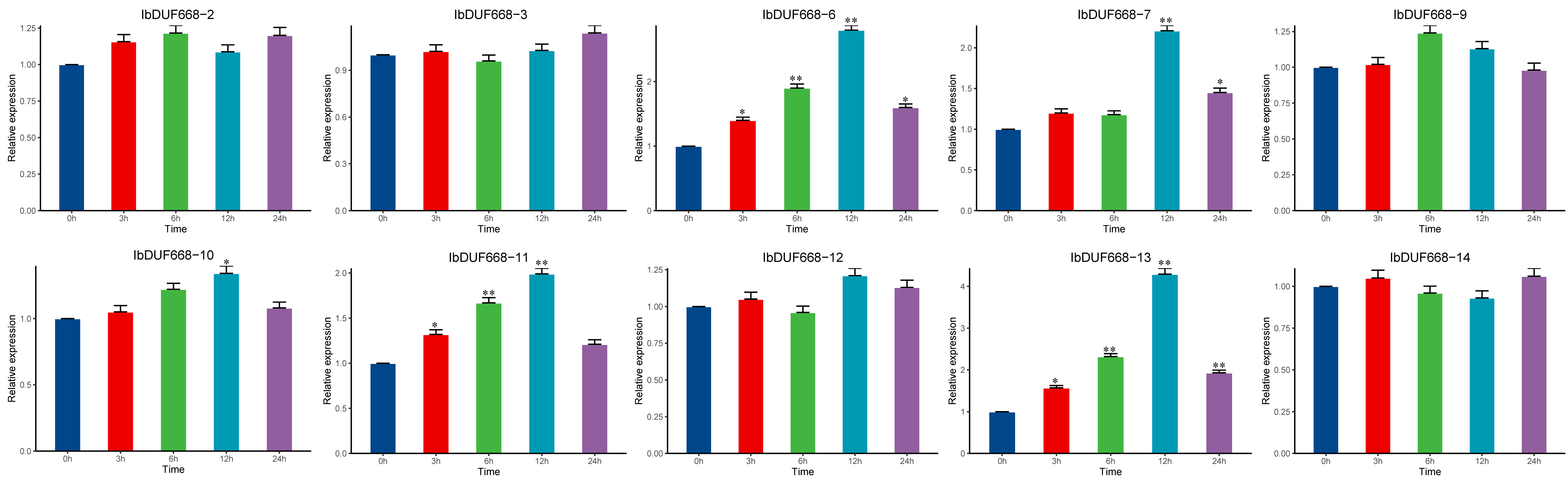
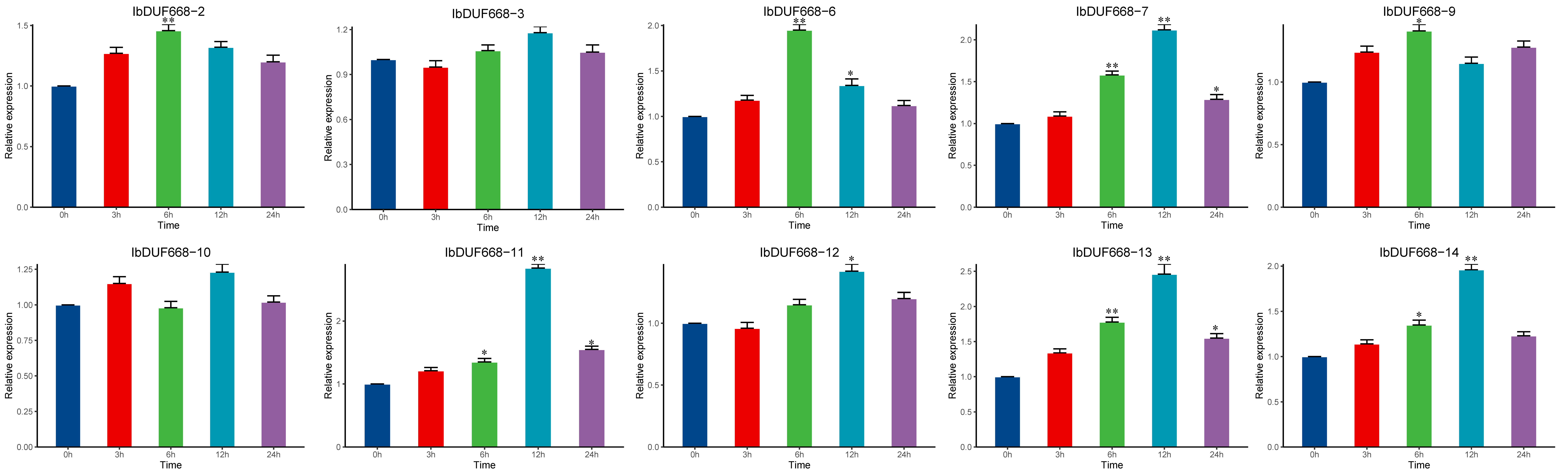
3.6. QRT–PCR of the Sweet Potato DUF668 Gene under Exogenous ABA, Drought and NaCl Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, H.; Zhang, P.; Zhu, Y.; Lou, Q.; He, S. Antioxidant and prebiotic activity of five peonidin-based anthocyanins extracted from purple sweet potato (Ipomoea batatas (L.) Lam.). Sci. Rep. 2018, 8, 5018. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Xie, Y.; Sun, H.; Bian, X.; Ke, Q.; Kim, H.S.; Ji, C.Y.; Jin, R.; Wang, W.; Zhang, C.; et al. IbINH positively regulates drought stress tolerance in sweetpotato. Plant Physiol. Biochem. 2020, 146, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Drought effects on root and tuber production: A meta-analysis. Agric. Water Manag. 2016, 176, 122–131. [Google Scholar] [CrossRef]
- Hong, Z.; Zhou, Y.Y.; Hong, Z.; He, S.Z.; Ning, Z.; Liu, Q.C. Transcriptome profiling reveals insights into the molecular mechanism of drought tolerance in sweetpotato. J. Integr. Agric. 2019, 18, 9–23. [Google Scholar]
- Biazin, B.; Low, J.W.; McEwan, M.A.; Brouwer, R.; Cherinet, M.; Aragaw, A. Unpacking the agroclimatic challenges and determinants of sweetpotato seed conservation and multiplication strategies by smallholder farmers in Southern Ethiopia. Agroecol. Sustain. Food Syst. 2022, 46, 294–315. [Google Scholar] [CrossRef]
- Huan, L.; Jin-Qiang, W.; Qing, L. Photosynthesis product allocation and yield in sweet potato with spraying exogenous hormones under drought stress. J. Plant Physiol. 2020, 253, 153265. [Google Scholar] [CrossRef]
- Yang, J.; Moeinzadeh, M.-H.; Kuhl, H.; Helmuth, J.; Xiao, P.; Haas, S.; Liu, G.; Zheng, J.; Sun, Z.; Fan, W.; et al. Haplotype-resolved sweet potato genome traces back its hexaploidization history. Nat. Plants 2017, 3, 696–703. [Google Scholar] [CrossRef]
- Zhong, H.; Zhang, H.; Guo, R.; Wang, Q.; Huang, X.; Liao, J.; Li, Y.; Huang, Y.; Wang, Z. Characterization and Functional Divergence of a Novel DUF668 Gene Family in Rice Based on Comprehensive Expression Patterns. Genes 2019, 10, 980. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, P.; Gao, W.; Long, Y.; Wang, Y.; Geng, S.; Su, X.; Jiao, Y.; Chen, Q.; Qu, Y. Genome-wide identification of the DUF668 gene family in cotton and expression profiling analysis of GhDUF668 in Gossypium hirsutum under adverse stress. BMC Genom. 2021, 22, 395. [Google Scholar] [CrossRef]
- Zhou, J.; Huang, L.; Lian, J.; Sheng, J.; Cai, J.; Xu, Z. Reconstruction of the UDP-N-acetylglucosamine biosynthetic pathway in cell-free system. Biotechnol. Lett. 2010, 32, 1481–1486. [Google Scholar] [CrossRef]
- Xu, F.; Liu, Z.; Xie, H.; Zhu, J.; Zhang, J.; Kraus, J.; Blaschnig, T.; Nehls, R.; Wang, H. Increased drought tolerance through the suppression of ESKMO1 gene and overexpression of CBF-related genes in Arabidopsis. PLoS ONE 2014, 9, e106509. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ye, T.; Xu, J.; Xie, C.; Gao, X.; Chen, R.; Xu, Z. Molecular characterization and function analysis of the rice OsDUF946 family. Biotechnol. Biotec. Eq. 2017, 31, 477–485. [Google Scholar] [CrossRef]
- Guo, C.; Luo, C.; Guo, L.; Li, M.; Guo, X.; Zhang, Y.; Wang, L.; Chen, L. OsSIDP366, a DUF1644 gene, positively regulates responses to drought and salt stresses in rice. J. Integr. Plant Biol. 2016, 58, 492–502. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Liang, Y.; He, X.; Shen, Y.; Huang, Z. A novel ABA-responsive TaSRHP gene from wheat contributes to enhanced resistance to salt stress in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2013, 31, 791–801. [Google Scholar] [CrossRef]
- Wang, L.; Shen, R.; Chen, L.T.; Liu, Y.G. Characterization of a novel DUF1618 gene family in rice. J. Integr. Plant Biol. 2014, 56, 151–158. [Google Scholar] [CrossRef]
- Yang, Q.; Niu, X.; Tian, X.; Zhang, X.; Cong, J.; Wang, R.; Zhang, G.; Li, G. Comprehensive genomic analysis of the DUF4228 gene family in land plants and expression profiling of ATDUF4228 under abiotic stresses. BMC Genom. 2020, 21, 12. [Google Scholar] [CrossRef]
- Zhou, X.; Zhu, X.; Shao, W.; Song, J.; Jiang, W.; He, Y.; Yin, J.; Ma, D.; Qiao, Y. Genome-Wide Mining of Wheat DUF966 Gene Family Provides New Insights Into Salt Stress Responses. Front. Plant Sci. 2020, 11, 569838. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Chang, T.H.; Wu, L.C.; Lee, T.Y.; Chen, S.P.; Huang, H.D.; Horng, J.T. EuLoc: A web-server for accurately predict protein subcellular localization in eukaryotes by incorporating various features of sequence segments into the general form of Chou’s PseAAC. J. Comput. Aided Mol. Des. 2013, 27, 91–103. [Google Scholar] [CrossRef]
- Kumar, S.; Nei, M.; Dudley, J.; Tamura, K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008, 9, 299–306. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Almeida, A.; Loy, A.; Hofmann, H. ggplot2 Compatible Quantile-Quantile Plots in R. R J. 2018, 10, 248. [Google Scholar] [CrossRef]
- Arocho, A.; Chen, B.; Ladanyi, M.; Pan, Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 2006, 15, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.-S.; Li, M.-J.; Ho, Y.-C.; Yu, C.-P.; Yang, T.-T.; Lin, Y.-J.; Hsing, H.-C.; Chen, T.-K.; Jhong, C.-M.; Li, W.-H.; et al. Rice transcription factor GAMYB modulates bHLH142 and is homeostatically regulated by TDR during anther tapetal and pollen development. J. Exp. Bot. 2021, 72, 4888–4903. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, S.R.; Basu, C. Comparative analyses of plant transcription factor databases. Curr. Genom. 2009, 10, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Sharif, R.; Xie, C.; Wang, J.; Cao, Z.; Zhang, H.; Chen, P.; Yuhong, L. Genome wide identification, characterization and expression analysis of HD-ZIP gene family in Cucumis sativus L. under biotic and various abiotic stresses. Int. J. Biol. Macromol. 2020, 158, 502–520. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.H.; Xia, D.N.; Xu, J.; Guo, D.; Li, H.L.; Wang, Y.; Peng, S.Q. Identification of the bHLH gene family in Dracaena cambodiana reveals candidate genes involved in flavonoid biosynthesis. Ind. Crop. Prod. 2020, 150, 112407. [Google Scholar] [CrossRef]
- Xu, Y.; Xu, H.; Wall, M.M.; Yang, J. Roles of transcription factor SQUAMOSA promoter binding protein-like gene family in papaya (Carica papaya) development and ripening. Genomics 2020, 112, 2734–2747. [Google Scholar] [CrossRef]
- Aviña-Padilla, K.; Ramírez-Rafael, J.A.; Herrera-Oropeza, G.E.; Muley, V.Y.; Valdivia, D.I.; Díaz-Valenzuela, E.; García-García, A.; Varela-Echavarría, A.; Hernández-Rosales, M. Evolutionary Perspective and Expression Analysis of Intronless Genes Highlight the Conservation of Their Regulatory Role. Front. Genet. 2021, 12, 654256. [Google Scholar] [CrossRef]
- Liu, H.; Lyu, H.M.; Zhu, K.; Van de Peer, Y.; Max Cheng, Z.M. The emergence and evolution of intron-poor and intronless genes in intron-rich plant gene families. Plant J. 2021, 105, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Morozov, S.Y.; Solovyev, A.G. Emergence of Intronless Evolutionary Forms of Stress Response Genes: Possible Relation to Terrestrial Adaptation of Green Plants. Front. Plant Sci. 2019, 10, 83. [Google Scholar] [CrossRef]
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar]
- Zhong, H.; Kong, W.; Gong, Z.; Fang, X.; Deng, X.; Liu, C.; Li, Y. Evolutionary Analyses Reveal Diverged Patterns of SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family in Oryza Genus. Front. Plant Sci. 2019, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Hu, W.; Zhou, Y.; Xiao, J.; Hu, R.; Wei, Q.; Zhang, Y.; Feng, J.; Sun, F.; Sun, J.; et al. TaASR1-D confers abiotic stress resistance by affecting ROS accumulation and ABA signalling in transgenic wheat. Plant Biotechnol. J. 2021, 19, 1588–1601. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Zhang, H.; Jin, Y.Y.; Wang, M.M.; Yang, H.Y.; Ma, H.Y.; Liang, Z.W. Abscisic acid primes rice seedlings for enhanced tolerance to alkaline stress by upregulating antioxidant defense and stress tolerance-related genes. Plant Soil 2019, 438, 39–55. [Google Scholar] [CrossRef]
- Batool, A.; Cheng, Z.G.; Akram, N.A.; Lv, G.C.; Xiong, J.L.; Zhu, Y.; Ashraf, M.; Xiong, Y.C. Partial and full root-zone drought stresses account for differentiate root-sourced signal and yield formation in primitive wheat. Plant Methods 2019, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Chakma, P.; Hossain, M.M.; Rabbani, M.G. Effects of salinity stress on seed germination and seedling growth of tomato: Salinity stress on seed germination and seedling growth. J. Bangladesh Agric. Univ. 2019, 17, 490–499. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, H.; Qin, H.; Li, Z.; Liu, H.; Wang, J.; Zhang, H.; Quan, R.; Huang, R.; Zhang, Z. The Synthesis of Ascorbic Acid in Rice Roots Plays an Important Role in the Salt Tolerance of Rice by Scavenging ROS. Int. J. Mol. Sci. 2018, 19, 3347. [Google Scholar] [CrossRef] [PubMed]
- Damaris, R.N.; Li, M.; Liu, Y.; Chen, X.; Murage, H.; Yang, P. A proteomic analysis of salt stress response in seedlings of two African rice cultivars. Biochim. Biophys. Acta 2016, 1864, 1570–1578. [Google Scholar] [CrossRef] [PubMed]
- Daszkowska-Golec, A. ABA is important not only under stress—revealed by the discovery of new ABA transporters. Trends Plant Sci. 2022, 27, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Ma, Z.; Hu, L.; Huang, K.; Zhang, M.; Zhang, S.; Jiang, W.; Wu, T.; Du, X. Involvement of rice transcription factor OsERF19 in response to ABA and salt stress responses. Plant Physiol. Biochem. 2021, 167, 22–30. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene ID | Open Reading Frame/bp | Protein Length/aa | Relative Molecular Weight (r)/kDa | Theoretical Isoelectric Point (pI) | Subcellular Localization |
|---|---|---|---|---|---|---|
| itb01g35230 | IbDUF668-1 | 1824 | 607 | 66.99 | 9.39 | nucleus |
| itb03g18020 | IbDUF668-2 | 1773 | 590 | 66.06 | 9.53 | nucleus |
| itb04g07990 | IbDUF668-3 | 1749 | 582 | 65.52 | 8.16 | nucleus |
| itb05g05910 | IbDUF668-4 | 1812 | 603 | 67.50 | 9.49 | nucleus |
| itb05g14710 | IbDUF668-5 | 1389 | 462 | 51.79 | 10.32 | nucleus |
| itb05g21500 | IbDUF668-6 | 1905 | 634 | 71.40 | 6.73 | nucleus |
| itb06g19930 | IbDUF668-7 | 1833 | 610 | 68.31 | 8.89 | nucleus |
| itb08g03240 | IbDUF668-8 | 1815 | 604 | 66.99 | 9.51 | nucleus |
| itb09g14900 | IbDUF668-9 | 1737 | 578 | 64.62 | 8.99 | endomembrane |
| itb13g18340 | IbDUF668-10 | 1731 | 576 | 65.24 | 7.92 | nucleus |
| itb13g19130 | IbDUF668-11 | 1947 | 648 | 71.84 | 9.93 | nucleus |
| itb13g24470 | IbDUF668-12 | 1779 | 592 | 66.68 | 7.67 | chloroplast |
| itb15g01020 | IbDUF668-13 | 1668 | 555 | 62.17 | 9.94 | organelle membrane |
| itb15g08500 | IbDUF668-14 | 1671 | 556 | 63.17 | 9.58 | chloroplast |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, E.; Li, Z.; Luo, Z.; Xu, L.; Jin, P.; Ji, S.; Zhou, G.; Wang, Z.; Zhou, Z.; Zhang, H. Genome-Wide Identification of DUF668 Gene Family and Expression Analysis under Drought and Salt Stresses in Sweet Potato [Ipomoea batatas (L.) Lam]. Genes 2023, 14, 217. https://doi.org/10.3390/genes14010217
Liu E, Li Z, Luo Z, Xu L, Jin P, Ji S, Zhou G, Wang Z, Zhou Z, Zhang H. Genome-Wide Identification of DUF668 Gene Family and Expression Analysis under Drought and Salt Stresses in Sweet Potato [Ipomoea batatas (L.) Lam]. Genes. 2023; 14(1):217. https://doi.org/10.3390/genes14010217
Chicago/Turabian StyleLiu, Enliang, Zhiqiang Li, Zhengqian Luo, Linli Xu, Ping Jin, Shun Ji, Guohui Zhou, Zhenyang Wang, Zhilin Zhou, and Hua Zhang. 2023. "Genome-Wide Identification of DUF668 Gene Family and Expression Analysis under Drought and Salt Stresses in Sweet Potato [Ipomoea batatas (L.) Lam]" Genes 14, no. 1: 217. https://doi.org/10.3390/genes14010217
APA StyleLiu, E., Li, Z., Luo, Z., Xu, L., Jin, P., Ji, S., Zhou, G., Wang, Z., Zhou, Z., & Zhang, H. (2023). Genome-Wide Identification of DUF668 Gene Family and Expression Analysis under Drought and Salt Stresses in Sweet Potato [Ipomoea batatas (L.) Lam]. Genes, 14(1), 217. https://doi.org/10.3390/genes14010217





