MYC Causes Multiple Myeloma Progression via Attenuating TP53-Induced MicroRNA-34 Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Patients
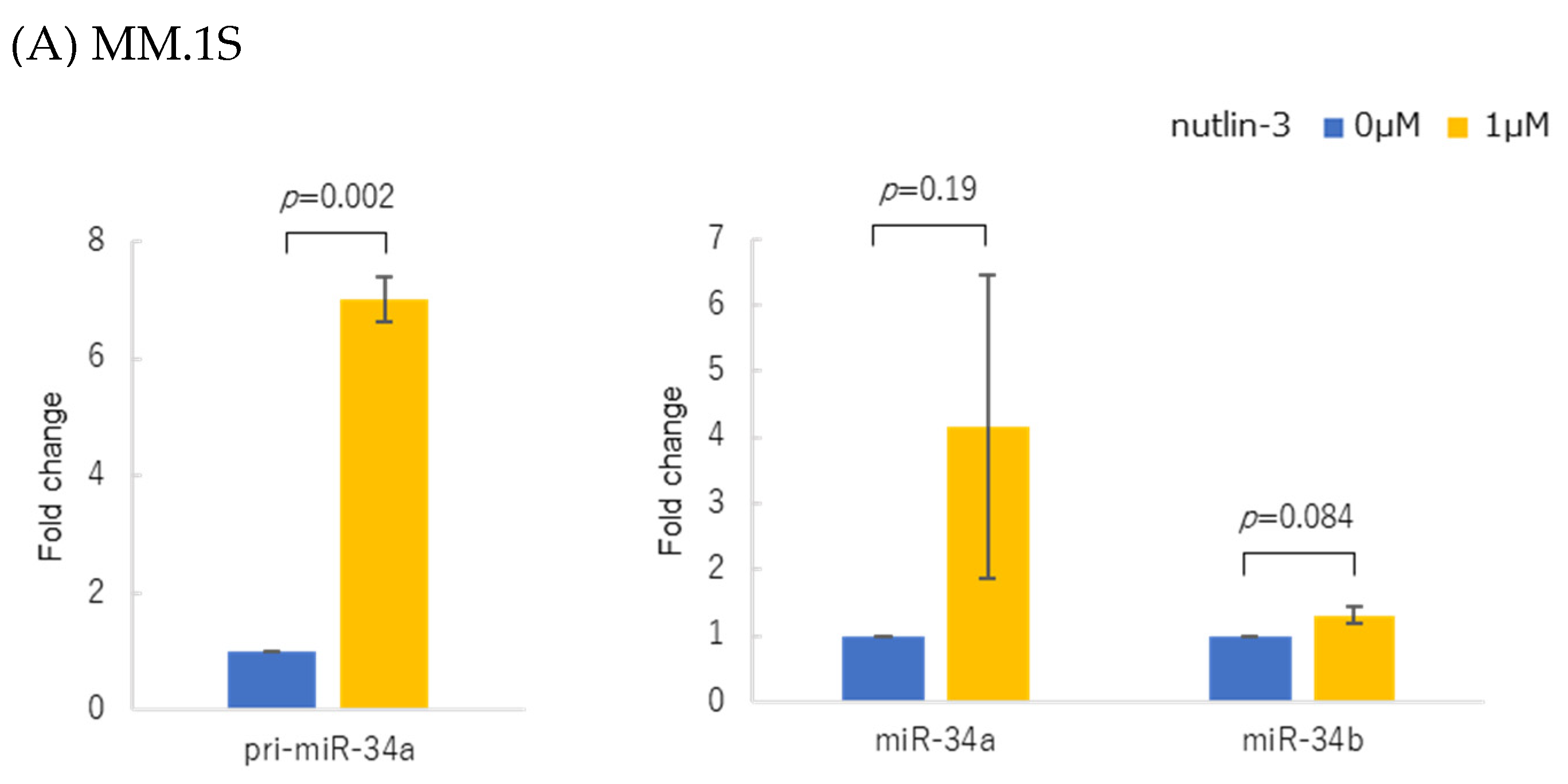
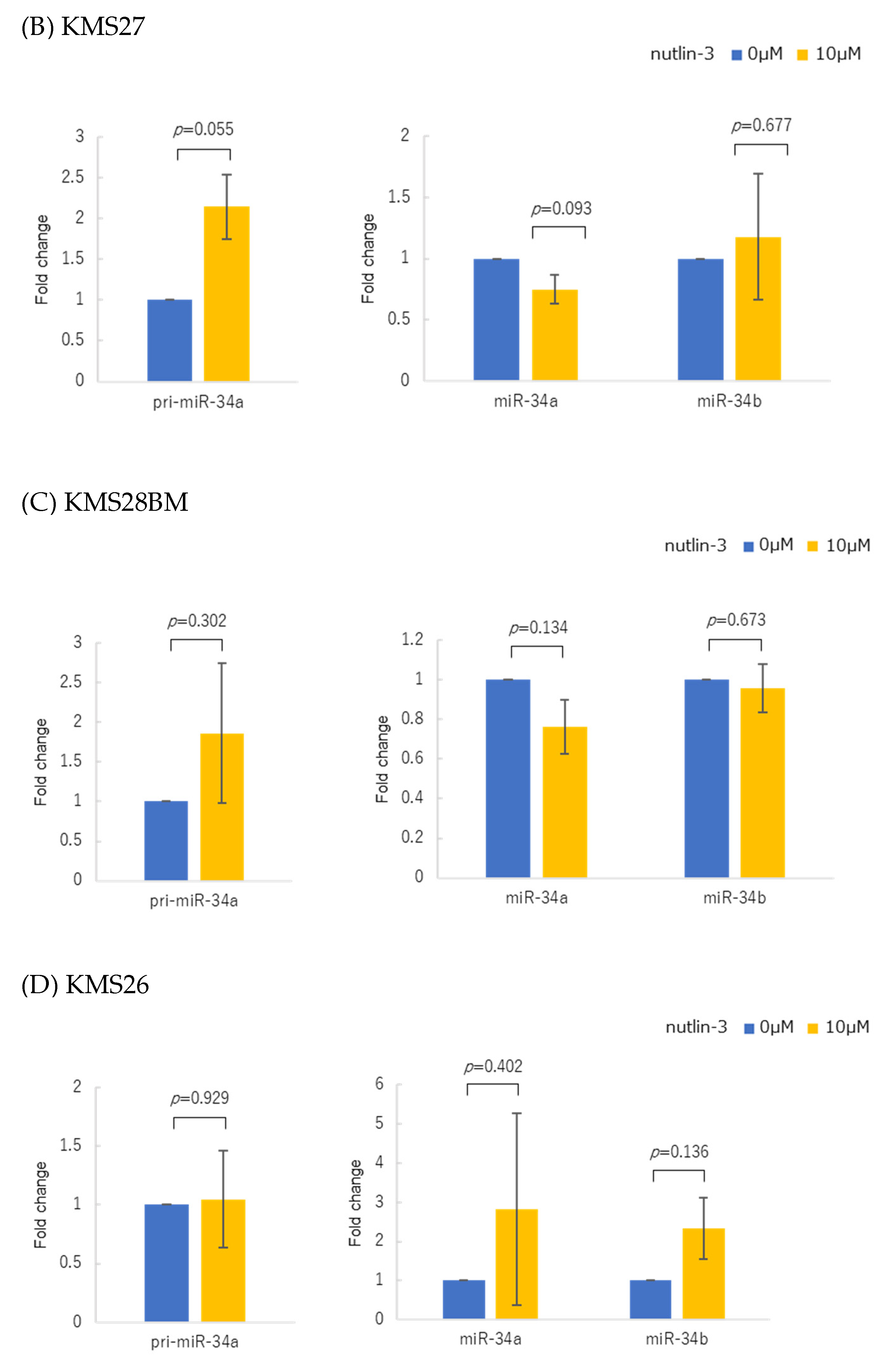
2.3. Treatment with MDM2 Inhibitor Nutlin-3
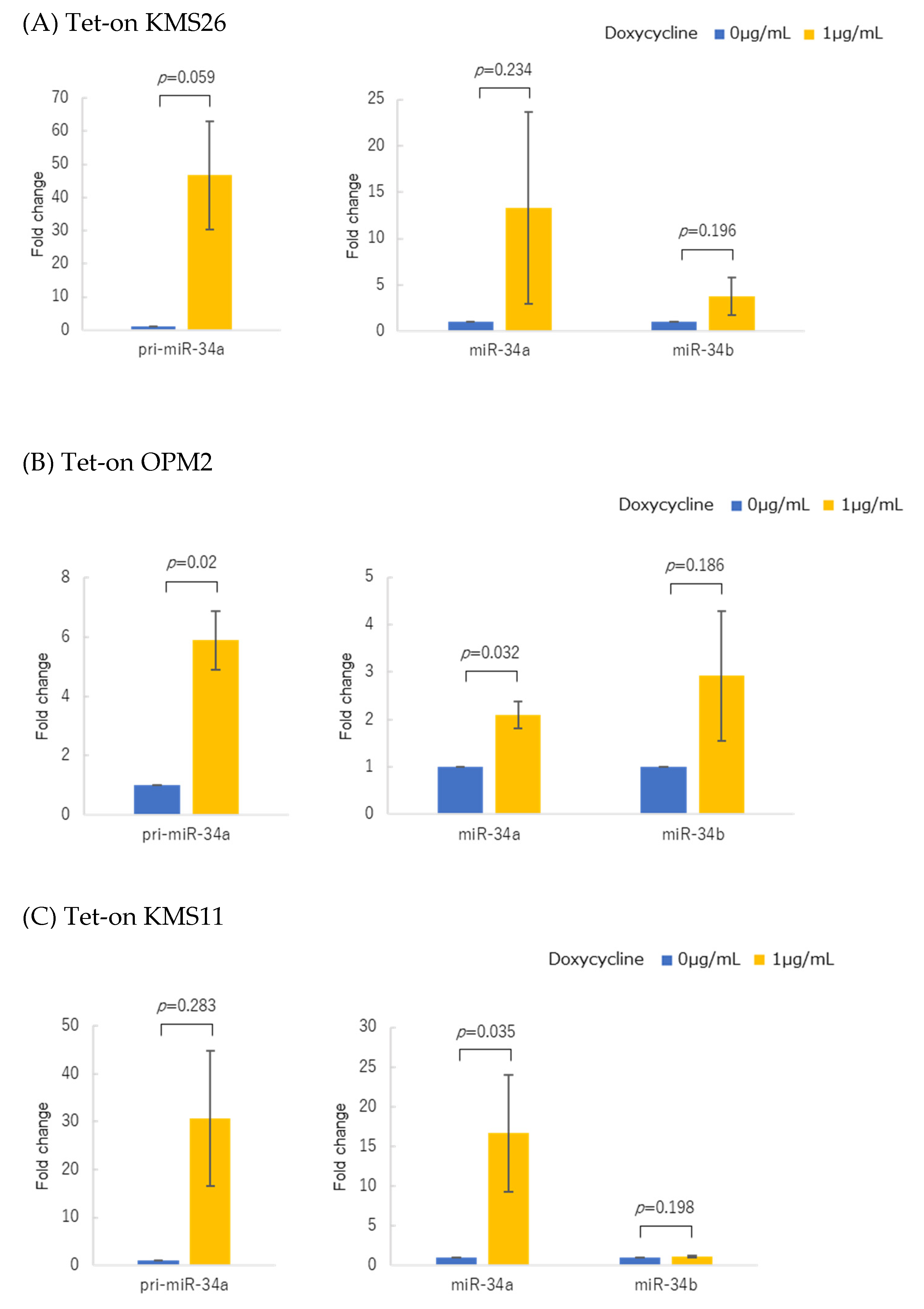
2.4. p53 Overexpression Using Tet-On System
2.5. Treatment with Myc Inhibitor
2.6. MYC Activation in MYC-ER Cell Lines
2.7. Isolation of Nucleic Acids
2.8. Real-Time PCR
2.9. Western Blot
2.10. Apoptosis Analysis
2.11. Statistical Analysis
3. Results
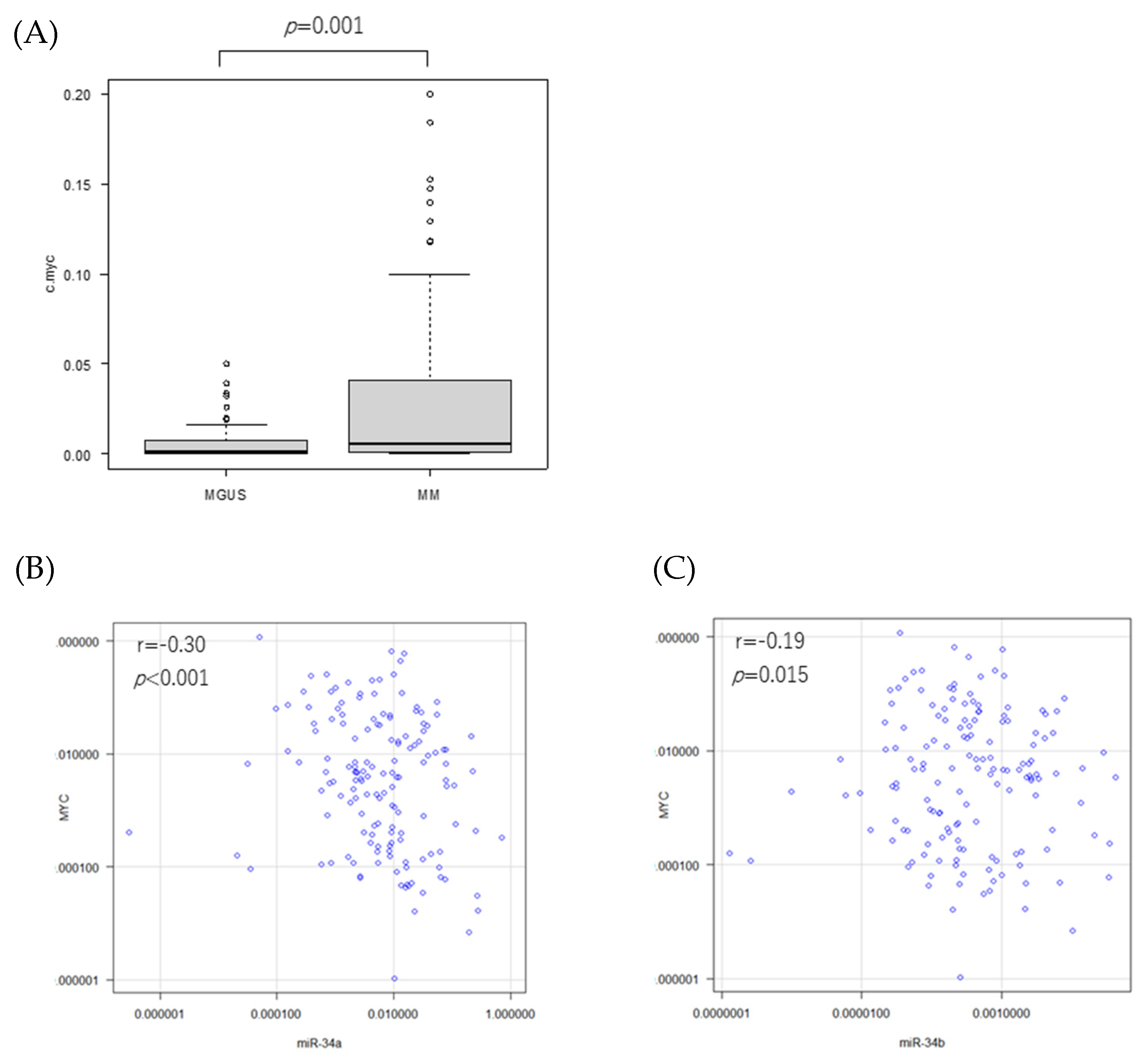
3.1. miR-34 Family and TP53 mRNA Expressions and Their Correlations in the Patients
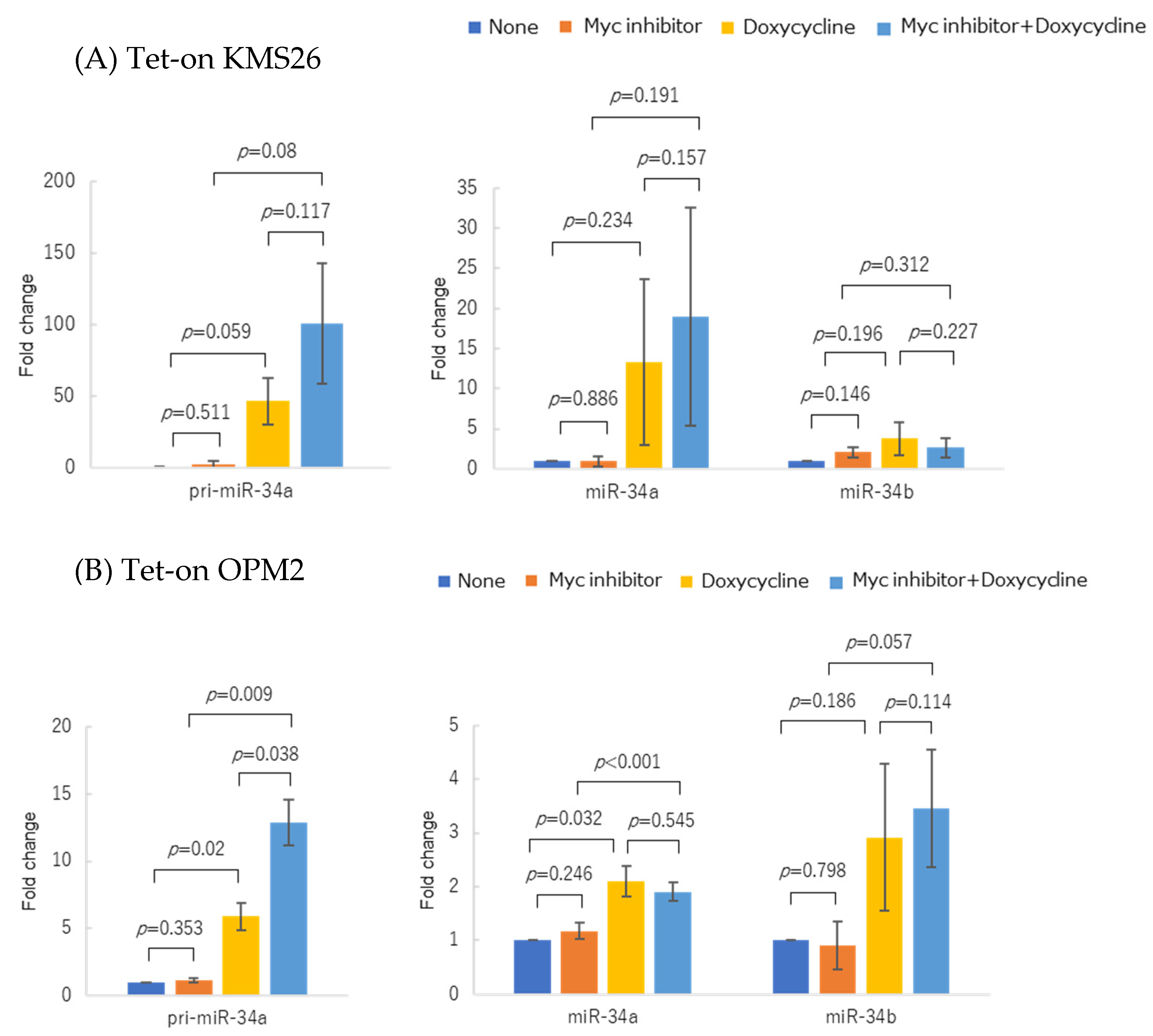
3.2. p53 Protein Accumulation and p53 Overexpression Upregulated Primary and Mature miR-34 in Human Multiple Myeloma Cell Lines (HMCLs)
3.3. MYC mRNA Expression in Patients
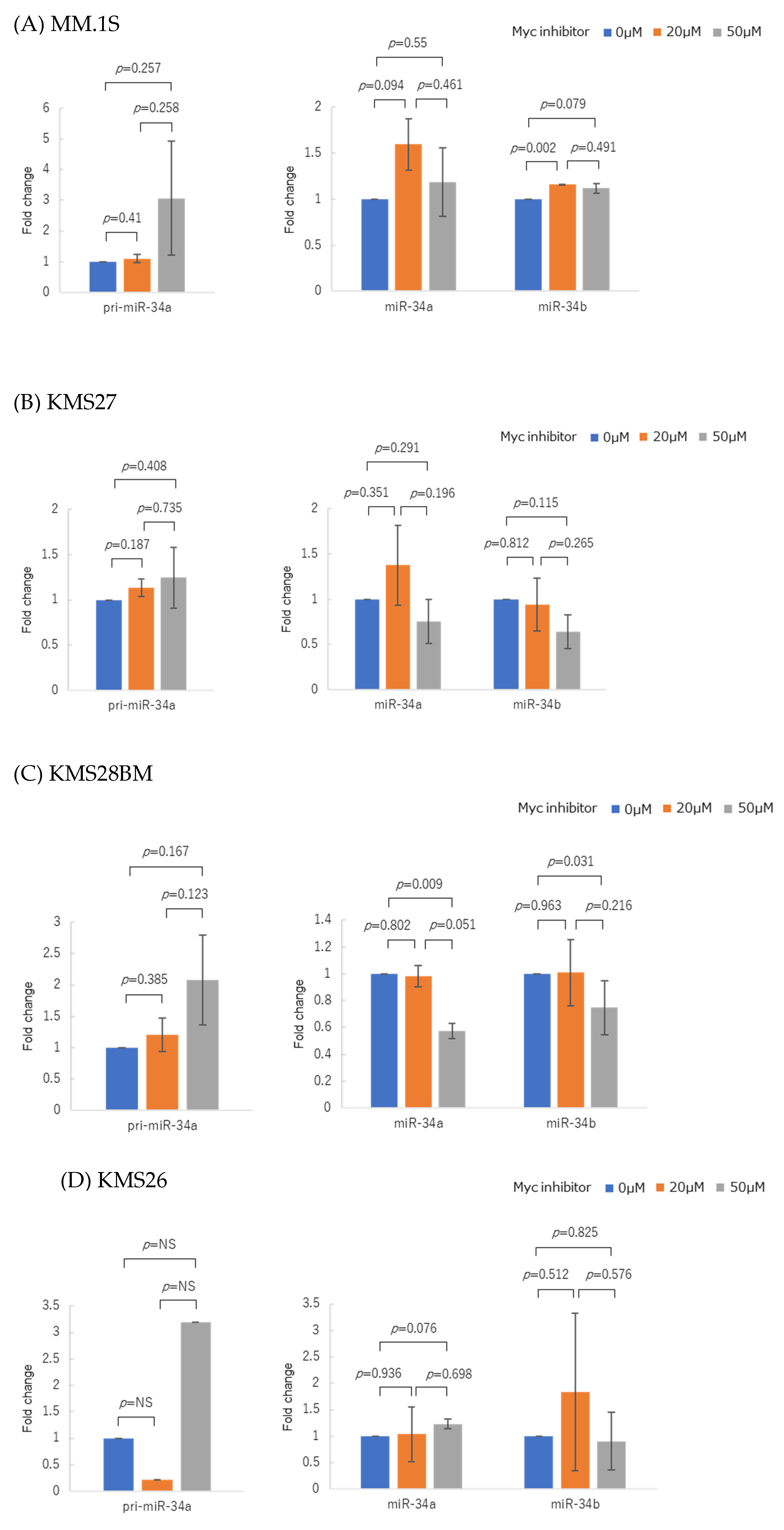
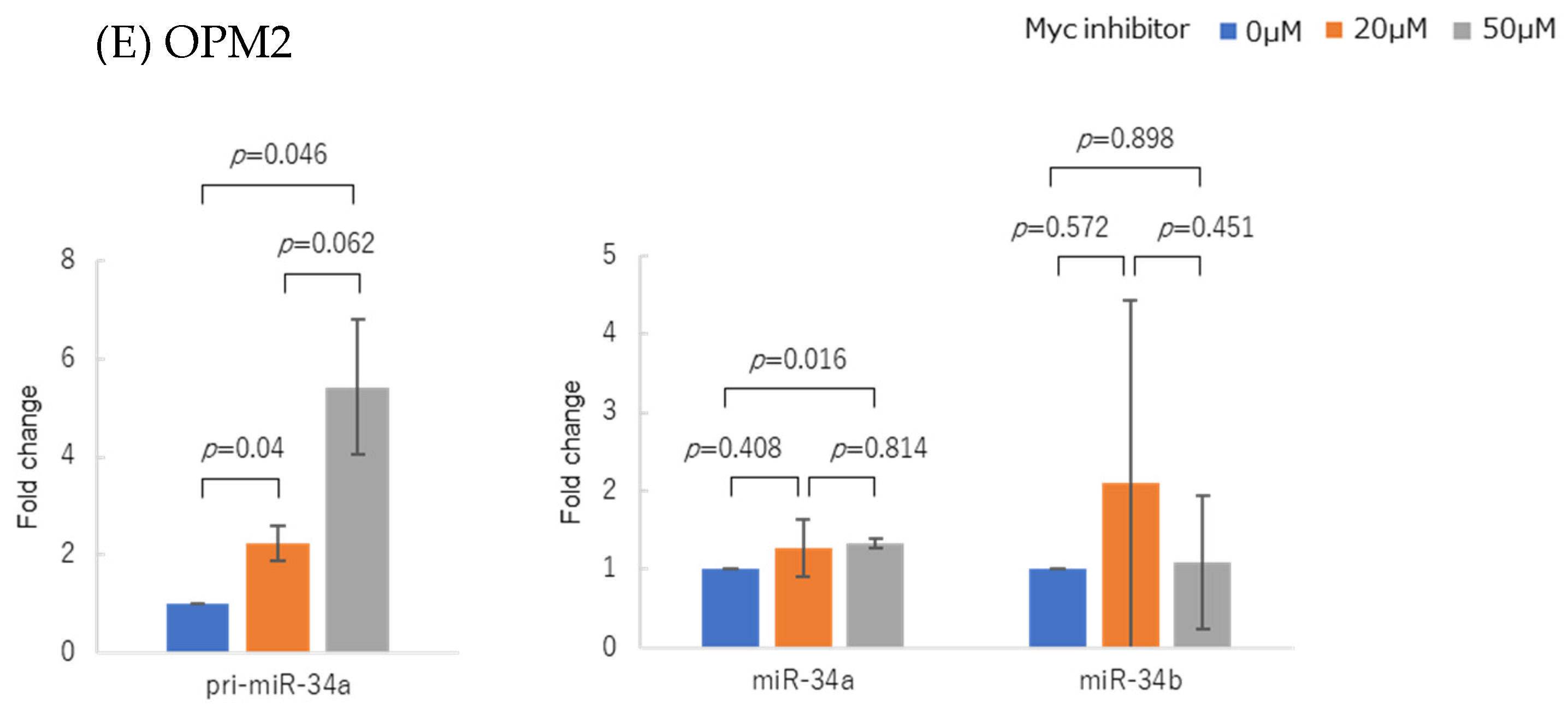
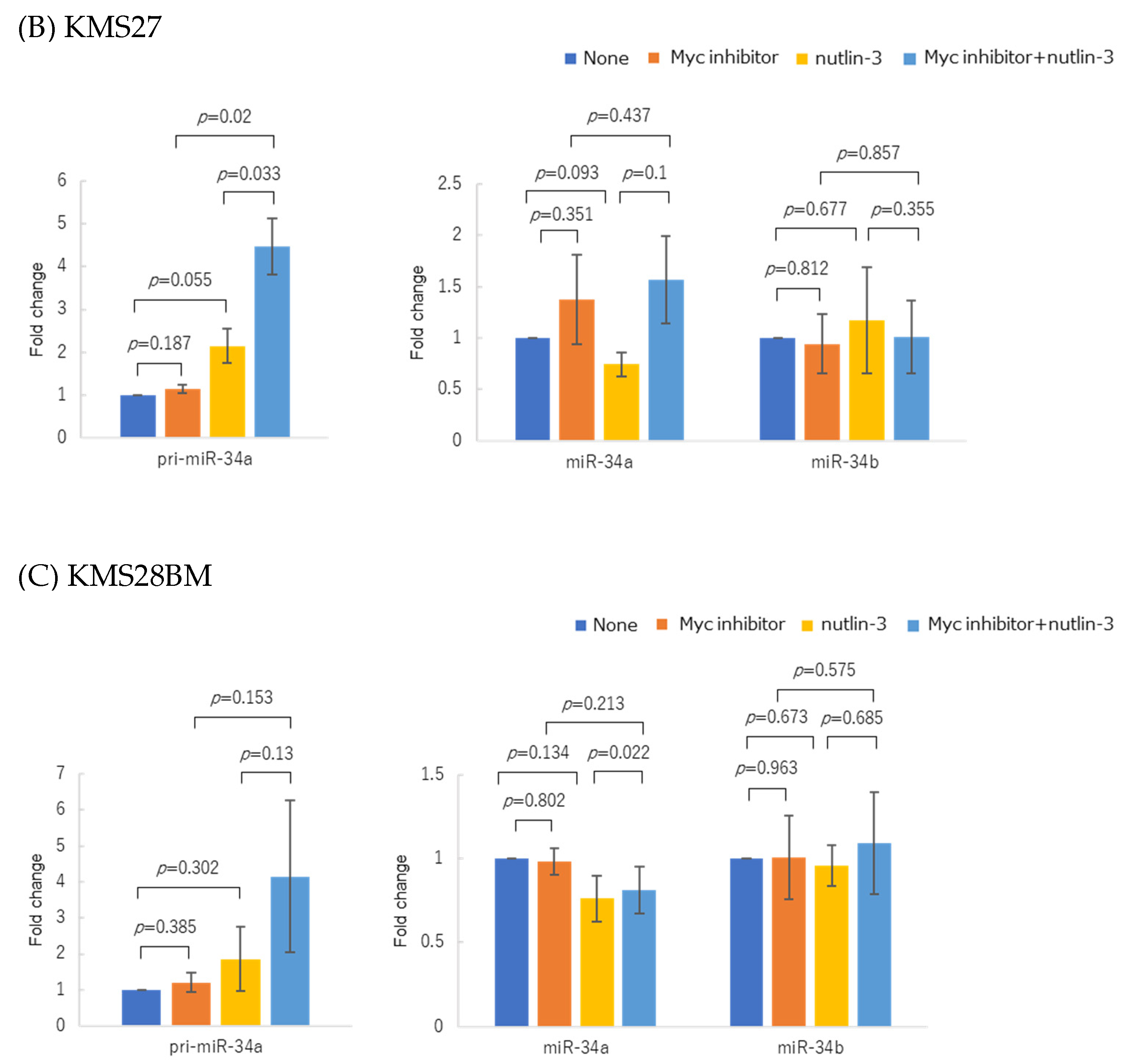
3.4. Myc Inhibitor Alone Did Not Change miR-34 Family Expression in Most HMCLs
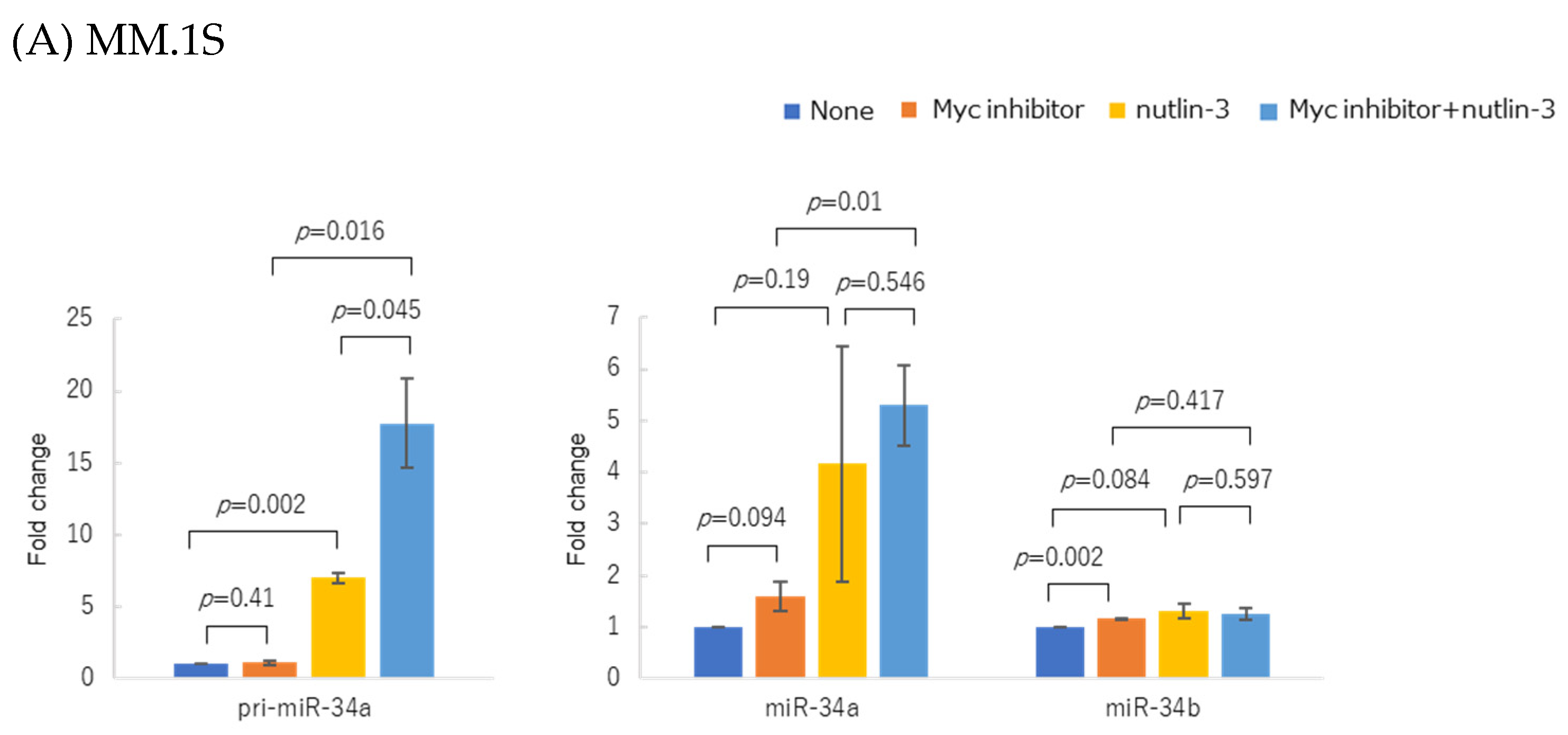
3.5. WT p53 Accumulation and Myc Inhibition Synergistically Upregulated miR-34 Expression
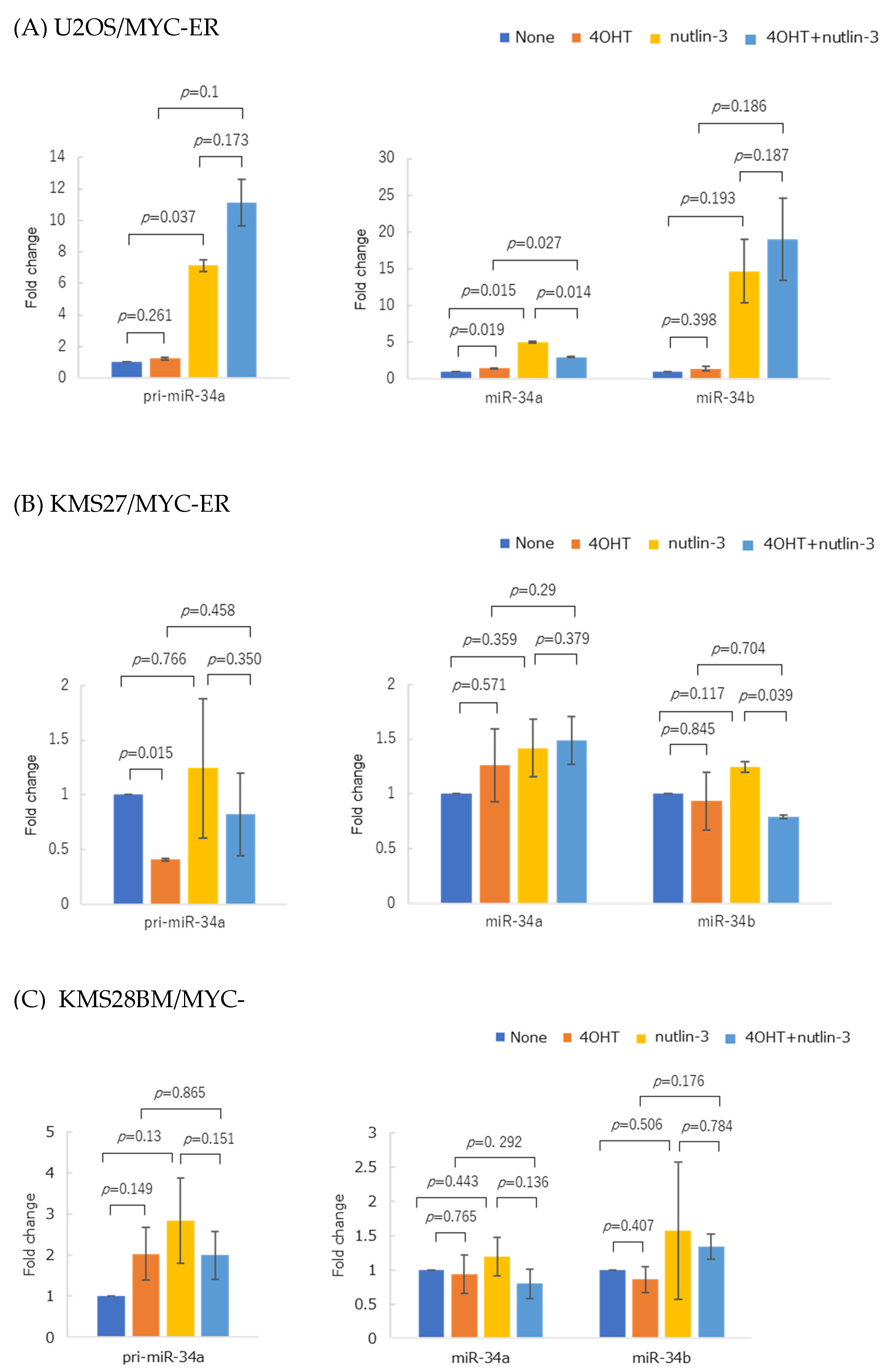
3.6. Forced MYC Activation Repressed p53-Mediated miR-34 Expression in MYC-ER Cell Lines
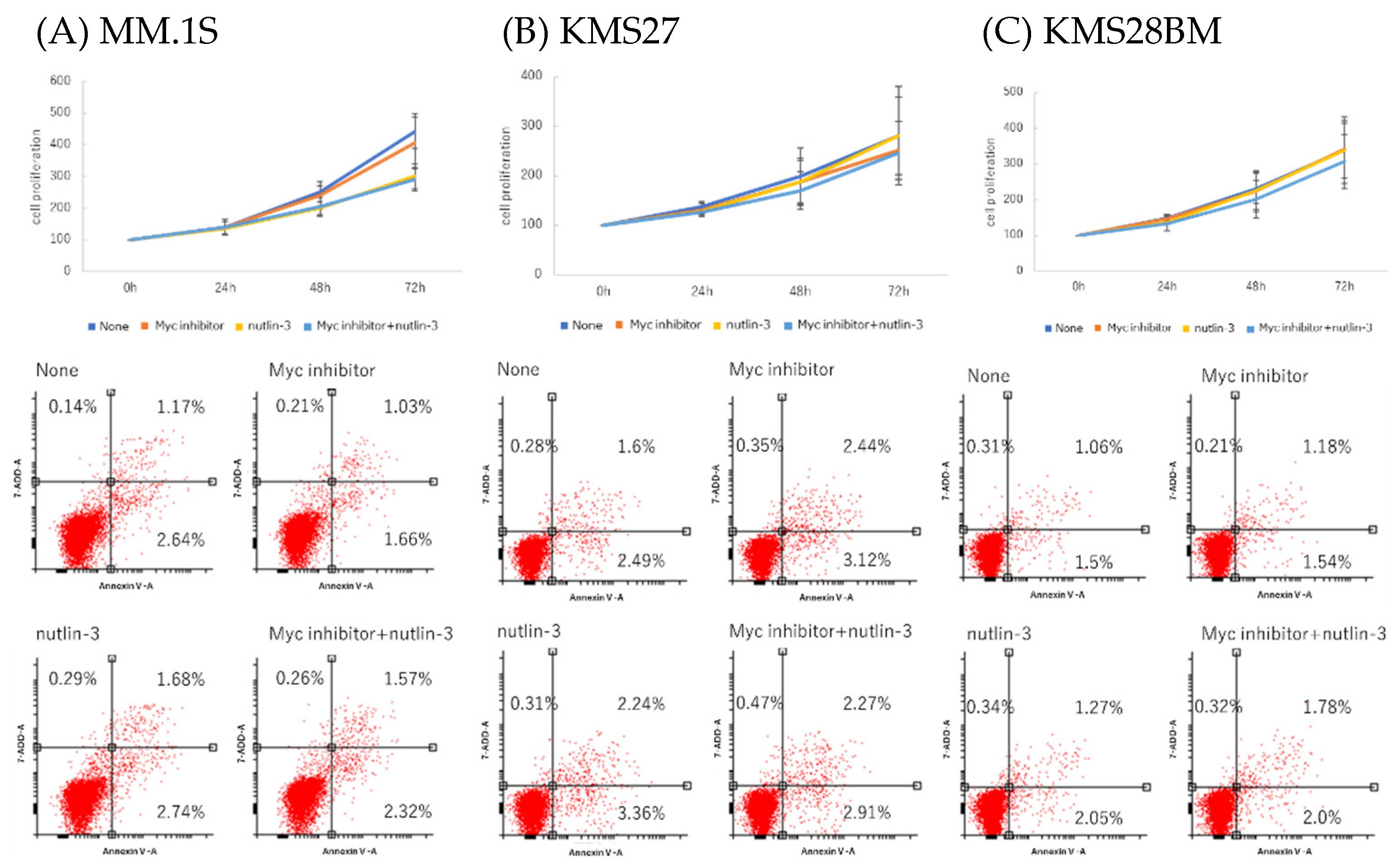
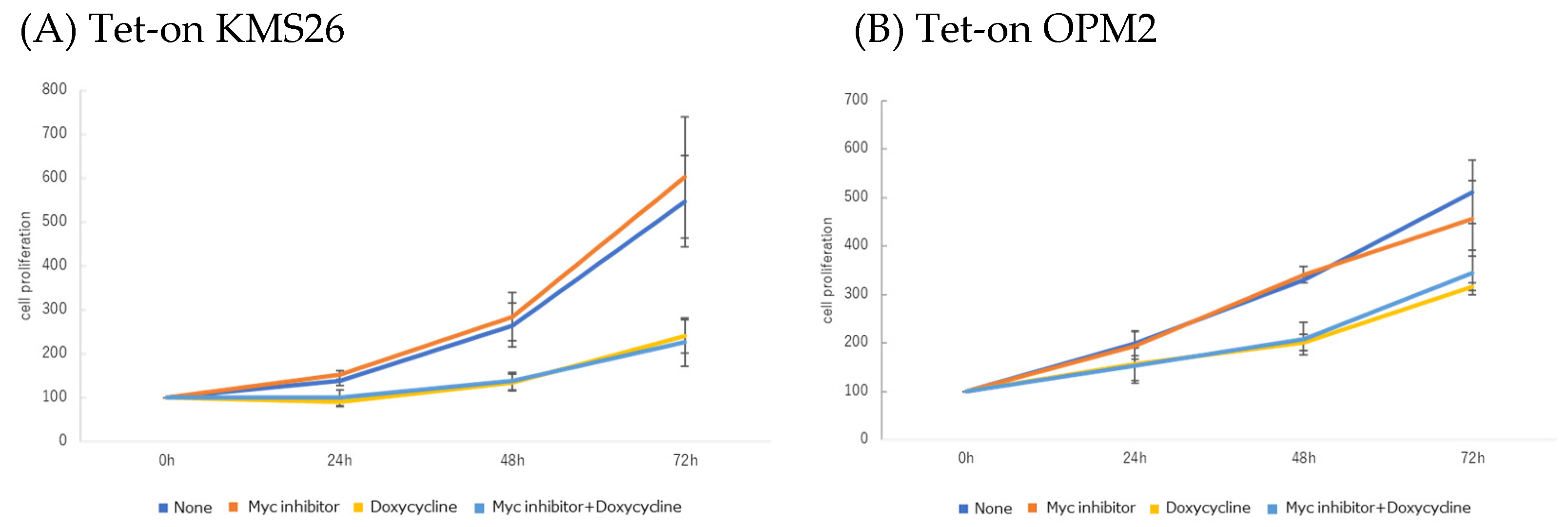
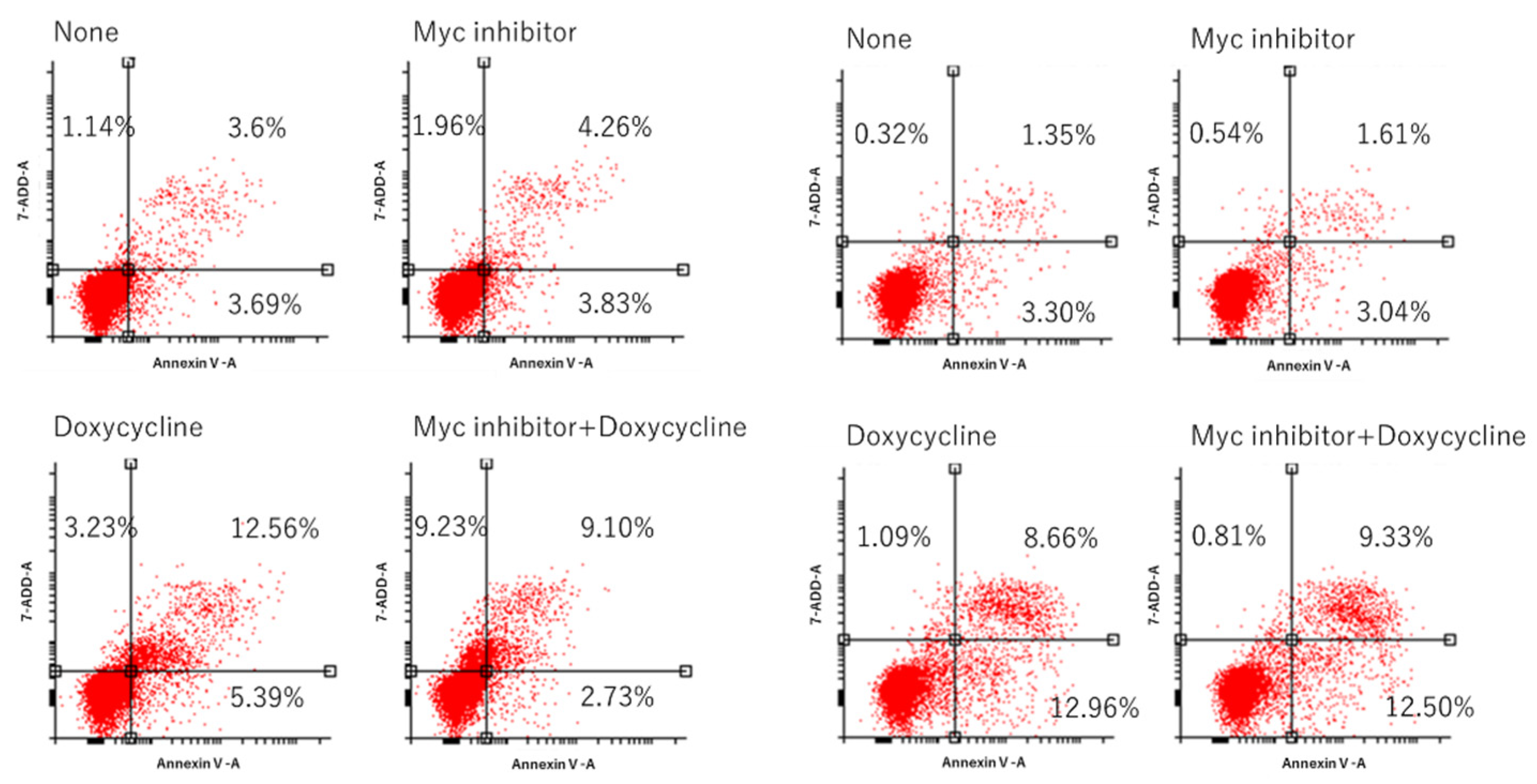
3.7. MM Cell Proliferation and Apoptosis after Co-Treatment with Nutlin-3 and Myc Inhibitor
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iranzo, J.; Martincorena, I.; Koonin, E.V. Cancer-mutation network and the number and specificity of driver mutations. Proc. Natl. Acad. Sci. USA 2018, 115, E6010–E6019. [Google Scholar] [CrossRef] [PubMed]
- Nebbioso, A.; Tambaro, F.P.; Dell’Aversana, C.; Altucci, L. Cancer epigenetics: Moving forward. PLoS Genet. 2018, 14, e1007362. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, R.; Gupta, K.; Gupta, S. Cancer epigenetics: An introduction. Methods Mol. Biol. 2015, 1238, 3–25. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.J.; Tay, Y. Noncoding RNA:RNA Regulatory Networks in Cancer. Int. J. Mol. Sci. 2018, 19, 1310. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Kyle, R.A.; Rajkumar, S.V. Multiple myeloma. Blood 2008, 111, 2962–2972. [Google Scholar] [CrossRef]
- Palumbo, A.; Anderson, K. Multiple myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef]
- Bergsagel, P.L.; Kuehl, W.M. Chromosome translocations in multiple myeloma. Oncogene 2001, 20, 5611–5622. [Google Scholar] [CrossRef]
- Caprio, C.; Sacco, A.; Giustini, V.; Roccaro, A.M. Epigenetic Aberrations in Multiple Myeloma. Cancers 2020, 12, 2996. [Google Scholar] [CrossRef]
- Adamia, S.; Abiatari, I.; Amin, S.B.; Fulciniti, M.; Minvielle, S.; Li, C.; Moreau, P.; Avet-Loiseau, H.; Munshi, N.C.; Anderson, K.C. The effects of MicroRNA deregulation on pre-RNA processing network in multiple myeloma. Leukemia 2020, 34, 167–179. [Google Scholar] [CrossRef]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Handa, H.; Murakami, Y.; Ishihara, R.; Kimura-Masuda, K.; Masuda, Y. The Role and Function of microRNA in the Pathogenesis of Multiple Myeloma. Cancers 2019, 11, 1738. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Kuroda, Y.; Masuda, Y.; Yamane, A.; Hattori, H.; Tahara, K.; Kaneko, A.; Suda, I.; Takahashi, N.; Gotoh, N.; et al. Loop regulation between microRNAs and epigenetics underlie microRNA dysregulation in multiple myeloma and is associated with the disease progression. Blood 2015, 126, 3013. [Google Scholar] [CrossRef]
- Handa, H.; Hattori, H.; Takahashi, N.; Sasaki, Y.; Saitoh, T.; Osaki, Y.; Tahara, K.; Koiso, H.; Mitsui, T.; Shimizu, H.; et al. Association between micro-RNA and epigenetic modifiers DNA methyltransferases (DNMTs), histone deacetylases (HDACs) in multiple myeloma (MM) and monoclonal gammopathy with undetermined significance (MGUS). Blood 2012, 120, 3942. [Google Scholar] [CrossRef]
- Nakamura, Y. Isolation of p53-target genes and their functional analysis. Cancer Sci. 2004, 95, 7–11. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, X.; Fiskus, W.; Lin, J.; Lwin, T.; Rao, R.; Zhang, Y.; Chan, J.C.; Fu, K.; Marquez, V.E.; et al. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell 2012, 22, 506–523. [Google Scholar] [CrossRef]
- Kurashima, K.; Sekimoto, T.; Oda, T.; Kawabata, T.; Hanaoka, F.; Yamashita, T. Polη, a Y-family translesion synthesis polymerase, promotes cellular tolerance of Myc-induced replication stress. J. Cell Sci. 2018, 131, jcs212183. [Google Scholar] [CrossRef]
- Oda, T.; Nakamura, R.; Kasamatsu, T.; Gotoh, N.; Okuda, K.; Saitoh, T.; Handa, H.; Murakami, H.; Yamashita, T. DNA-double strand breaks enhance the expression of major histocompatibility complex class II through the ATM-NF-κΒ-IRF1-CIITA pathway. Cancer Gene. Ther. 2022, 29, 225–240. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef]
- Pichiorri, F.; Suh, S.S.; Rocci, A.; De Luca, L.; Taccioli, C.; Santhanam, R.; Zhou, W.; Benson, D.M.; Hofmainster, C.; Alder, H.; et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 Impairs the p53/MDM2 Autoregulatory Loop in Multiple Myeloma Development. Cancer Cell 2016, 30, 349–351. [Google Scholar] [CrossRef]
- Saha, M.N.; Jiang, H.; Chang, H. Molecular mechanisms of nutlin-induced apoptosis in multiple myeloma: Evidence for p53-transcription-dependent and -independent pathways. Cancer Biol. Ther. 2010, 10, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Dalgard, C.L.; Gonzalez, M.; de Niro, J.E.; O’Brien, J.M. Differential microRNA-34a expression and tumor suppressor function in retinoblastoma cells. Invest. Ophthalmol. Vis. Sci. 2009, 50, 4542–4551. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, Z.; Chen, C.; Luo, X.; Xiao, J.; Dong, D.; Lu, Y.; Yang, B.; Wang, Z. Transcriptional and post-transcriptional mechanisms for oncogenic overexpression of ether à go-go K+ channel. PLoS ONE 2011, 6, e20362. [Google Scholar] [CrossRef]
- Krug, U.; Ganser, A.; Koeffler, H.P. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene 2002, 21, 3475–3495. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Chng, W.J.; Huang, G.F.; Chung, T.H.; Ng, S.B.; Gonzalez-Paz, N.; Troska-Price, T.; Mulligan, G.; Chesi, M.; Bergsagel, P.L.; Fonseca, R. Clinical and biological implications of MYC activation: A common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 2011, 25, 1026–1035. [Google Scholar] [CrossRef]
- Min, D.J.; Ezponda, T.; Kim, M.K.; Will, C.M.; Martinez-Garcia, E.; Popovic, R.; Basrur, V.; Elenitoba-Johnson, K.S.; Licht, J.D. MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia 2013, 27, 686–694. [Google Scholar] [CrossRef]
- Chang, T.C.; Zeitels, L.R.; Hwang, H.W.; Chivukula, R.R.; Wentzel, E.A.; Dews, M.; Jung, J.; Gao, P.; Dang, C.V.; Beer, M.A.; et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. USA 2009, 106, 3384–3389. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Tchernyshyov, I.; Chang, T.C.; Lee, Y.S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, X.; Lin, J.; Lwin, T.; Wright, G.; Moscinski, L.C.; Dalton, W.S.; Seto, E.; Wright, K.; Sotomayor, E.; et al. Myc represses miR-15a/miR-16-1 expression through recruitment of HDAC3 in mantle cell and other non-Hodgkin B-cell lymphomas. Oncogene 2012, 31, 3002–3008. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.J.; Cheng, Y.C.; Liu, C.R.; Lin, S.; Liu, H.E. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 2006, 34, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Craig, V.J.; Cogliatti, S.B.; Imig, J.; Renner, C.; Neuenschwander, S.; Rehrauer, H.; Schlapbach, R.; Dirnhofer, S.; Tzankov, A.; Müller, A. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood 2011, 117, 6227–6236. [Google Scholar] [CrossRef] [PubMed]
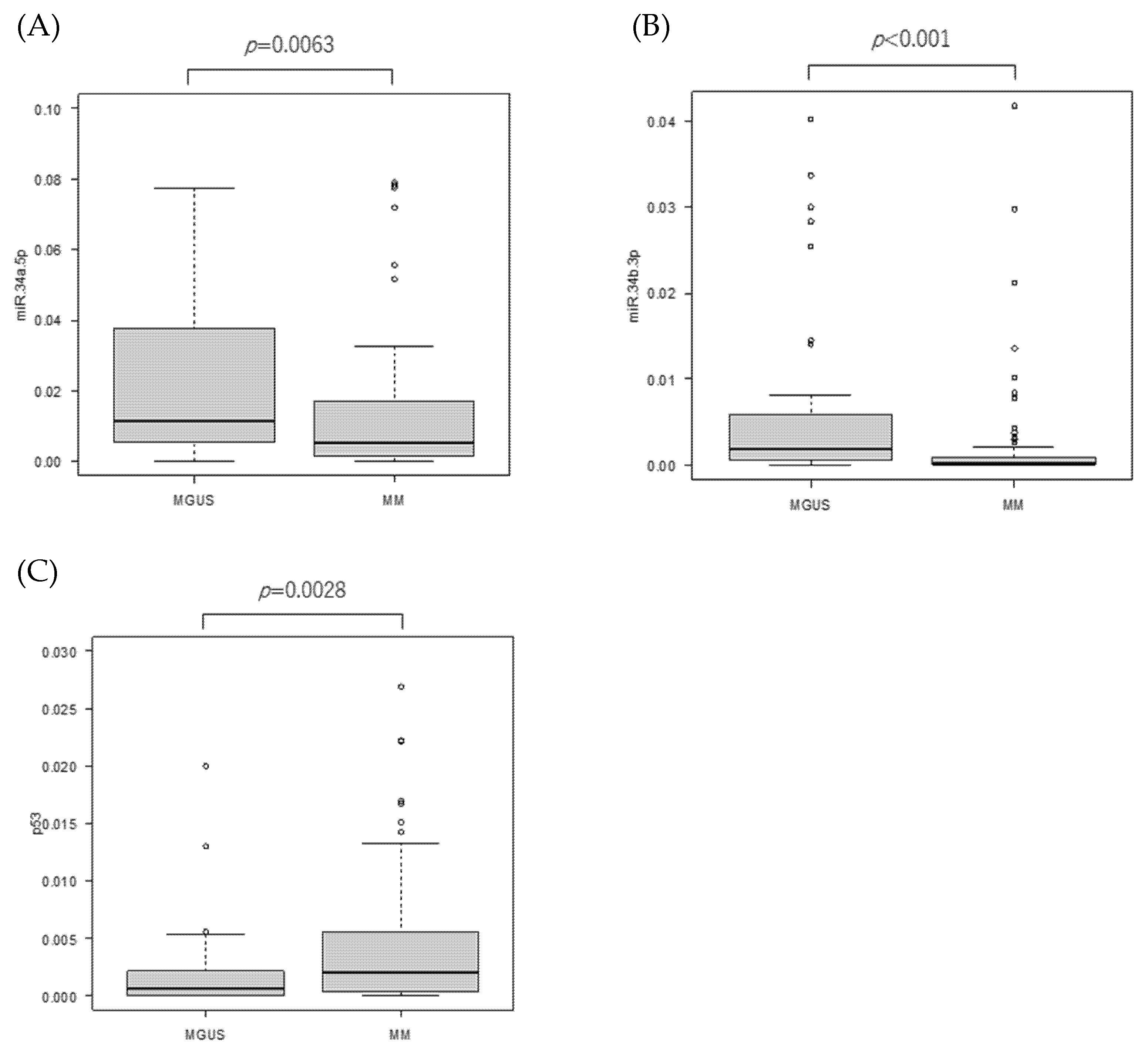


| Group | MGUS | MM | |
|---|---|---|---|
| n | 64 | 109 | |
| age | 71 (38–88) | 69.5 (44–88) | |
| Gender (%) | F | 37 (58.7) | 53 (49.1) |
| M | 26 (41.3) | 55 (50.9) | |
| IgH (%) | BJ | 2 (3.5) | 19 (17.6) |
| IgG | 40 (70.2) | 63 (58.3) | |
| IgA | 11 (19.3) | 22 (20.4) | |
| IgD | 0 (0.0) | 2 (1.9) | |
| IgM | 2 (3.5) | 0 (0.0) | |
| unknown | 2 (3.5) | 2 (1.9) | |
| IgL (%) | κ | 31 (54.4) | 60 (55.6) |
| λ | 24 (42.1) | 46 (42.6) | |
| unknown | 2 (3.5) | 2 (1.9) | |
| ISS (%) | 1 | NA | 22 (21.0) |
| 2 | NA | 45 (42.9) | |
| 3 | NA | 38 (36.2) | |
| R.ISS (%) | 1 | NA | 12 (12.1) |
| 2 | NA | 72 (72.7) | |
| 3 | NA | 15 (15.2) | |
| Cytogenetics.Risk (%) | High | NA | 35 (34.3) |
| Standard | NA | 67 (65.7) | |
| Cytogenetics.Karyotype (%) | del 17p | NA | 12 (12.5) |
| t (11; 14) | NA | 21 (21.9) | |
| t (14; 16) | NA | 2 (2.1) | |
| t (4; 14) | NA | 16 (16.7) | |
| trisomy11 | NA | 18 (18.8) | |
| NP | NA | 27 (28.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murakami, Y.; Kimura-Masuda, K.; Oda, T.; Matsumura, I.; Masuda, Y.; Ishihara, R.; Watanabe, S.; Kuroda, Y.; Kasamatsu, T.; Gotoh, N.; et al. MYC Causes Multiple Myeloma Progression via Attenuating TP53-Induced MicroRNA-34 Expression. Genes 2023, 14, 100. https://doi.org/10.3390/genes14010100
Murakami Y, Kimura-Masuda K, Oda T, Matsumura I, Masuda Y, Ishihara R, Watanabe S, Kuroda Y, Kasamatsu T, Gotoh N, et al. MYC Causes Multiple Myeloma Progression via Attenuating TP53-Induced MicroRNA-34 Expression. Genes. 2023; 14(1):100. https://doi.org/10.3390/genes14010100
Chicago/Turabian StyleMurakami, Yuki, Kei Kimura-Masuda, Tsukasa Oda, Ikuko Matsumura, Yuta Masuda, Rei Ishihara, Saki Watanabe, Yuko Kuroda, Tetsuhiro Kasamatsu, Nanami Gotoh, and et al. 2023. "MYC Causes Multiple Myeloma Progression via Attenuating TP53-Induced MicroRNA-34 Expression" Genes 14, no. 1: 100. https://doi.org/10.3390/genes14010100
APA StyleMurakami, Y., Kimura-Masuda, K., Oda, T., Matsumura, I., Masuda, Y., Ishihara, R., Watanabe, S., Kuroda, Y., Kasamatsu, T., Gotoh, N., Takei, H., Kobayashi, N., Saitoh, T., Murakami, H., & Handa, H. (2023). MYC Causes Multiple Myeloma Progression via Attenuating TP53-Induced MicroRNA-34 Expression. Genes, 14(1), 100. https://doi.org/10.3390/genes14010100







