Evaluating the Genetic Effects of Gut Microbiota on the Development of Neuroticism and General Happiness: A Polygenic Score Analysis and Interaction Study Using UK Biobank Data
Abstract
1. Introduction
2. Materials and Methods
2.1. UK Biobank Dataset
2.2. Phenotypes Definition
2.3. GWAS Summaries of Gut Microbiota
2.4. PRS Analysis
2.5. Statistical Analysis
2.6. Gene Set Enrichment Analyses
3. Result
3.1. Basic Characteristics of Study Samples
3.2. Interaction Analysis of Gut Microbiota with Mental Traits
3.3. GO Enrichment Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Antunes, A.; Frasquilho, D.; Azeredo-Lopes, S.; Neto, D.; Silva, M.; Cardoso, G.; Caldas-de-Almeida, J.M. Disability and common mental disorders: Results from the World Mental Health Survey Initiative Portugal. Eur. Psychiatry 2018, 49, 56–61. [Google Scholar] [CrossRef] [PubMed]
- James, S.L.; Abate, D.; Abate, K.H.; Abay, S.M.; Abbafati, C.; Abbasi, N.; Abbastabar, H.; Abd-Allah, F.; Abdela, J.; Abdelalim, A.; et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [Google Scholar] [CrossRef] [PubMed]
- Ormel, J.; Bastiaansen, A.; Riese, H.; Bos, E.H.; Servaas, M.; Ellenbogen, M.; Rosmalen, J.G.M.; Aleman, A. The biological and psychological basis of neuroticism: Current status and future directions. Neurosci. Biobehav. Rev. 2013, 37, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Etkin, A. The genetics of happiness. Sci. Transl. Med. 2016, 8, 359ec159. [Google Scholar] [CrossRef]
- Ormel, J.; Jeronimus, B.F.; Kotov, R.; Riese, H.; Bos, E.H.; Hankin, B.; Rosmalen, J.G.M.; Oldehinkel, A.J. Neuroticism and common mental disorders: Meaning and utility of a complex relationship. Clin. Psychol. Rev. 2013, 33, 686–697. [Google Scholar] [CrossRef] [PubMed]
- Gigantesco, A.; Stazi, M.A.; Alessandri, G.; Medda, E.; Tarolla, E.; Fagnani, C. Psychological well-being (PWB): A natural life outlook? An Italian twin study on heritability of PWB in young adults. Psychol. Med. 2011, 41, 2637–2649. [Google Scholar] [CrossRef]
- Hudson, A.; Wekerle, C.; Goldstein, A.L.; Ellenbogen, S.; Waechter, R.; Thompson, K.; Stewart, S.H. Gender differences in emotion-mediated pathways from childhood sexual abuse to problem drinking in adolescents in the child welfare system. J. Child Adolesc. Trauma 2017, 10, 19–28. [Google Scholar] [CrossRef]
- Rosenthal, M.Z.; Cheavens, J.S.; Lejuez, C.W.; Lynch, T.R. Thought suppression mediates the relationship between negative affect and borderline personality disorder symptoms. Behav. Res. Ther. 2005, 43, 1173–1185. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.R.; Jia, W.; Pettersson, S. Host-Gut Microbiota Metabolic Interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell 2012, 148, 1258–1270. [Google Scholar] [CrossRef]
- Bosch, J.A.; Nieuwdorp, M.; Zwinderman, A.H.; Deschasaux, M.; Radjabzadeh, D.; Kraaij, R.; Davids, M.; de Rooij, S.R.; Lok, A. The gut microbiota and depressive symptoms across ethnic groups. Nat. Commun. 2022, 13, 7129. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-H.; Zhou, C.-H.; Yu, H.; Wu, W.-J.; Wang, Y.-W.; Liu, L.; Hu, G.-T.; Li, B.-J.; Peng, Z.-W.; Wang, H.-N. Gut microbial signatures and differences in bipolar disorder and schizophrenia of emerging adulthood. CNS Neurosci. Ther. 2022, 00, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Mayer, E.A.; Knight, R.; Mazmanian, S.K.; Cryan, J.F.; Tillisch, K. Gut Microbes and the Brain: Paradigm Shift in Neuroscience. J. Neurosci. 2014, 34, 15490–15496. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T.R.; Mazmanian, S.K. Control of Brain Development, Function, and Behavior by the Microbiome. Cell Host Microbe 2015, 17, 565–576. [Google Scholar] [CrossRef]
- Hughes, D.A.; Bacigalupe, R.; Wang, J.; Rühlemann, M.C.; Tito, R.Y.; Falony, G.; Joossens, M.; Vieira-Silva, S.; Henckaerts, L.; Rymenans, L.; et al. Genome-wide associations of human gut microbiome variation and implications for causal inference analyses. Nat. Microbiol. 2020, 5, 1079–1087. [Google Scholar] [CrossRef]
- Visscher, P.M.; Brown, M.A.; McCarthy, M.I.; Yang, J. Five years of GWAS discovery. Am. J. Hum. Genet. 2012, 90, 7–24. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G. Use of allele scores as instrumental variables for Mendelian randomization. Int. J. Epidemiol. 2013, 42, 1134–1144. [Google Scholar] [CrossRef]
- Chen, Y.; Meng, P.; Cheng, S.; Jia, Y.; Wen, Y.; Yang, X.; Yao, Y.; Pan, C.; Li, C.E.; Zhang, H.; et al. Assessing the effect of interaction between C-reactive protein and gut microbiome on the risks of anxiety and depression. Mol. Brain 2021, 14, 133. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, L.; Cheng, S.; Jia, Y.; Wen, Y.; Yang, X.; Meng, P.; Li, C.e.; Pan, C.; Chen, Y.; et al. Assessing the joint effects of brain aging and gut microbiota on the risks of psychiatric disorders. Brain Imaging Behav. 2022, 16, 1504–1515. [Google Scholar] [CrossRef]
- Sudlow, C.; Gallacher, J.; Allen, N.E.; Beral, V.; Burton, P.R.; Danesh, J.; Downey, P.; Elliott, P.; Green, J.; Landray, M.J. UK biobank: An open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015, 12, e1001779. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; Oconnell, J.; Cortes, A. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Canelaxandri, O.; Rawlik, K.; Tenesa, A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018, 50, 1593–1599. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.A.M.; Vattikuti, S.; Purcell, S.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.; Luo, H.; Huo, P.; Wang, Z.; Zhang, S.; He, Z.; Wu, Y.; Zhao, L.; Liu, J.; Guo, J.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Yun, Y.; Ryu, S.; Chang, Y.; Kwon, M.J.; Cho, J.; Shin, H.; Kim, H.L. Correlation between gut microbiota and personality in adults: A cross-sectional study. Brain Behav. Immun. 2018, 69, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Michels, N.; Van de Wiele, T.; Fouhy, F.; O’Mahony, S.; Clarke, G.; Keane, J. Gut microbiome patterns depending on children’s psychosocial stress: Reports versus biomarkers. Brain Behav. Immun. 2019, 80, 751–762. [Google Scholar] [CrossRef]
- Lee, S.-H.; Yoon, S.-H.; Jung, Y.; Kim, N.; Min, U.; Chun, J.; Choi, I. Emotional well-being and gut microbiome profiles by enterotype. Sci. Rep. 2020, 10, 20736. [Google Scholar] [CrossRef]
- Park, E.; Yun, K.E.; Kim, M.-H.; Kim, J.; Chang, Y.; Ryu, S.; Kim, H.-L.; Kim, H.-N.; Jung, S.-C. Correlation between Gut Microbiota and Six Facets of Neuroticism in Korean Adults. J. Pers. Med. 2021, 11, 1246. [Google Scholar] [CrossRef]
- Yang, X.; Yu, B.; Song, C.; Feng, C.; Zhang, J.; Wang, X.; Cheng, G.; Yang, R.; Wang, W.; Zhu, Y. The Effect of Long-Term Moderate Static Magnetic Field Exposure on Adult Female Mice. Biology 2022, 11, 1585. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Chen, Y.; Zhu, H.; Wang, L.; Qian, X.; Zou, R.; Zhao, J.; Zhang, H.; Qian, L.; Wang, Q.; et al. Bifidobacterium breve CCFM1025 attenuates major depression disorder via regulating gut microbiome and tryptophan metabolism: A randomized clinical trial. Brain Behav. Immun. 2022, 100, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Han, B.; Ding, M.; Wen, Y.; Ma, M.; Zhang, L.; Qi, X.; Cheng, B.; Li, P.; Kafle, O.P.; et al. Identifying psychiatric disorder-associated gut microbiota using microbiota-related gene set enrichment analysis. Brief. Bioinform. 2020, 21, 1016–1022. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, L.; Fang, X.; Guo, Z.; Wang, X.; Shi, B.; Meng, Q. Host-microbiota interaction-mediated resistance to inflammatory bowel disease in pigs. Microbiome 2022, 10, 115. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Cai, J.; Zhou, L.; Xing, L.; Zhang, W. Dietary oxidized beef protein alters gut microbiota and induces colonic inflammatory damage in C57BL/6 mice. Front. Nutr. 2022, 9, 980204. [Google Scholar] [CrossRef] [PubMed]
- van de Wouw, M.; Boehme, M.; Lyte, J.M.; Wiley, N.; Strain, C.; O’Sullivan, O.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol. 2018, 596, 4923–4944. [Google Scholar] [CrossRef]
- Yan, F.; Xia, L.; Xu, L.; Deng, L.; Jin, G. A comparative study to determine the association of gut microbiome with schizophrenia in Zhejiang, China. BMC Psychiatry 2022, 22, 731. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Zheng, C.; Xu, X.; Jin, R.; Huang, F.; Shi, M.; He, Z.; Luo, Y.; Liu, L.; Liu, Z.; et al. Clostridium butyricum potentially improves inflammation and immunity through alteration of the microbiota and metabolism of gastric cancer patients after gastrectomy. Front. Immunol. 2022, 13, 1076245. [Google Scholar] [CrossRef]
- Wu, J.; Wang, J.; Lin, Z.; Liu, C.; Zhang, Y.; Zhang, S.; Zhou, M.; Zhao, J.; Liu, H.; Ma, X. Clostridium butyricum alleviates weaned stress of piglets by improving intestinal immune function and gut microbiota. Food Chem. 2023, 405, 135014. [Google Scholar] [CrossRef]
- Peirce, J.M.; Alviña, K. The role of inflammation and the gut microbiome in depression and anxiety. J. Neurosci. Res. 2019, 97, 1223–1241. [Google Scholar] [CrossRef]
- Kong, Q.; Chen, Q.; Mao, X.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Bifidobacterium longum CCFM1077 Ameliorated Neurotransmitter Disorder and Neuroinflammation Closely Linked to Regulation in the Kynurenine Pathway of Autistic-like Rats. Nutrients 2022, 14, 1615. [Google Scholar] [CrossRef] [PubMed]
- Accettulli, A.; Corbo, M.R.; Sinigaglia, M.; Speranza, B.; Campaniello, D.; Racioppo, A.; Altieri, C.; Bevilacqua, A. Psycho-Microbiology, a New Frontier for Probiotics: An Exploratory Overview. Microorganisms 2022, 10, 2141. [Google Scholar] [CrossRef] [PubMed]
- Friedman, H.S. Neuroticism and health as individuals age. Pers. Disord. 2019, 10, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Terracciano, A.; Tanaka, T.; Sutin, A.R.; Sanna, S.; Deiana, B.; Lai, S.; Uda, M.; Schlessinger, D.; Abecasis, G.R.; Ferrucci, L.; et al. Genome-wide association scan of trait depression. Biol. Psychiatry 2010, 68, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Ino, H. Immunohistochemical Characterization of the Orphan Nuclear Receptor RORα in the Mouse Nervous System. J. Histochem. Cytochem. 2004, 52, 311–323. [Google Scholar] [CrossRef]
- Le-Niculescu, H.; Patel, S.D.; Bhat, M.; Kuczenski, R.; Faraone, S.V.; Tsuang, M.T.; McMahon, F.J.; Schork, N.J.; Nurnberger Jr, J.I.; Niculescu Iii, A.B. Convergent functional genomics of genome-wide association data for bipolar disorder: Comprehensive identification of candidate genes, pathways and mechanisms. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2009, 150B, 155–181. [Google Scholar] [CrossRef]
- Soria, V.; Martínez-Amorós, È.; Escaramís, G.; Valero, J.; Pérez-Egea, R.; García, C.; Gutiérrez-Zotes, A.; Puigdemont, D.; Bayés, M.; Crespo, J.M.; et al. Differential Association of Circadian Genes with Mood Disorders: CRY1 and NPAS2 are Associated with Unipolar Major Depression and CLOCK and VIP with Bipolar Disorder. Neuropsychopharmacology 2010, 35, 1279–1289. [Google Scholar] [CrossRef]
- Sato, T.K.; Panda, S.; Miraglia, L.J.; Reyes, T.M.; Rudic, R.D.; McNamara, P.; Naik, K.A.; FitzGerald, G.A.; Kay, S.A.; Hogenesch, J.B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004, 43, 527–537. [Google Scholar] [CrossRef]
- Coles, M.E.; Schubert, J.R.; Nota, J.A. Sleep, Circadian Rhythms, and Anxious Traits. Curr. Psychiatry Rep. 2015, 17, 73. [Google Scholar] [CrossRef]
- Boukhtouche, F.; Vodjdani, G.; Jarvis, C.I.; Bakouche, J.; Staels, B.; Mallet, J.; Mariani, J.; Lemaigre-Dubreuil, Y.; Brugg, B. Human retinoic acid receptor-related orphan receptor alpha1 overexpression protects neurones against oxidative stress-induced apoptosis. J. Neurochem. 2006, 96, 1778–1789. [Google Scholar] [CrossRef]
- Sands, T.T.; Miceli, F.; Lesca, G.; Beck, A.E.; Sadleir, L.G.; Arrington, D.K.; Schönewolf-Greulich, B.; Moutton, S.; Lauritano, A.; Nappi, P.; et al. Autism and developmental disability caused by KCNQ3 gain-of-function variants. Ann. Neurol. 2019, 86, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Kaminsky, Z.; Jones, I.; Verma, R.; Saleh, L.; Trivedi, H.; Guintivano, J.; Akman, R.; Zandi, P.; Lee, R.S.; Potash, J.B. DNA methylation and expression of KCNQ3 in bipolar disorder. Bipolar Disord. 2015, 17, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Gao, Z.; Wang, W.; Li, M. Activation of Kv7 (KCNQ) voltage-gated potassium channels by synthetic compounds. Trends Pharmacol. Sci. 2008, 29, 99–107. [Google Scholar] [CrossRef]
- Wulff, H.; Castle, N.A.; Pardo, L.A. Voltage-gated potassium channels as therapeutic targets. Nat. Rev. Drug Discov. 2009, 8, 982–1001. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.S.; Pan, Z.; Shi, W.; Brown, B.S.; Wymore, R.S.; Cohen, I.S.; Dixon, J.E.; McKinnon, D. KCNQ2 and KCNQ3 potassium channel subunits: Molecular correlates of the M-channel. Science 1998, 282, 1890–1893. [Google Scholar] [CrossRef] [PubMed]
- Costi, S.; Morris, L.S.; Kirkwood, K.A.; Hoch, M.; Corniquel, M.; Vo-Le, B.; Iqbal, T.; Chadha, N.; Pizzagalli, D.A.; Whitton, A.; et al. Impact of the KCNQ2/3 Channel Opener Ezogabine on Reward Circuit Activity and Clinical Symptoms in Depression: Results From a Randomized Controlled Trial. Am. J. Psychiatry 2021, 178, 437–446. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, Z.; Ni, J.; Zhang, Y.; Chen, M.; Cai, J.; Li, X.; Zhang, W.; Zhang, C. Genetic variant in NDUFS1 gene is associated with schizophrenia and negative symptoms in Han Chinese. J. Hum. Genet. 2015, 60, 11–16. [Google Scholar] [CrossRef]
- de Moor, M.H.M.; Berg, S.M.V.D.; Verweij, K.J.H.; Krueger, R.F.; Luciano, M.; Vasquez, A.A.; Matteson, L.K.; Derringer, J.; Esko, T.; Amin, N.; et al. Meta-analysis of Genome-wide Association Studies for Neuroticism, and the Polygenic Association With Major Depressive Disorder. JAMA Psychiatry 2015, 72, 642–650. [Google Scholar] [CrossRef]
- Lopez-Fabuel, I.; Le Douce, J.; Logan, A.; James, A.M.; Bonvento, G.; Murphy, M.P.; Almeida, A.; Bolaños, J.P. Complex I assembly into supercomplexes determines differential mitochondrial ROS production in neurons and astrocytes. Proc. Natl. Acad. Sci. USA 2016, 113, 13063–13068. [Google Scholar] [CrossRef]
- Glessner, J.T.; Wang, K.; Cai, G.; Korvatska, O.; Kim, C.E.; Wood, S.; Zhang, H.; Estes, A.; Brune, C.W.; Bradfield, J.P. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature 2009, 459, 569–572. [Google Scholar] [CrossRef]
- Bernardini, L.; Alesi, V.; Loddo, S.; Novelli, A.; Bottillo, I.; Battaglia, A.; Digilio, M.C.; Zampino, G.; Ertel, A.; Fortina, P. High-resolution SNP arrays in mental retardation diagnostics: How much do we gain? Eur. J. Hum. Genet. 2010, 18, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Lesch, K.P.; Timmesfeld, N.; Renner, T.J.; Halperin, R.; Röser, C.; Nguyen, T.T.; Craig, D.W.; Romanos, J.; Heine, M.; Meyer, J.; et al. Molecular genetics of adult ADHD: Converging evidence from genome-wide association and extended pedigree linkage studies. J. Neural Transm. 2008, 115, 1573–1585. [Google Scholar] [CrossRef] [PubMed]
- Vrijenhoek, T.; Buizervoskamp, J.E.; Der Stelt, I.V.; Strengman, E.; Sabatti, C.; Van Kessel, A.G.; Brunner, H.G.; Ophoff, R.A.; Veltman, J.A. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am. J. Hum. Genet. 2008, 83, 504–510. [Google Scholar] [CrossRef]
- Desmedt, A.; Marighetto, A.; Richter-Levin, G.; Calandreau, L. Adaptive emotional memory: The key hippocampal-amygdalar interaction. Stress 2015, 18, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Van‘t Ent, D.; den Braber, A.; Baselmans, B.M.L.; Brouwer, R.M.; Dolan, C.V.; Hulshoff Pol, H.E.; de Geus, E.J.C.; Bartels, M. Associations between subjective well-being and subcortical brain volumes. Sci. Rep. 2017, 7, 6957. [Google Scholar] [CrossRef]
- Bis, J.C.; DeCarli, C.; Smith, A.V.; van der Lijn, F.; Crivello, F.; Fornage, M.; Debette, S.; Shulman, J.M.; Schmidt, H.; Srikanth, V.; et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat. Genet. 2012, 44, 545–551. [Google Scholar] [CrossRef]
- Hibar, D.P.; Adams, H.H.H.; Jahanshad, N.; Chauhan, G.; Stein, J.L.; Hofer, E.; Renteria, M.E.; Bis, J.C.; Arias-Vasquez, A.; Ikram, M.K.; et al. Novel genetic loci associated with hippocampal volume. Nat. Commun. 2017, 8, 13624. [Google Scholar] [CrossRef]
- Wiegert, O.; Joëls, M.; Krugers, H.J. Corticosteroid hormones, synaptic strength and emotional memories: Corticosteroid modulation of memory—A cellular and molecular perspective. Prog. Brain Res. 2008, 167, 269–271. [Google Scholar]
- Behesti, H.; Fore, T.R.; Wu, P.; Horn, Z.; Leppert, M.; Hull, C.; Hatten, M.E. ASTN2 modulates synaptic strength by trafficking and degradation of surface proteins. Proc. Natl. Acad. Sci. USA 2018, 115, E9717–E9726. [Google Scholar] [CrossRef]
- Kim, S.E.; Kim, H.-N.; Yun, Y.-J.; Heo, S.G.; Cho, J.; Kwon, M.-J.; Chang, Y.; Ryu, S.; Shin, H.; Shin, C.; et al. Meta-analysis of genome-wide SNP- and pathway-based associations for facets of neuroticism. J. Hum. Genet. 2017, 62, 903–909. [Google Scholar] [CrossRef]
- Fan, Y.; Abrahamsen, G.; Mills, R.; Calderón, C.C.; Tee, J.Y.; Leyton, L.; Murrell, W.; Cooper-White, J.; McGrath, J.J.; Mackay-Sim, A. Focal adhesion dynamics are altered in schizophrenia. Biol. Psychiatry 2013, 74, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Kamimura, K.; Maeda, N. Glypicans and Heparan Sulfate in Synaptic Development, Neural Plasticity, and Neurological Disorders. Front. Neural Circuits 2021, 15, 595596. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Goemans, C.V.; Wirbel, J.; Kuhn, M.; Eberl, C.; Pruteanu, M.; Müller, P.; Garcia-Santamarina, S.; Cacace, E.; Zhang, B.; et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature 2021, 599, 120–124. [Google Scholar] [CrossRef] [PubMed]
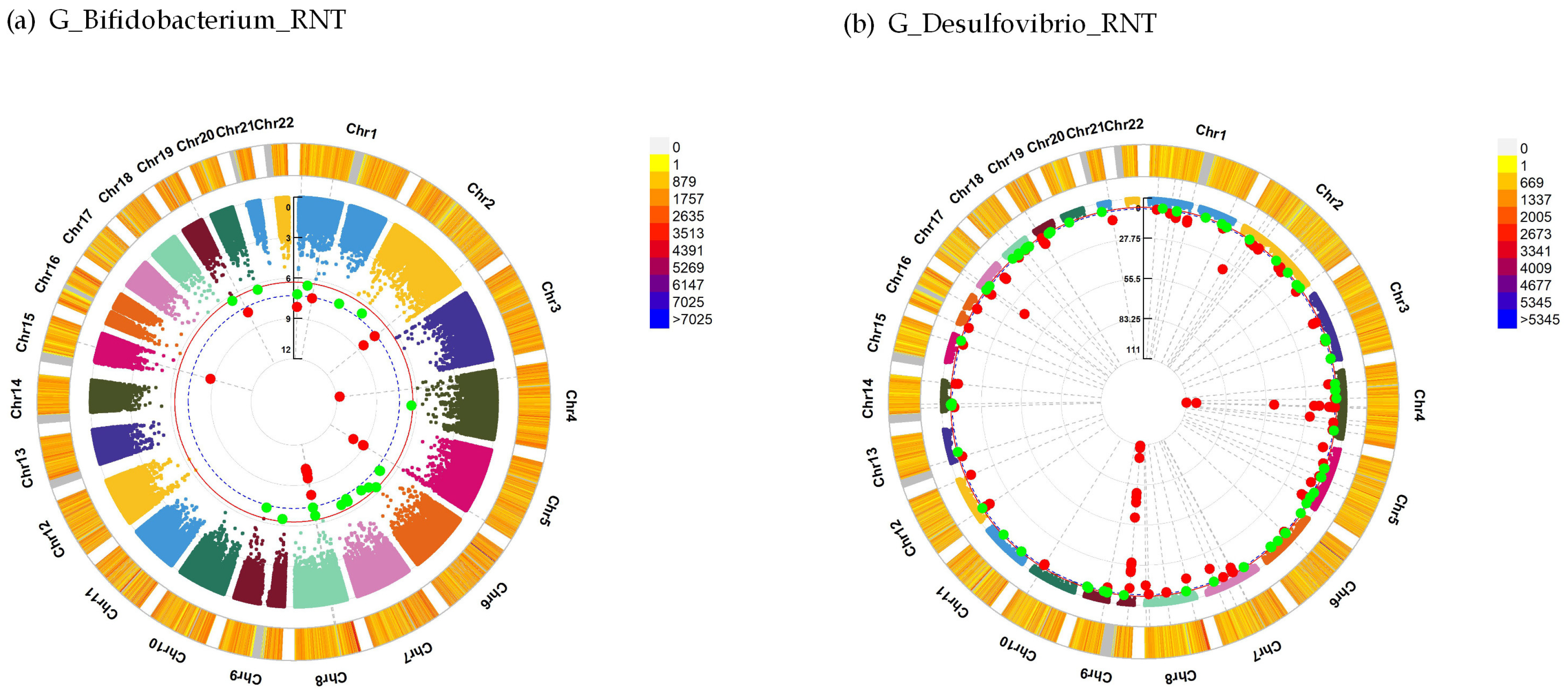
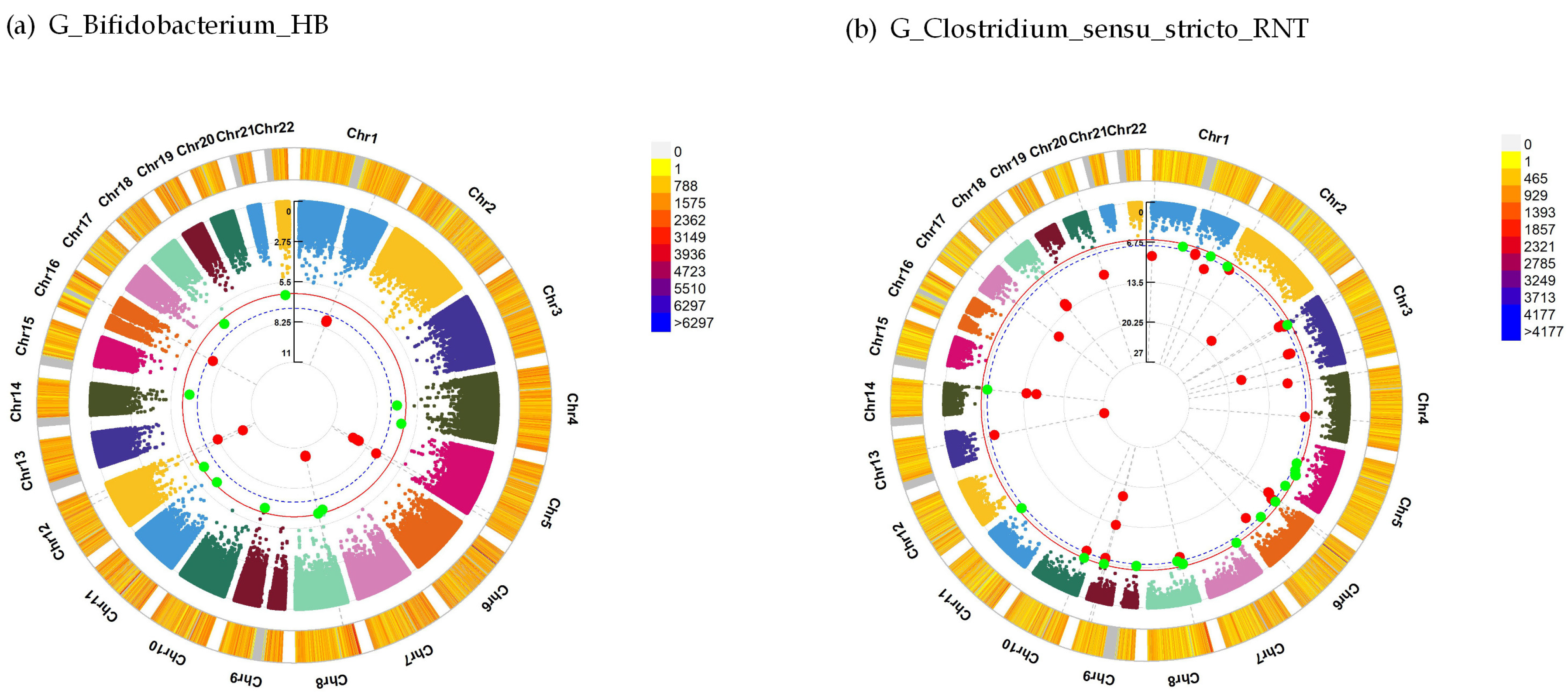
| Mental Traits | Gut Microbiota Traits | Sample Size | Sex (Female) | Age ± SD | Beta | SE | R2 | P |
|---|---|---|---|---|---|---|---|---|
| Neuroticism | G_Bifidobacterium_RNT | 306,161 | 161,977 | 56.81 ± 7.93 | −1.10 | 0.38 | 4.16 × 10−3 | 4.16 × 10−3 |
| G_Desulfovibrio_RNT | 153,483 | 81,310 | 56.80 ± 7.93 | 0.54 | 0.20 | 7.46 × 10−3 | 7.46 × 10−3 | |
| General happiness | G_Bifidobacterium_HB | 89,206 | 49,806 | 56.27 ± 7.62 | 1.99 | 0.71 | 5.23 × 10−3 | 5.24 × 10−3 |
| G_Clostridium_sensu_stricto_RNT | 115,013 | 64,229 | 56.28 ± 7.61 | 1.26 | 0.49 | 9.27 × 10−3 | 9.27 × 10−3 |
| Chromosome | Gene | Beta | P | |
|---|---|---|---|---|
| G_Bifidobacterium_RNT | 5 | GRK6 | −19.43 | 1.10 × 10−10 |
| 2 | NDUFS1 | −15.60 | 3.71 × 10−8 | |
| 5 | NSD1 | −6.28 | 7.91 × 10−10 | |
| 15 | RORA | −45.00 | 1.82 × 10−9 | |
| G_Desulfovibrio_RNT | 15 | ARNT2 | −6.40 | 7.16 × 10−10 |
| 14 | CCDC85C | −12.94 | 7.27 × 10−10 | |
| 4 | CCNG2 | −9.75 | 2.73 × 10−8 | |
| 19 | CERS4 | −7.55 | 1.21 × 10−10 | |
| 13 | FLT3 | −4.94 | 8.55 × 10−10 | |
| 8 | KCNQ3 | −14.79 | 2.62 × 10−8 | |
| 4 | LAMTOR3 | −13.25 | 2.09 × 10−50 | |
| 4 | MANBA | −9.25 | 1.18 × 10−11 | |
| 8 | MSR1 | 5.15 | 1.47 × 10−15 | |
| 4 | NR3C2 | −13.59 | 1.49 × 10−8 | |
| 12 | PPFIBP1 | −3.43 | 3.27 × 10−13 | |
| 1 | PRSS38 | −6.85 | 2.76 × 10−9 | |
| 2 | RAMP1 | −7.73 | 6.59 × 10−11 | |
| 12 | RIMBP2 | −5.15 | 4.32 × 10−11 | |
| 4 | SLC9B1 | −10.70 | 3.01 × 10−12 | |
| 7 | TPST1 | −15.90 | 5.34 × 10−10 | |
| 4 | UBE2D3 | −9.78 | 3.03 × 10−12 | |
| 19 | ZNF317 | −12.96 | 1.01 × 10−12 |
| Chromosome | Gene | Beta | P | |
|---|---|---|---|---|
| G_Bifidobacterium_HB | 5 | DCTN4 | 41.95 | 9.23 × 10−10 |
| 12 | MAP1LC3B2 | −15.80 | 6.11 × 10−9 | |
| 5 | MYOZ3 | 43.12 | 4.74 × 10−10 | |
| 16 | PRSS54 | −11.58 | 2.38 × 10−8 | |
| 1 | RGS21 | 14.30 | 1.67 × 10−8 | |
| G_Clostridium_sensu_stricto_RNT | 9 | ASTN2 | 19.15 | 3.37 × 10−8 |
| 1 | ATF6 | 8.28 | 2.54 × 10−8 | |
| 6 | BMP6 | 7.55 | 1.98 × 10−9 | |
| 1 | CAMTA1 | 9.60 | 8.80 × 10−10 | |
| 3 | CCDC14 | 13.67 | 1.72 × 10−9 | |
| 3 | ECT2 | 14.69 | 2.17 × 10−18 | |
| 2 | FMNL2 | 7.78 | 1.66 × 10−19 | |
| 1 | LAMB3 | 10.86 | 4.61 × 10−10 | |
| 3 | MYLK | 13.61 | 3.95 × 10−9 | |
| 17 | RHBDL3 | 8.91 | 3.35 × 10−16 | |
| 9 | SVEP1 | 12.88 | 5.36 × 10−19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, Y.; Cheng, S.; Liu, L.; Cheng, B.; Liang, C.; Ye, J.; Chu, X.; Yao, Y.; Wen, Y.; Kafle, O.P.; et al. Evaluating the Genetic Effects of Gut Microbiota on the Development of Neuroticism and General Happiness: A Polygenic Score Analysis and Interaction Study Using UK Biobank Data. Genes 2023, 14, 156. https://doi.org/10.3390/genes14010156
Jia Y, Cheng S, Liu L, Cheng B, Liang C, Ye J, Chu X, Yao Y, Wen Y, Kafle OP, et al. Evaluating the Genetic Effects of Gut Microbiota on the Development of Neuroticism and General Happiness: A Polygenic Score Analysis and Interaction Study Using UK Biobank Data. Genes. 2023; 14(1):156. https://doi.org/10.3390/genes14010156
Chicago/Turabian StyleJia, Yumeng, Shiqiang Cheng, Li Liu, Bolun Cheng, Chujun Liang, Jing Ye, Xiaomeng Chu, Yao Yao, Yan Wen, Om Prakash Kafle, and et al. 2023. "Evaluating the Genetic Effects of Gut Microbiota on the Development of Neuroticism and General Happiness: A Polygenic Score Analysis and Interaction Study Using UK Biobank Data" Genes 14, no. 1: 156. https://doi.org/10.3390/genes14010156
APA StyleJia, Y., Cheng, S., Liu, L., Cheng, B., Liang, C., Ye, J., Chu, X., Yao, Y., Wen, Y., Kafle, O. P., & Zhang, F. (2023). Evaluating the Genetic Effects of Gut Microbiota on the Development of Neuroticism and General Happiness: A Polygenic Score Analysis and Interaction Study Using UK Biobank Data. Genes, 14(1), 156. https://doi.org/10.3390/genes14010156







