Abstract
Gomphus purpuraceus (Iwade) Yokoyama is a species of wild fungi that grows in southwest China, considered an edible and medicinal fungus with potential commercial prospects. However, the detailed mechanisms related to the development of mycelium and the formation of the fruiting body are unclear. To obtain a comprehensive overview of genetic features, whole-genome and comparative genomics analyses of G. purpuraceus were performed. High-quality DNA was extracted from the mycelium, which was isolated from a fresh fruiting body of G. purpuraceus. The DNA sample was subjected to sequencing using Illumina and Oxford Nanopore sequencing platforms. A genome assembly totaling 40.15 Mb in 50 contigs with an N50 length of 2.06 Mb was generated, and 8705 putative predicted genes were found. Subsequently, phylogenetic analysis revealed a close evolutionary relationship between G. purpuraceus and Gomphus bonarii. Moreover, a total of 403 carbohydrate-active enzymes (CAZymes) were identified in G. purpuraceus, which included 147 glycoside hydrolases (GHs), 85 glycosyl transferases (GTs), 8 polysaccharide lyases (PLs), 76 carbohydrate esterases (CEs), 57 auxiliary activities (AAs) and 30 carbohydrate-binding modules (CBMs). Compared with the other 13 fungi (Laccaria bicolor, Russula virescens, Boletus edulis, etc.), the number and distribution of CAZymes in G. purpuraceus were similar to other mycorrhizal fungi. Furthermore, the optimization of culture medium for G. purpuraceus showed the efficient utilization of disaccharides such as sucrose and maltose. The genome of G. purpuraceus provides new insights into its niche, food applications and potential artificial domestication.
1. Introduction
Wild mushrooms are a vital source of income and nutrition for rural communities and recreational foragers. At present, a total of 2006 species from 99 countries can be consumed safely [1]. The wild edible mushroom has always been favored by people for its unique taste, flavor and nutritional value [2]. Of these wild edible fungi, some have been used as medications due to their special compounds, such as terpenes, phenolic substances, ceramides, lactones, etc., which are beneficial to the body [3,4]. The huge land areas in China provide highly diverse habitats for macrofungi, especially in northeast and southwest China and Tibet. Wu et al. considered 1020 and 692 taxa of 1662 in total to be edible and medicinal mushrooms, respectively [5]. In recent years, mushroom cultivation has become the fifth-largest agricultural industry in China. Around 100 edible mushrooms have been domesticated, of which 60 have been commercially cultivated [6]. However, many known edible or medicinal fungi have been unsuccessfully cultivated due to the uncertain ecological niches (symbiotic or saprotrophic).
G.purpuraceus (Iwade) Yokoyama (Figure 1A, B), an ancient and endangered species, is a kind of wild edible mushroom [7,8]. Although the fruiting bodies of the genus Gomphus are thought to be indigestible [9,10], G. purpuraceus is considered to be edible and do not cause physical discomfort, according to local consumers. Wu et al. has also classified G. purpuraceus as an edible mushroom without known toxicity [5]. The fruiting bodies of G. purpuraceus have been popular at wild mushroom markets in southwest and central China for hundreds of years, especially in Guizhou, Hubei and Sichuan Provinces, with different local names such as “matijun” (horse’s hoof) and “zituoluo”. Owing to the outstanding flavor and old health myths, some researchers have begun to focus on the extraction and functional assessment of the bioactive compounds in G. purpuraceus. Thus far, only two alliacane sesquiterpenes were found in the fruiting bodies of G. purpuraceus [11,12]. According to previous reports, terpenoids have some health-promoting properties, such as protecting the liver, detoxification, decreasing blood pressure, lowering blood fat and other medicinal functions [13,14,15]. Moreover, terpenoids have been demonstrated to effectively inhibit the DNA synthesis in carcinoma [16,17]. Therefore, G. purpuraceus has significant prospects for edible and medicinal development.

Figure 1.
Fruiting bodies of G. purpuraceus. (A) Habitat. (B) Basidiomata.
The genus Gomphus is classified as a part of the ectomycorrhizal (ECM) fungal lineage/ramaria-gautieria [18]. According to the existing ecological survey, G. purpuraceus fungi usually grow in stony yellow-brown soil under Quercus and Pinus trees, which indicates a possible symbiotic relationship with these trees [19]. However, there is still a lack of clear evidence of the mycorrhizal niche of G. purpuraceus.
As sequencing technology advances, genome sequencing has been widely applied to understand the ecological niches of wild fungi [20,21]. Comparative genomes can provide insights into the lignocellulosic decay capabilities of fungi [22,23]. In saprotrophic fungi genomes, there are abundant genes related to oxidoreductases and carbohydrate-active enzymes (CAZymes), which can help these fungi to degrade lignin (peroxidases, DyP, laccases s.s.), crystalline cellulose (GH6, GH7, AA9) and other carbohydrates (GH43, GH74) [24,25,26]. Furthermore, white-rot fungi also have abundant copies of the cellulose-binding module 1 (CBM1) to facilitate the attachment of enzymes to crystalline cellulose [27], and some oxidoreductases, especially in class II peroxidases (class II PODs), give white-rot fungi the ability to degrade lignin [20]. ECM fungi mainly obtain carbon sources from plants due to the reduction in genes encoding plant cell wall-degrading enzymes (PCWDEs), as compared to their ancestral wood decayers [28]. Nevertheless, certain ECM fungi have been reported to decompose PCWDEs existing in soil organic matter (SOM) according to transcriptome analysis [29] and the occasional discovery of fruiting bodies from Russulaceae on rotting wood and trunks of trees [30,31,32]. Comparative genomic analysis of ECM fungi also confirmed that certain ECM fungi have retained a unique array of PCWDEs, indicating that they possess diverse abilities to decompose lignocellulose [33,34]. To better understand fungi, the Fungal Genomics Program was held by the U.S. Department of Energy (DOE) Joint Genome Institute (JGI), which focuses on bioenergy and environmental microbiome processes [22]. Furthermore, the Mycorrhizal Genomics Initiative was put forward to reveal the symbiosis mechanisms between ECM fungi and related trees [35]. Thus, the high-quality whole genomic sequence for G. purpuraceus can greatly provide insights into its mycelial growth, fruiting body formation and metabolic mechanism.
Here, the whole genome of G. purpuraceus was de novo sequenced and assembled with a combination of Illumina and Oxford Nanopore sequencing platforms. We also performed the gene functional annotation, which provides the possibility of finding genes involved in secondary metabolism and carbohydrate metabolism. In addition, the genes encoding membrane transport proteins and CAZymes were also analyzed, and the reaction of G. purpuraceus mycelia in different carbon sources was also investigated, which could provide support for microbial food resources and strain propagation. On the whole, the aim of the first genome-scale assembly for G. purpuraceus is to develop foundational genomic and genetic resources for food application, artificial domestication and species conservation.
2. Materials and Methods
2.1. Fungal Strains
In this study, fruiting bodies of G. purpuraceus were collected in July 2020 from a mixed forest with scattered Pinus massoniana Lamb in Xiazai village, Longli County, Guizhou Province, China (26.5690° N, 106.8556° E, alt. 1149 m). The fruiting body samples were quickly transported at 4 °C to the laboratory for mycelial isolation with low glucose (2.5 g/L) modified Melin-Norkrans medium (MMN) [29]. The specimen voucher was deposited in the herbarium of the Guizhou Academy of Agriculture Sciences, with accession number GZAAS22-0001 (GAF-20071601). The strain was derived from a cylindrical fruiting body. Then, the purity of the strain was molecularly identified by ITS analysis. The pure cultures were produced by liquid culture (15 g/L fresh potato tissue, 5.0 g/L glucose, 2.0 g/L peptone, 0.5 g/L KH2PO4, 0.15 g/L MgSO4·7H2O, 2.5 mL of 10 g/L NaCl, 5.0 mL of 10 g/L CaCl2, 1.5 mL of 10 g/L FeCl3, 1 mg/L thiamine-HCl; pH 6.5) for 60 days at 25 °C, stirring with 165 rpm in darkness. Thereafter, the mycelium was collected from the liquid medium by vacuum filtration under aseptic conditions, frozen in liquid nitrogen. Finally, the samples were sent to Beijing Biomarker Technologies Co., Ltd. (Beijing, China) under dry ice conditions for DNA extraction and genome sequencing.
2.2. Library Construction, Sequencing and Assembly
Genome sequencing requires high-quality DNA [36]. The purity, integrity and concentration of the extracted DNA from G. purpuraceus were checked by Nanodrop, Qubit and 0.7% agarose gel electrophoresis. Genome survey sequences were obtained by the Illumina HiSeq X Ten sequencing platform, which can provide rapid and accurate basial information such as the genome size, heterozygosity and repeat content by the k-mer method [37]. Using the software “kmer_freq_stat” independently developed by Beijing Biomarker Technologies, the heterozygosity rate of G. purpuraceus genome was estimated to be 0.01% (Figure S1), which met the requirements for whole-genome construction. Then, the whole genome was sequenced by the Oxford Nanopore sequencing platform. About 15.13 Gb of raw data were generated, and the quality control of data was performed with the sequence length ≥ 2000 bp, which was subsequently corrected by Canu v1.5 [38]. Then, the corrected reads were used for genome assembly and adjusted with wtdbg2 and Racon [39]. Finally, the highly accurate genome was achieved by the correction and optimization performed with Pilon v1.14 combined with the second-generation sequencing data [40]. Genome assembly integrity was assessed through BWA and BUSCO v4.1.2 software [41,42].
2.3. Genomic Prediction and Genome Annotation
Through the construction and classification of the repeated sequence database, the repeated sequence rate was predicted by RepeatMasker v4.0.6 [43]. Protein coding sequences were mainly predicted by the alignment of ab initio predictions, homologous proteins and transcriptional data, and then integrated (EVM v1.1.1) and modified (PASA v2.0.2). According to the structural characteristics of non-coding RNA, the genes involved in transfer RNAs (tRNAs) were predicted by tRNAscan-SE [44]. The sequences of ribosomal RNAs (rRNAs) and other ncRNA were identified by Infernal v1.1 software based on the Rfam database [45,46]. Pseudogenes were identified by finding the termination codon or frameshift mutation using GeneWise [47]. In addition, gene clusters were predicted by antiSMASH v6.0 [48].
All predicted gene models were annotated to the functional databases, namely, Clusters of Orthologous Groups (KOG) (http://www.ncbi.nlm.nih.gov/COG, accessed on 7 August 2021), Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/, accessed on 7 August 2021), Swiss-Prot, TrEMBL (http://www.expasy.org/sprot/ and http://www.ebi.ac.uk/swissprot/, accessed on 7 August 2021) and Non-Redundant Protein Database databases (NR) (ftp://ftp.ncbi.nlm.nih.gov/blast/db/, accessed on 7 August 2021) by the similar alignment [49]. Based on annotation results from the NR database, the genes were mapped to gene ontology (GO) (http://www.ebi.ac.uk/GOA, accessed on 7 August 2021) using Blast2GO [50]. The functional annotation of the Pfam database (http://pfam.xfam.org/, accessed on 7 August 2021) used HMMER software [51]. Transporter Classification Database for membrane transport protein analyses (TCDB) (http://www.tcdb.org, accessed on 7 August 2021) and Cytochrome P450 Engineering Database (CYPED) (http://drnelson.utmem.edu/CytochromeP450.html, accessed on 7 August 2021) were also used to analyze related genes and their functional information. Pathogen–host interaction (PHI) (http://www.phi-base.org/, accessed on 7 August 2021) and the database of fungal virulence factor (DFVF) (http://sysbio.unl.edu/DFVF/, accessed on 7 August 2021) were used to evaluate the edible safety of the fruiting body of G. purpuraceus. Annotation of CAZymes for the G. purpuraceus genome was performed by HMMER based on the CAZy database (http://www.cazy.org/, accessed on 7 August 2021), which can provide insights into the way G. purpuraceus obtains carbon compounds, as in the study of Kohler et al. [28]. In addition, subcellular localization information is one of the key features of protein function research. Signal peptides, transmembrane proteins and secreted proteins were predicted by SignalP v4.0 (DTU Health Tech, Lyngby, Denmark), tmhmm (DTU Health Tech, Lyngby, Denmark) and EffectorP software (CSIRO, Campbell, Australia), respectively.
Mating systems are responsible for the degree of selfing/outcrossing in natural populations and impact gene flow, the accumulation of deleterious alleles and adaptability [52]. Therefore, the identification and structuring of the mating-type (MAT) genes were performed based on the genomic prediction. The MAT-A genes, including homeodomain type 1 and homeodomain type 2 MAT genes (HD1 and HD2 genes) in the G. purpuraceus genome, were identified using TBLASTN. For MAT-B genes, the pheromone receptor genes in G. purpuraceus were identified by the Swissprot annotation with the keyword “pheromone receptor”. The sequence length of a pheromone precursor is so short (usually 50–100 amino acid) that it could not be predicted in the normal genome annotation procedure. Transdecoder (https://transdecoder.github.io/, accessed on 7 August 2021) software with Pfam search was used to annotate the pheromone precursor by searching in the ~20 kb flanking sequence of pheromone receptor genes.
2.4. Comparative Genomics Analysis
The widely studied genomes of ECM edible fungi, L. bicolor [53], Lactarius deliciosus [54], Lactarius hatsudake [54], R. virescens [54], Russula griseocarnosa [55], B. edulis [33] and Tuber melanosporum [56], were selected for comparison with the genomic data of G. purpuraceus and G. bonarii [54]. Four saprotrophic fungi, Lentinula edodes [57], Flammulina velutipes [58], Pleurotus ostreatus [59] and Morchella importuna [60], were also added to the comparative genomics analysis, thought to have a unique ability to degrade lignin, cellulose and other phenolic compounds [24]. In addition, due to the saprotrophic properties on artificial substrates and the association with plants as an ECM symbiont, the genome sequence of Phlebopus portentosus was selected as the reference genome for the saprophytes and ECM fungi mentioned above [61,62]. The sequences of all selected genomic proteins were obtained from the National Center for Biotechnology Information at https://www.ncbi.nlm.nih.gov/ (accessed on 8 November 2021) and the U.S. Department of Energy (DOE) Joint Genome Institute (JGI) at https://genome.jgi.doe.gov/ (accessed on 8 November 2021) (Table S1). Because the annotation information of the selected partial genome is not available, the required information is annotated through the relevant public database in this paper.
Based on the protein sequences of G. purpuraceus predicted by the OrthoMCL-v2.0.3 software, gene family clustering, including cluster gene, single-copy ortholog gene families and unique gene families, was performed by comparison with selected fungi. The single-copy ortholog genes in each genome were aligned by MAFFT v7.205 with the default parameters. Every gene family was used to build the Hidden Markov Model by HMMER v3.0, and the genes with the highest scores were identified as homologous genes [63]. The conserved region was selected to construct phylogenetic trees by removing poorly aligned regions using phyML software (Microsoft Corporation, Redmond, WA, USA).
2.5. Optimization of Culture Medium for G. purpuraceus
The medium was optimized based on the analysis of the CAZyme genes involved in the α-amylase family and the starch and sucrose metabolism pathway (ko00500) for suitable carbon source selection. The solid medium was adopted with 2.0 g/L peptone, 0.2 g/L NH4Cl, 0.5 g/L KH2PO4, 0.15 g/L MgSO4·7H2O, 2.5 mL of 10 g/L NaCl, 5.0 mL of 10 g/L CaCl2, 1.5 mL of 10 g/L FeCl3 and 18 g/L agar powder. All optimization experiments were carried out in 7 cm diameter Petri dishes added with 10 g/L of different carbon sources. A 6 mm holepunch was used to pick out the culture for inoculation. Each medium formulation had 30 plates. All samples were cultivated at 25 °C in the dark for 60 days, and the measurement of colony diameters was conducted by the cross-over method [64]. The standard error was calculated by EXCEL 2016. The statistical analysis was performed using SPSS 19.0 software (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Analysis of the Genome Assembly and Gene Prediction
The analysis of the assembly scaffold and genome of G. purpuraceus is shown in Table 1. A total of 15,126,272,717 bp raw data were generated by the Nanopore sequencing platforms, while 13,941,417,498 bp clean reads passed the quality control check. After genome assembly, correction and optimization, a total sequence length of 40,153,731 bp was assembled into 50 contigs with an N50 length of 2,068,469 bp. The maximum contig length was 3,972,391 bp. The GC content of the assembled contig was 47.74%. The genome assembly integrity of the mapped rate and complete BUSCO was 76.84% and 252 (86.90%), respectively. The predicted result of the total repeated sequence length was 15,467,010 bp, covering 38.52% of the genomic length. Based on the alignment and integration of ab initio predictions, homologous proteins and transcriptional data, the G. purpuraceus genome includes 8705 coding protein genes, and the average length of protein-coding genes was 2125.01 bp. In addition, the non-coding RNA, including 11 rRNA (family number 4), 78 tRNA (family number 46) and 18 other ncRNA (family number 10), was predicted, and a total of 119 pseudogenes were found in the G. purpuraceus genome by GeneWise (https://www.ebi.ac.uk/Tools/psa/genewise/, accessed on 7 August 2021).

Table 1.
The analysis of the gene assembly and genome of G. purpuraceus.
3.2. Gene General Annotation
Genome functional annotations of the 8705 predicted protein-coding genes using the public databases of GO, KEGG, KOG, Pfam, SwissProt, TrEMBL and NR are shown in Table S2. The 4258 G. purpuraceus protein-coding genes were annotated in the GO database, accounting for only 48.91% of the total predicted coding genes, which were divided into three major classes and 43 functional groups (Figure 2A). Among them, 14, 14 and 15 subclasses exist in the cellular component, molecular function and biological process, respectively (Table S3). A total of 3066 genes had matches in the KEGG database, which involved 109 KEGG pathways. The classification chart of the KEGG pathways is shown in Table S4. Among the 109 KEGG pathways, ribosome (107 genes), biosynthesis of amino acids (102 genes) and RNA transport (100 genes) have more abundant gene numbers than other pathways (Figure 2B).
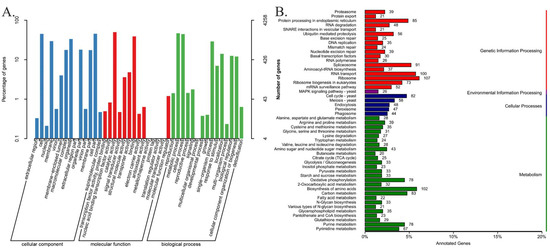
Figure 2.
The mainly functional annotation of the G. purpuraceus genome. (A) Gene Ontology (GO) functional annotation; (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotation.
In addition, there are 4797 genes annotated in the KOG database, shown in Table S5. The protein functions are mainly concentrated in the general function prediction only (800 genes), post-translation modification, protein turnover, chaperones (532 genes), signal transduction mechanisms (405 genes) and other aspects. The numbers of annotated genes in the Pfam, Swissprot and TrEMBL databases were 6145, 5231 and 7971, accounting for 70.59%, 60.09% and 91.57%, respectively. The largest number of genes was annotated by the NR database (94.18%), including 8198 genes (Table S6). Overall, 8212 genes were successfully annotated in the database mentioned above, accounting for 94.34% of the 8705 predicted protein-coding genes.
In the genome of G. purpuraceus, the two MAT-A-related genes on the contig00001 were found by BLAST homology search (Table S7). The open reading frames (ORFs) of two genes (EVM0008346.1 and EVM0002788.1) are in opposite directions, and the interval between them is 138 bp. The protein domain of the two genes was the homeobox domain. Meanwhile, HD1 (2269 bp) consists of 436 amino acids, containing three exons. HD2 (encoding 811 amino acids) has a physical length of 2486 bp on the genome, containing two exons. The mitochondrial intermediate peptidase (762 amino acids) was found to be located in close proximity with the two MAT-A-related genes on the same contig00001, which is much like that of other Agaricomycetes. For MAT-B genes, two potential pheromone receptor genes (EVM0003467.2 and EVM0007013.3) were identified and clustered on the contig00006. Like in other Agaricomycetes, the two genes are homologous to the STE3 gene (pheromone, a factor receptor). For pheromone genes, we searched the ~20 kb flanking region of pheromone receptor genes, but found no pheromone precursor genes in the proximity of these receptors.
3.3. Gene Special Annotation
The summary statistics of gene special annotation in CAZymes, P450, TCDB, PHI and DFVF are shown in Table S8.
CAZymes of the genome can provide an insight into the metabolism of complex carbohydrates in the studied fungus. The genome of G. purpuraceus was compared with the CAZy database to study the CAZymes of the strain. In this database, 376 candidate CAZyme genes were identified (Figure 3A). This included 147 glycoside hydrolases (GHs, 36.47%), 85 glycosyl transferases (GTs, 21.09%), 8 polysaccharide lyases (PLs, 1.98%), 76 carbohydrate esterases (CEs, 18.85%), 57 auxiliary activities (AAs, 14.14%) and 30 carbohydrate-binding modules (CBMs, 7.44%). It is worth noting that 10 genes of G. purpuraceus involved in class II lignin-modifying peroxidases (AA2) were found in the CAZy database.
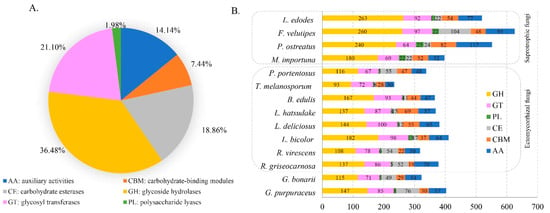
Figure 3.
Distribution and number of carbohydrate-active enzyme genes (CAZymes) genes in G. purpuraceus and other 13 fungi. (A) Distribution of CAZymes in G. purpuraceus. (B) Comparison of CAZymes in the 14 fungi.
The antiSMASH software can accurately identify gene clusters involved in secondary metabolites of a wide range of known chemical classes. In the genome of G. purpuraceus, a total of 22 gene clusters were predicted by antiSMASH software. The gene cluster total length was 780,051 bp, and the gene cluster mean length was 35,456 bp. These gene clusters were distributed over 13 gene clusters. Among them, there are nine gene regions associated with terpenes, six gene regions involved in non-ribosomal peptide synthase (NRPS) or NRPS-like, five gene regions related to polyketide synthase (PKS) or PKS-like and one region associated with siderophore. In addition, one gene region was identified as both terpene and NRPS-like. The summary of predicted metabolite gene clusters is shown in Table S9.
A total of 76 genes were annotated in the TCDB database (Table S10), accounting for 0.87% of the predicted genes. About 25.56% of the predicted protein-coding genes in the G. purpuraceus genome were annotated in the PHI database, totaling 2225 genes (Table S11). Moreover, 1632 genes were found in the DFVF database, accounting for 18.75% of the predicted genes (Table S12).
In addition, studies have found that a class of secreted effector protein with less than 300 amino acids was related to the mycorrhizal formation [65,66,67]. The predicted results of signal peptides, transmembrane proteins, secreted proteins and effector proteins are shown in Table S13. The transmembrane proteins were found to have the highest number (1795), followed by signal peptides (673), secreted proteins (400) and effector proteins (32).
Finally, a detailed circular genome diagram of G. purpuraceus is displayed in Figure 4. The outermost circle is labeled with the size of the genome, and each scale is 50 Kb. From the outside to the inside (Figure 4A), the second and third loops represent the characteristics of the whole genome, including the positive chain gene and negative chain gene, where different colors represent different KOG functional classifications (Figure 4B). The fourth circle indicated the repeated sequence. The fifth loop exhibited the ncRNA, including tRNA (blue part) and rRNA (purple part). The sixth circle is the GC content; the outward light yellow part indicates that the GC content in this region is higher than the average GC content in the genome. The higher the peak value, the greater the difference from the average GC content. The inward blue part is the opposite. The innermost layer is the GC skew (the specific algorithm: G−C/G+C), where the outward dark gray part indicates that the content of C in this area is lower than that of G, and the inward red part is the opposite.
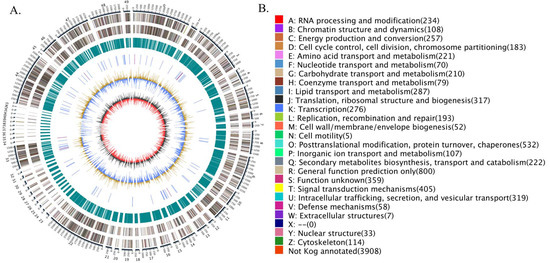
Figure 4.
The G. purpuraceus circular genome diagram. (A) The circular genome diagram. (B) The KOG functional classification in circular diagram.
3.4. Comparative Genomics Analysis
3.4.1. Gene Family Analysis
The gene families of G. purpuraceus, L. bicolor, L. deliciosus, L. hatsudake, R. griseocarnosa, R. virescens, G. bonarii, B. edulis, T. melanosporum, L. edodes, F. velutipes, P. ostreatus, M. importuna and P. portentosus were subjected to cluster analysis. The Venn diagram of common and unique gene families among 14 fungi is shown in Figure 5. The numbers of total genes, total gene families and cluster genes are listed in Table 2. A total of 22,569 gene families from 14 species were obtained by OrthoMCL. There were 6590 gene families found in the G. purpuraceus genome, of which 1533 were core orthologous gene families among 14 fungi genomes and 120 were unique gene families found in G. purpuraceus (Figure 5). Among these genomes, the number of common gene families with single-copy genes was 573.
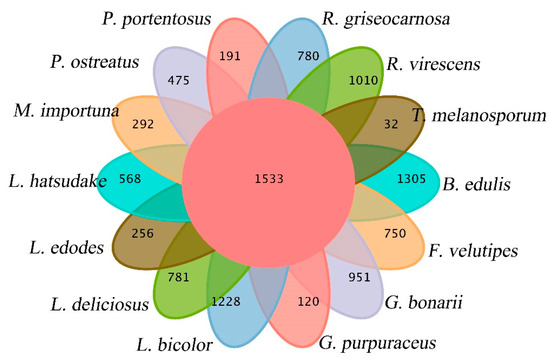
Figure 5.
Venn diagram of orthologous gene families.

Table 2.
Statistical results of the gene families.
3.4.2. Phylogenetic Relationship of G. purpuraceus
According to the results of 573 single-copy genes identified by gene family clustering, a phylogenetic tree was constructed, as shown in Figure 6. The value on a branch of the phylogenetic tree represents the branch reliability, and the number close to 100 represents high reliability. The bootstrap value was 100% of highly supported internal branches, which accurately reflects the availability of the datasets and evolutionary relationship of these 14 fungi. In the phylogenetic tree, most species of the same genus are clustered in the same branch, except for some saprophytes. In the NCBI taxonomy, L. deliciosus and L. hatsudake, R. griseocarnosa and R. virescens were found to belong to the genus Lactarius and Russula, which was also confirmed by phylogenetic analysis. In addition, L. bicolor is in the same evolutionary branch as P. ostreatus, and the evolutionary distance of P. ostreatus is greater based on the branch length. As far as the target fungus is concerned, phylogenetic analysis indicated that G. purpuraceus was most related to G. bonarii.
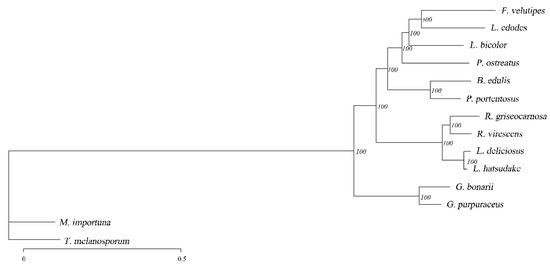
Figure 6.
Maximum likelihood phylogenetic tree based on single-copy ortholog genes.
The phylogenetic analysis was based on different lifestyle fungi, and these different trophic types of fungi were selected for comparative genomic analysis. This provides a foundation for the functional genomics analysis of G. purpuraceus.
3.4.3. Comparative Analysis of CAZymes
The CAZyme composition characteristics in G. purpuraceus were used to compare with those of 13 other fungi, including eight ECM, four saprotrophic fungi and Phlebopus portentosus considered as a facultative saprotrophic fungus. The global statistics and comparisons are shown in Figure 4B and Table S14, respectively. Compared with other saprotrophic species, G. purpuraceus had few CAZyme protein families, especially GHs, showing that it is similar to ECM fungi such as B. edulis, L. deliciosus, R. griseocarnosa and G. bonarii. The dominant GH families are mainly involved in the degradation of plant cell wall polysaccharides, such as GH6, GH7, GH12, GH45, GH61 and AA9 (LPMOs) in the degradation of cellulose. Apart from G. purpuraceus (GH6, two genes; GH7, one gene) and G. bonarii (GH6, two genes; GH7, two genes), these two CAZyme families were completely absent from these ECM fungi genomes, showing that the ECM fungi have little ability to degrade crystalline cellulose [20]. In the AA9 CAZyme family, the number of LPMOs (auxiliary activity family 9-type) in the G. purpuraceus genome was much lower than that of saprotrophic fungi, as well as the other six ECM fungi, except for L. deliciosus and L. hatsudake, which oxidatively attack cellulose chains [68]. In these CAZyme families involved in the degradation of hemicellulose, such as GH3, GH10 and GH115, the G. purpuraceus genome also appeared to encode relatively few enzymes involved in the degradation of hemicellulose, like other ECM fungi. Compared with these ECM fungi, the CAZyme families involved in the digestion of hemicellulose were increased in the four saprotrophic fungi. In addition, the carbohydrate-binding module belonging to family 1 (CBM1) as an additional domain that promotes the absorption of cellulose was found to have three genes in the G. purpuraceus genome, which is indispensable for the efficiency of CBH-type catalytic domains (i.e., processive cellulases) in depolymerizing crystalline regions of cellulose [64]. Furthermore, the pectin-degrading enzymes (GH43, GH51, GH93, PL1, PL3 and PL4) in G. purpuraceus and other symbiotic fungi were also exhibiting a similar pattern. For the AA1 family, there may be four genes in the G. purpuraceus genome through the CAZy database annotation, which is widely present in fungi and is currently divided into three subfamilies, laccases, ferroxidases and laccase-like multicopper oxidases (http://www.cazy.org/AA1.html, accessed on 8 November 2021). For the CAZyme genes encoding ligninolytic enzymes, focusing on class II peroxidases (AA2), G. purpuraceus had a maximum of 10 genes encoding class II PODs. However, this does not prove that G. purpuraceus can degrade lignin since the class II PODs-encoding genes were found in a phylogenetically wide range of ECM fungi [69]. Considering the highly efficient ability of P. portentosus to degrade starch [70], these CAZyme families encoding α-amylase enzymes (e.g., GH13, GH70, GH77, GH57, GH119 families) were investigated. Similar to the P. portentosus genome (Table S14), the main α-amylase enzyme clan was mainly in GH13, while GH70, GH77, GH57 and GH119 were all absent in the selected genomes. A total of 33 genes found in the G. purpuraceus genome were involved in the starch and sucrose metabolism pathway.
The results of the comparative analysis of CAZymes showed that G. purpuraceus may have lost the capacity to completely degrade plant cell walls, though it still retains some genes for degrading PCWDEs, similar to R. griseocarnosa [55]. However, the abundant genes in GH 13, similar to P. Portentosus, suggest that G. purpuraceus may have a certain ability to degrade starch, which needs further confirmation.
3.4.4. Comparative Analysis of Secondary Metabolisms
Fungi are a rich source of bioactive secondary metabolites, and mushroom-forming fungi are especially known for the synthesis of numerous bioactive and often cytotoxic sesquiterpenoid secondary metabolites [71]. The comparison of secondary metabolisms of G. purpuraceus with those of 13 other fungi was performed by genome mining and searching based on previous studies. The results are shown in Table 3. From only 14 selected fungal genomes, most basidiomycetes appear to have a greater number of terpene-involved genes than ascomycetes. However, more statistical data are required to support this conclusion in both taxa. Meanwhile, there was no obvious difference between ECM fungi and saprotrophic fungi.

Table 3.
Comparison of secondary metabolism of G. purpuraceus with 13 other fungi.
In plants and fungi, terpenoids, or modified terpenes, are an important group of natural bioactive products [72,73]. For ectomycorrhizas in nature, terpenoids have been recognized as key compounds, especially sesquiterpenes. ECM fungi can promote lateral root formation by releasing sesquiterpenes [74]. In the genome of G. purpuraceus, there are seven gene clusters associated with terpene biosynthesis. Through checking the G. purpuraceus genes in the “terpenoid backbone biosynthesis (map00900)” pathway, 16 key genes were identified, shown in Table 4. Just like most fungi, terpenoid backbone biosynthesis in G. purpuraceus can also proceed through the mevalonate pathway (Figure S2) [75].

Table 4.
Putative genes involved in terpenoid backbone biosynthesis.
In addition, NRPSs are considered candidates for roles in fungal pathogenesis, because the products of several NRPSs have already been proven as virulence factors, such as the virulence of Fusarium avenaceum on potatoes, HC-toxin for Cochliobolus carbonum race 1 on corn and AM-toxin for Alternaria alternata on apple [76]. In the genome of G. purpuraceus, a total of 10 NRPS and NRPS-like were identified (Table 3). Through comparative analysis, the number of NPRS and NRPS-like of G. purpuraceus is not too much, even lower than L. Edodes and P. ostreatus, which are common edible mushrooms. Even so, the pathogenicity of G. purpuraceus still needs to be confirmed through a series of experiments.
3.5. Optimization of Culture Medium
From the genome annotation results, G. purpuraceus exhibited a limited ability to degrade lignocellulose. Comparative genomics analysis showed that the gene families associated with starch and sucrose metabolism were abundant, which is similar to P. Portentosus. Therefore, seven different carbon sources were further screened to find the most suitable culture medium for food fermentation and strain propagation. Measurements of the colony diameter, the growth parameter and morphology of G. purpuraceus are shown in Figure 7. From the colony diameter (Figure 7A), the fastest growth of G. purpuraceus is glucose, reaching 2.66 cm, followed by sucrose (2.65 cm), malt extract powder (2.24 cm), maltose (2.23 cm), potato extract powder (1.35 cm) and lactose (1.34 cm). It is confusing that G. purpuraceus cannot utilize the corn extract powder, where the colony diameter still showed the original 0.6 cm. The results showed that sucrose and maltose were efficiently utilized by G. purpuraceus, except for lactose. In the medium added with potato and corn extract powder containing a lot of starch, the ability of G. purpuraceus to utilize starch was not as efficient as P. Portentosus [70]. As for colony diameter, the utilization of sucrose by G. purpuraceus was comparable to that of glucose, and there was no significant difference between them (p > 0.05). The utilization of maltose or malt extract powder was second only to glucose and sucrose, with a significant difference (p < 0.01). In addition, the morphology of G. purpuraceus in malt extract powder exhibited denser mycelium and healthy growth (Figure 7B-5). The colony diameters and morphology of G. purpuraceus cultured on different carbon sources provided a reference for the selection of cheap carbon sources for the large-scale production of strains, such as sugarcane juice usually used in yeast fermentation.
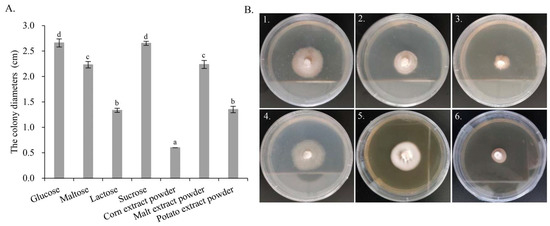
Figure 7.
The colony diameters and morphology of G. purpuraceus cultured on different carbon sources. (A) Colony diameters. Different letters on the column indicate a significant difference (p < 0.01). (B) Morphology of G. purpuraceus on different carbon sources after 60 days of cultivation: (B1) glucose, (B2) maltose, (B3) lactose, (B4) sucrose, (B5) malt extract powder, (B6) potato extract powder.
4. Discussion
G. purpuraceus is a potential edible and medicinal wild fungus. For the purpose of artificial domestication, we sequenced the whole genome of G. purpuraceus and compared it with the genomes of 13 edible fungi, including white-rot, brown-rot and ECM edible fungi. Among the 14 fungal genomes, the average number of CAZymes showed a publicly recognized phenomenon: white-rot fungi were larger than brown-rot fungi, and brown-rot fungi were larger than symbiotic fungi, which may be related to the role of different ecological niches of edible fungi in the environment [20,21,22]. In the genome of G. purpuraceus, CAZyme protein families exhibited similarity to those of the ECM fungi, and the global numbers of GHs, GTs, PLs, CEs, CBMs and AAs of G. purpuraceus were less than those of typical white/brown-rot fungi. Some species of Gomphus are ectomycorrhizal, and the mycorrhizal system between Gomphus floccosus and Abies religiosa has been successfully synthesized under controlled conditions [10], which indicates that G. purpuraceus may also be an ECM fungus.
The PCWDEs are a major pool of organic carbon, so the ability to decompose PCWDEs is crucial to the artificial domestication of wild edible mushrooms. For white-rot fungi, the extracellular oxidative enzymes, particularly class II PODs, give white-rot fungi the ability to completely degrade plant cell wall components, including cellulose, hemicellulose and lignin [77]. Moreover, glycoside hydrolases (GHs) in white-rot fungi can break down crystalline cellulose. The comparative genomics analysis also showed that the numbers of GH, CE, AA and CBM family members are far greater than other niches of fungi, which provides the genetic basis for the degradation ability of white-rot fungi. In addition to white-rot fungi, brown-rot fungi are also important components of plant-degrading filamentous fungi. Due to the absence of a series of oxidative enzymes related to lignin degradation, especially in class II PODs, GH6, GH7 and family 1 carbohydrate-binding module (CBM1), brown-rot fungi only cause complete degradation of polysaccharides and partial degradation of lignin [78,79]. In brown-rot fungi, the initial decomposition of lignocellulose takes place through a nonenzymatic step including hydroxyl radicals generated by the Fenton reaction [25]. This is also demonstrated by the statistics and comparisons of CAZymes among 14 fungi. Owing to these key genes involved in the degradation of lignocellulose, many white/brown-rot mushrooms can be successfully domesticated artificially, such as L. edodes, F. velutipes, P. ostreatus, etc. Another large group of fungi found in nature is ECM fungi. ECM fungi mainly obtain carbon sources from tree roots by forming ectomycorrhiza [28,80]. However, obtaining carbon sources from plants does not mean that the fungi have completely lost the ability to degrade lignocellulose that accumulates in wood and SOM [29,34]. The ability of certain ECM fungi to degrade lignocellulose has long been observed in fruiting bodies found on rotting wood and trunks of trees [31,32], which was also confirmed by comparative genomic analysis [33,34]. In particular, P. Portentosus has been successfully domesticated in China and Thailand, despite being considered an ECM fungus. Therefore, the artificial domestication of wild edible fungi should not be restricted by the niches of ectomycorrhizal fungi.
Considering the artificial domestication of G. purpuraceus, genes encoding CAZymes were analyzed. In the G. purpuraceus genome, GH6 (two genes) and GH7 (one gene) were closely related to the degradation of plant cell wall polysaccharides, which were completely absent from eight other ECM fungi. Therefore, the results suggested that G. purpuraceus fungi have a certain ability to degrade crystalline cellulose. For LPMOs (AA9) involved in attacking cellulose chains, there were only two genes in the genome of G. purpuraceus, even less than some ECM fungi, such as R. griseocarnosa, L. bicolor and B. edulis. Meanwhile, three genes associated with CBM1 that promote the absorption of cellulose were found. In addition, 10 genes encoding class II PODs were found, which are the most abundant in the 14 fungi. Furthermore, laccases (multicopper oxidases) are widely distributed in plants, fungi, bacteria and insects, which are important to biosynthesis and lignin degradation, morphogenesis and pigment biosynthesis, among others [81]. For G. purpuraceus, only four genes existed in the AA1 family, which was lower than ECM and saprotrophic fungi, except for G. bonarii, M. importuna and F. velutipes. The few genes encoding laccase reduced the ability of G. purpuraceus to utilize lignocellulose existing in rotting wood or SOM. On the whole, the discovery of key genes encoding CAZymes indicated that G. purpuraceus may have a certain ability to degrade lignocellulose. However, whether these key genes involved in lignocellulosic degradation can meet the demand for the carbon source in the life cycle of G. purpuraceus must be further explored. Additionally, genes involved in the α-amylase family and starch and sucrose metabolism pathway were abundant in the G. purpuraceus genome, just like P. portentosus. To preliminarily investigate the ability to utilize different carbon sources, especially in starch, the optimization of culture medium for G. purpuraceus was performed. The results showed that starch from potato and corn extract powder could not be efficiently utilized by G. purpuraceus. The maximum colony diameter in sucrose indicated that G. purpuraceus could efficiently utilize sucrose, which was confirmed by 13 potential genes encoding glycosidase. According to the description of malt powder product ingredients, malt extract powder contains a variety of carbohydrates, proteins, peptides, amino acids, purines and pyrimidines, as well as various vitamins, etc. Considering the addition of peptone in the basial formulation, the growth of G. purpuraceus requires the direct supply of purines, pyrimidines and vitamins, in addition to carbon sources, rather than obtaining them from complex substances. Therefore, we posit that the morphology of G. purpuraceus would exhibit healthy growth in the medium containing sucrose/maltose, peptides, amino acids, purines, pyrimidines and various vitamins. Of course, the hypothesis needs verification in further research, and the formulation still must be further investigated.
In addition to focusing on the ability of G. purpuraceus to acquire carbon sources, the mating type genes that are crucial in the sexual development of mushroom-forming have also been preliminarily identified. Mating is governed by mating genes located at distinct loci. In most basidiomycetes, there are two complex MATs (MAT-A and B) that control sexual compatibility in the monokaryons and regulate the maintenance of the dikaryotic state [53]. The typical MAT-A locus contains one or more pairs of genes for two types of homeodomain transcription factors, which regulate clamp-cell formation and conjugate nuclear division. Meanwhile, the MAT-B regulates nuclear migration and clamp-cell fusion [82]. Generally, the homeodomain-encoding genes and the flanking genes in the MAT-A locus are highly conserved in most mushroom-forming fungi, while the MAT-Bs of different mushrooms vary in the genomic organization. Overall, additional research is needed to elucidate the fruiting body formation of G. purpuraceus by MAT genes.
5. Conclusions
In summary, we performed Nanopore sequencing and de novo assembly of the G. purpuraceus genome and provided a 40.15 Mb genome sequence consisting of 50 scaffolds with an N50 length of 2.06 Mb. The G. purpuraceus genome allowed us to predict the gene functions and study the biosynthesis of active compounds. The comparative genomics analysis of gene family, CAZymes and secondary metabolism may enhance our understanding of the survival mechanisms and saprotrophic capacity of G. purpuraceus. Moreover, optimization of the culture medium for G. purpuraceus also provided a foundation for food fermentation and strain propagation. The elucidation of the G. purpuraceus genome in this study provides the foundational information to further study the niche, biosynthesis of pharmacologically active compounds and functional food applications of G. purpuraceus by molecular biology techniques.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13091628/s1, Figure S1: Kmer distribution of G. purpuraceus genome; Figure S2: The genes involved in the terpenoid backbone biosynthesis pathway identified from the G. purpuraceus genome (shown in blue); Table S1: Fungi and the origin of their genomes used in the study; Table S2: Summary statistics of functional annotation of predicted genes in public databases; Table S3: Gene Ontology (GO) functional classification of the G. purpuraceus genome; Table S4: Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotation of the G. purpuraceus genome; Table S5: Clusters of Orthologous Groups (KOG) functional classification of the G. purpuraceus genome; Table S6: Pfam, Swissprot and TrEMBL and NR functional annotation of the G. purpuraceus genome; Table S7: Homologue genes identified in the mating-type loci of G. purpuraceus; Table S8: Summary statistics of functional annotation of predicted genes in public databases; Table S9: The secondary metabolites gene clusters in G. purpuraceus; Table S10: TCDB annotation of the G. purpuraceus genome; Table S11: PHI annotation of the G. purpuraceus genome; Table S12: DFVF annotation of the G. purpuraceus genome; Table S13: The subcellular localization information of the G. purpuraceus genome; Table S14: Distribution of CAZyme in G. purpuraceus and 13 other fungi.
Author Contributions
Conceptualization, Y.G. and L.Q.; methodology, Y.G.; software, Y.G.; formal analysis, Y.G. and S.Z.; investigation, Y.G., S.Z. and N.Y.; writing—original draft preparation, Y.G.; writing—review and editing, L.Q.; visualization, Y.G.; supervision, L.Q.; project administration, Y.G.; funding acquisition, Y.G. The authors state that all data necessary for confirming the conclusions presented in the article are presented fully within the article and the Supplementary Materials. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the Major Scientific and Technological Special Project of Guizhou Province (No. 20193006-2) and the Science and Technology Program of Guizhou Province (No. 2022-General 112).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials. The datasets used and/or analysed during the current study are available from the corresponding author. All the sequences were submitted on NCBI. For raw data, the accession numbers are SRR18321074. This Whole Genome Shotgun project has been deposited into DDBJ/ENA/GenBank under the accession JALBUN000000000. The bioproject accession is PRJNA815479, and the biosample accession is SAMN26589593.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Li, H.; Tian, Y.; Menolli, N.; Ye, L.; Karunarathna, S.C.; Perez-Moreno, J.; Rahman, M.M.; Rashid, H.; Phengsintham, P.; Rizal, L.; et al. Reviewing the world’s edible mushroom species: A new evidence-based classification system. Compr. Rev. Food. Sci. Food Saf. 2021, 20, 1982–2014. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Wang, G.; Zhang, Y.; Liao, G.; Ge, C. Analysis of flavor-related compounds in four edible wild mushroom soups. Microchem. J. 2020, 159, 105548. [Google Scholar] [CrossRef]
- Jacinto-Azevedo, B.; Valderrama, N.; Henríquez, K.; Aranda, M.; Aqueveque, P. Nutritional value and biological properties of Chilean wild and commercial edible mushrooms. Food Chem. 2021, 356, 129651. [Google Scholar] [CrossRef] [PubMed]
- De Silva, D.D.; Rapior, S.; Sudarman, E.; Stadler, M.; Xu, J.; Alias, S.A.; Hyde, K.D. Bioactive metabolites from macrofungi: Ethnopharmacology, biological activities and chemistry. Fungal Divers. 2013, 62, 1–40. [Google Scholar] [CrossRef]
- Wu, F.; Zhou, L.-W.; Yang, Z.-L.; Bau, T.; Li, T.-H.; Dai, Y.-C. Resource diversity of Chinese macrofungi: Edible, medicinal and poisonous species. Fungal Divers. 2019, 98, 1–76. [Google Scholar] [CrossRef]
- Zhang, J.X.; Chen, Q.; Huang, C.Y.; Gao, W.; Qu, J. History, current situation and trend of edible mushroom industry development. Mycosystema 2015, 34, 524–540. (In Chinese) [Google Scholar] [CrossRef]
- Agerer, R. Descriptions of ectomycorrhizae. Mycologist 1996, 16, 83–84. [Google Scholar] [CrossRef]
- Giachini, A.J.; Castellano, M.A. A new taxonomic classification for species in Gomphus sensu lato. Mycotaxon 2011, 115, 183–201. [Google Scholar] [CrossRef]
- Cantrell, C.L.; Case, B.P.; Mena, E.E.; Kniffin, T.M.; Duke, S.O.; Wedge, D.E. Isolation and identification of antifungal fatty acids from the basidiomycete Gomphus floccosus. J. Agric. Food Chem. 2008, 56, 5062–5068. [Google Scholar] [CrossRef]
- Lamus, V.; Franco, S.; Montoya, L.; Endara, A.R.; Caballero, L.A.; Bandala, V. Mycorrhizal synthesis of the edible mushroom Turbinellus floccosus with Abies religiosa from central Mexico. Mycoscience 2015, 56, 622–626. [Google Scholar] [CrossRef]
- Jiang, M.-Y.; Yang, X.-L.; Fang, L.-Z.; Zhang, L.; Dong, Z.-J.; Liu, J.-K. Purpuracolide: A new alliacane sesquiterpene from the basidiomycete Gomphus purpuraceus. Z. Naturforsch. B 2008, 63, 1012–1014. [Google Scholar] [CrossRef]
- He, Y.; Wan, Y.; Zhou, Y.; Cai, X.; Guo, Z.; Hu, F.; Qin, Y.; Tan, A.; Deng, Z. Two new alliacane sesquiterpenes from the fruiting bodies of edible mushroom Gomphus purpuraceus. Phytochem. Lett. 2022, 50, 85–88. [Google Scholar] [CrossRef]
- Grayer, R.J.; Thabrew, M.I.; Hughes, R.D.; Bretherton, S.; Lever, A.; Veitch, N.C.; Kite, G.C.; Lelli, R.; Simmonds, M.S. Phenolic and terpenoid constituents from the SriLankan medicinal plant Osbeckia aspera. Pharm. Biol. 2008, 46, 154–161. [Google Scholar] [CrossRef][Green Version]
- Ulubelen, A.; Öksüz, S.; Kolak, U.; Birman, H.; Voelter, W. Cardioactive terpenoids and a new rearranged diterpene from Salvia syriaca. Planta Med. 2000, 66, 627–629. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhang, H.; Sun, X.; Zhao, H.; Wu, L.; Zhu, D.; Yang, G.; Shao, Y.; Zhang, X.; Mao, X.; et al. A comprehensive review of the structure elucidation and biological activity of triterpenoids from Ganoderma spp. Molecules 2014, 19, 17478–17535. [Google Scholar] [CrossRef]
- Kupka, J.; Anke, T.; Giannetti, B.-M.; Steglich, W. Antibiotics from basidiomycetes. Arch. Microbiol. 1981, 130, 223–227. [Google Scholar] [CrossRef]
- Sharma, S.H.; Thulasingam, S.; Nagarajan, S. Terpenoids as anti-colon cancer agents-A comprehensive review on its mechanistic perspectives. Eur. J. Pharmacol. 2017, 795, 169–178. [Google Scholar] [CrossRef]
- Tedersoo, L.; May, T.W.; Smith, M.E. Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 2010, 20, 217–263. [Google Scholar] [CrossRef]
- Tan, A.H.; Liu, F.Z. First report of the characteristic form and growing up environment of Gomphus purpuraceus. Edible Fungi. China. 2002, 21, 6–7. (In Chinese) [Google Scholar] [CrossRef]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef]
- Floudas, D.; Held, B.W.; Riley, R.; Nagy, L.G.; Koehler, G.; Ransdell, A.S.; Younus, H.; Chow, J.; Chiniquy, J.; Lipzen, A.; et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet. Biol. 2015, 76, 78–92. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.G.; Riley, R.; Tritt, A.; Adam, C.; Daum, C.; Floudas, D.; Sun, H.; Yadav, J.S.; Pangilinan, J.; Larsson, K.-H.; et al. Comparative genomics of early-diverging mushroom-forming fungi provides insights into the origins of lignocellulose decay capabilities. Mol. Biol. Evol. 2016, 33, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Almási, É.; Sahu, N.; Krizsán, K.; Balint, B.; Kovács, G.M.; Kiss, B.; Cseklye, J.; Drula, E.; Henrissat, B.; Nagy, I.; et al. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. New Phytol. 2019, 224, 902–915. [Google Scholar] [CrossRef]
- Baldrian, P. Enzymes of saprotrophic basidiomycetes. Br. Mycol. Soc. Symp. 2008, 28, 19–41. [Google Scholar] [CrossRef]
- Martinez, D.; Challacombe, J.; Morgenstern, I.; Hibbett, D.; Schmoll, M.; Kubicek, C.P.; Ferreira, P.; Ruiz-Duenas, F.J.; Martinez, A.T.; Kersten, P.; et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. USA 2009, 106, 1954–1959. [Google Scholar] [CrossRef] [PubMed]
- Eastwood, D.C.; Floudas, D.; Binder, M.; Majcherczyk, A.; Schneider, P.; Aerts, A.; Asiegbu, F.O.; Baker, S.E.; Barry, K.; Bendiksby, M.; et al. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 2011, 333, 762–765. [Google Scholar] [CrossRef] [PubMed]
- Boraston, A.B.; Bolam, D.; Gilbert, H.J.; Davies, G. Carbohydrate-binding modules: Fine-tuning polysaccharide recognition. Biochem. J. 2004, 382, 769–781. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A.; et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef]
- Shah, F.; Nicolás, C.; Bentzer, J.; Ellström, M.; Smits, M.M.; Rineau, F.; Canbäck, B.; Floudas, D.; Carleer, R.; Lackner, G.; et al. Ectomycorrhizal fungi decompose soil organic matter using oxidative mechanisms adapted from saprotrophic ancestors. New Phytol. 2016, 209, 1705–1719. [Google Scholar] [CrossRef]
- Kropp, B.R. Fungi from decayed wood as ectomycorrhizal symbionts of western hemlock. Can. J. For. Res. 1982, 21, 36–39. [Google Scholar] [CrossRef]
- Roberts, C.; Ceska, O.; Kroeger, P.; Kendrick, B. Macrofungi from six habitats over five years in Clayoquot Sound, Vancouver Island. Can. J. Bot. 2004, 82, 1518–1538. [Google Scholar] [CrossRef]
- Lõster, H.; Punzel, M. Systematics of pleurotoid Russulaceae from Guyana and Japan, with notes on their ectomycorrhizal status. Mycologia 2000, 92, 1119–1132. [Google Scholar] [CrossRef]
- Miyauchi, S.; Kiss, E.; Kuo, A.; Drula, E.; Kohler, A.; Sánchez-García, M.; Morin, E.; Andreopoulos, B.; Barry, K.W.; Bonito, G.; et al. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat. Commun. 2020, 11, 5125. [Google Scholar] [CrossRef] [PubMed]
- Looney, B.P.; Meidl, P.; Piatek, M.J.; Miettinen, O.; Martin, F.M.; Matheny, P.B.; Labbé, J.L. Russulaceae: A new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytol. 2018, 218, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Plettm, J.M.; Martin, F. Blurred boundaries: Lifestyle lessons from ectomycorrhizal fungal genomes. Trends Genet. 2011, 27, 14–22. [Google Scholar] [CrossRef]
- Cali, D.S.; Kim, J.S.; Ghose, S.; Alkan, C.; Mutlu, O. Nanopore sequencing technology and tools for genome assembly: Computational analysis of the current state, bottlenecks and future directions. Brief. Bioinform. 2019, 20, 1542–1559. [Google Scholar] [CrossRef]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Ruan, J.; Li, H. Fast and accurate long-read assembly with wtdbg2. Nat. Methods. 2020, 17, 155–158. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.S. Using repeatmasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2004, 4, 4–10. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef]
- Awrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.; Eddy, S.; Floden, E.; Gardner, P.P.; Jones, T.A.; Tate, J.; et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2014, 43, 130–137. [Google Scholar] [CrossRef]
- Birney, E.; Clamp, M.; Durbin, R. GeneWise and Genomewise. Genome Res. 2004, 14, 988–995. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Boeckmann, B. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Foulongne-Oriol, M.; Taskent, O.; Kües, U.; Sonnenberg, A.; van Peer, A.; Giraud, T. Mating-type locus organization and mating-type chromosome differentiation in the bipolar edible button mushroom Agaricus Bisporus. Genes 2021, 12, 1079. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Aerts, A.; Ahrén, D.; Brun, A.; Danchin, E.G.J.; Duchaussoy, F.; Gibon, J.; Kohler, A.; Lindquist, E.; Pereda, V.; et al. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 2008, 452, 88–92. [Google Scholar] [CrossRef]
- Li, H.; Wu, S.; Ma, X.; Chen, W.; Zhang, J.; Duan, S.; Gao, Y.; Kui, L.; Huang, W.; Wu, P.; et al. The Genome Sequences of 90 Mushrooms. Sci. Rep. 2018, 8, 9982. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Song, J.; Liang, J.; Wang, S.; Lu, J. Whole genome sequencing and genome annotation of the wild edible mushroom, Russula griseocarnosa. Genomics 2020, 112, 603–614. [Google Scholar] [CrossRef]
- Martin, F.; Kohler, A.; Murat, C.; Balestrini, R.; Coutinho, P.M.; Jaillon, O.; Montanini, B.; Morin, E.; Noel, B.; Percudani, R.; et al. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature 2010, 464, 1033–1038. [Google Scholar] [CrossRef]
- Gong, W.-B.; Li, L.; Zhou, Y.; Bian, Y.-B.; Kwan, H.-S.; Cheung, M.-K.; Xiao, Y. Genetic dissection of fruiting body-related traits using quantitative trait loci mapping in Lentinula edodes. Appl. Microbiol. Biotechnol. 2016, 100, 5437–5452. [Google Scholar] [CrossRef]
- Park, Y.-J.; Baek, J.H.; Lee, S.; Kim, C.; Rhee, H.; Kim, H.; Seo, J.-S.; Park, H.-R.; Yoon, D.-E.; Nam, J.-Y.; et al. Whole genome and global gene expression analyses of the model mushroom Flammulina velutipes reveal a high capacity for lignocellulose degradation. PLoS ONE 2014, 9, e93560. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Zhao, M.; Hsiang, T.; Feng, X.; Zhang, J.; Huang, C. Identification and characterization of small noncoding RNAs in genome sequences of the edible fungus Pleurotus ostreatus. BioMed Res. Int. 2016, 7, 2503023. [Google Scholar] [CrossRef]
- Liu, W.; Chen, L.; Cai, Y.; Zhang, Q.; Bian, Y. Opposite polarity monospore genome de novo sequencing and comparative analysis reveal the possible heterothallic life cycle of Morchella importuna. Int. J. Mol. Sci. 2018, 19, 2525. [Google Scholar] [CrossRef]
- Cao, Y.; Zhang, Y.; Yu, Z.; Wang, P.; Tang, X.; He, X.; Mi, F.; Liu, C.; Yang, D.; Xu, J. Genome sequence of Phlebopus portentosus strain PP33, a cultivated Bolete. Genome Announc. 2015, 3, e00326-15. [Google Scholar] [CrossRef]
- Kumla, J.; Hobbie, E.A.; Suwannarach, N.; Lumyong, S. The ectomycorrhizal status of a tropical black bolete, Phlebopus portentosus, assessed using mycorrhizal synthesis and isotopic analysis. Mycorrhiza 2016, 26, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. A new generation of homology search tools based on probabilistic inference. Genome Inf. 2009, 23, 205–211. [Google Scholar] [CrossRef]
- Gu, C.B.; Jiang, L.L.; Wang, K.Y.; Shi, X.B.; Duan, H.M.; Lin, C.H. Induction and Characteristics of Fusarium oxysporum f. sp. fragariae ZY-W Resistant to Tebuconazole. Sci. Agric. Sin. 2010, 43, 2897–2904. [Google Scholar] [CrossRef]
- Kang, H.; Chen, X.; Kemppainen, M.; Pardo, A.G.; Veneault-Fourrey, C.; Kohler, A.; Martin, F.M. The small secreted effector protein MiSSP7.6 of Laccaria bicolor is required for the establishment of ectomycorrhizal symbiosis. Environ. Microbiol. 2020, 22, 1435–1446. [Google Scholar] [CrossRef]
- Plett, J.M.; Kemppainen, M.; Kale, S.D.; Kohler, A.; Legué, V.; Brun, A.; Tyler, B.M.; Pardo, A.G.; Martin, F. A secreted effector protein of Laccaria bicolor is required for symbiosis development. Curr. Biol. 2011, 21, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Pellegrin, C.; Daguerre, Y.; Ruytinx, J.; Guinet, F.; Kemppainen, M.; Frey, N.F.D.; Puech-Pagès, V.; Hecker, A.; Pardo, A.G.; Martin, F.M.; et al. Laccaria bicolor MiSSP8 is a small-secreted protein decisive for the establishment of the ectomycorrhizal symbiosis. Environ. Microbiol. 2019, 21, 3765–3779. [Google Scholar] [CrossRef]
- Kojima, Y.; Várnai, A.; Eijsink, V.G.H.; Yoshida, M. The Role of Lytic Polysaccharide Monooxygenases in Wood Rotting Basidiomycetes, Trends Glycosci. Glycotechnol 2020, 32, 135–143. [Google Scholar] [CrossRef]
- Bödeker, I.T.M.; Nygren, C.M.R.; Taylor, A.F.S.; Lindahl, B.D. ClassII peroxidase-encoding genes are present in a phylogenetically wide range of ectomycorrhizal fungi. ISME J. 2009, 3, 1387–1395. [Google Scholar] [CrossRef]
- Wan, J.-N.; Li, Y.; Guo, T.; Ji, G.-Y.; Luo, S.-Z.; Ji, K.-P.; Cao, Y.; Tan, Q.; Bao, D.-P.; Yang, R.-H. Whole-genome and transcriptome sequencing of Phlebopus portentosus reveals its associated ectomycorrhizal niche and conserved pathways involved in fruiting body development. Front. Microbiol. 2021, 12, 732458. [Google Scholar] [CrossRef]
- Agger, S.; Lopez-Gallego, F.; Schmidt-Dannert, C. Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol. Microbiol. 2009, 72, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Wawrzyn, G.T.; Held, M.A.; Bloch, S.E.; Schmidt-Dannert, C. Genome mining for fungal secondary metabolic gene clusters. In Biosynthesis and Molecular Genetics of Fungal Secondary Metabolites; Springer: New York, NY, USA, 2015; Volume 2, pp. 43–65. [Google Scholar] [CrossRef]
- Quin, M.B.; Flynn, C.M.; Schmidtdannert, C. Traversing the fungal terpenome. Nat. Prod. Rep. 2014, 31, 1449–1473. [Google Scholar] [CrossRef] [PubMed]
- Ditengou, F.A.; Müller, A.; Rosenkranz, M.; Felten, J.; Lasok, H.; Van Doorn, M.M.; Legué, V.; Palme, K.; Schnitzler, J.-P.; Polle, A. Volatile signalling by sesquiterpenes from ectomycorrhizal fungi reprogrammes root architecture. Nat. Commun. 2015, 6, 6279. [Google Scholar] [CrossRef] [PubMed]
- Yap, H.-Y.Y.; Chooi, Y.-H.; Firdaus-Raih, M.; Fung, S.-Y.; Ng, S.-T.; Tan, C.-S.; Tan, N.-H. The genome of the tiger milk mushroom, Lignosus rhinocerotis, provides insights into the genetic basis of its medicinal properties. BMC Genom. 2014, 15, 635. [Google Scholar] [CrossRef]
- Yoder, O.C.; Turgeon, B.G. Fungal genomics and pathogenicity. Curr. Opin. Struct. Biol. 2001, 4, 315–321. [Google Scholar] [CrossRef]
- Wymelenberg, A.V.; Gaskell, J.; Mozuch, M.; Sabat, G.; Ralph, J.; Skyba, O.; Mansfield, S.D.; Blanchette, R.A.; Martinez, D.; Grigoriev, I.; et al. Comparative transcriptome and secretome analysis of wood decay fungi Postia placenta and Phanerochaete chrysosporium. Appl. Environ. Microbiol. 2010, 76, 3599–3610. [Google Scholar] [CrossRef]
- Yelle, D.J.; Ralph, J.; Lu, F.; Hammel, K.E. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ. Microbiol. 2008, 10, 1844–1849. [Google Scholar] [CrossRef]
- Hori, C.; Gaskell, J.; Igarashi, K.; Samejima, M.; Hibbett, D.; Henrissat, B.; Cullen, D. Genomewide analysis of polysaccharides degrading enzymes in 11 white- and brown-rot Polyporales provides insight into mechanisms of wood decay. Mycologia 2013, 105, 1412–1427. [Google Scholar] [CrossRef]
- Martin, F.; Kohler, A.; Murat, C.; Veneault-Fourrey, C.; Hibbett, D.S. Unearthing the roots of ectomycorrhizal symbioses. Nat. Rev. Microbiol. 2016, 14, 760–773. [Google Scholar] [CrossRef]
- Rivera-Hoyos, C.M.; Morales-Alvarez, E.D.; Poutou-Pinales, R.A.; Pedroza-Rodriguez, A.M.; Rodriguez-Vazquez, R.; Del-gado-Boada, J.M. Fungal laccases. Fungal Biol. Rev. 2013, 27, 67–82. [Google Scholar] [CrossRef]
- Gong, W.; Wang, Y.; Xie, C.; Zhou, Y.; Zhu, Z.; Peng, Y. Whole genome sequence of an edible and medicinal mushroom, Hericium erinaceus (Basidiomycota, Fungi). Genomics 2020, 112, 2393–2399. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
