Comprehensive Analyses of Simple Sequence Repeat (SSR) in Bamboo Genomes and Development of SSR Markers with Peroxidase Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and SSR Identification
2.2. Primer Design for SSR Markers
2.3. Functional Annotation and Identification of Peroxidase Family Genes
2.4. Evolution Analysis of Peroxidase Gene Family
2.5. Ka/Ks Calculation and Divergent Time Prediction
2.6. Plant Materials, Genomic DNA Isolation, and Detection
2.7. SSR Polymorphism Assessment
3. Results
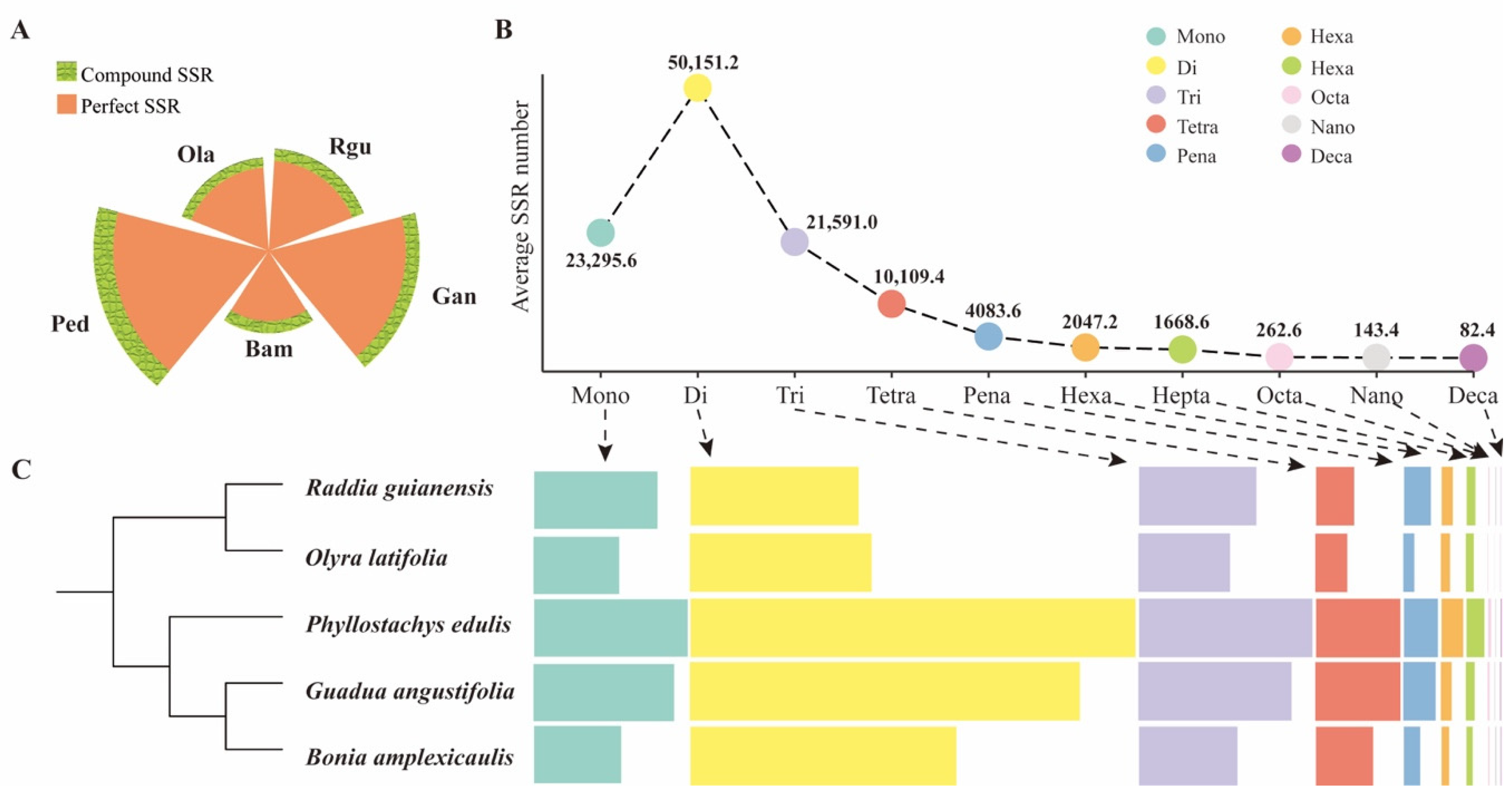
3.1. Comprehensive SSR Identification
3.2. Comparison of SSR Characteristics in Different Bamboo Species
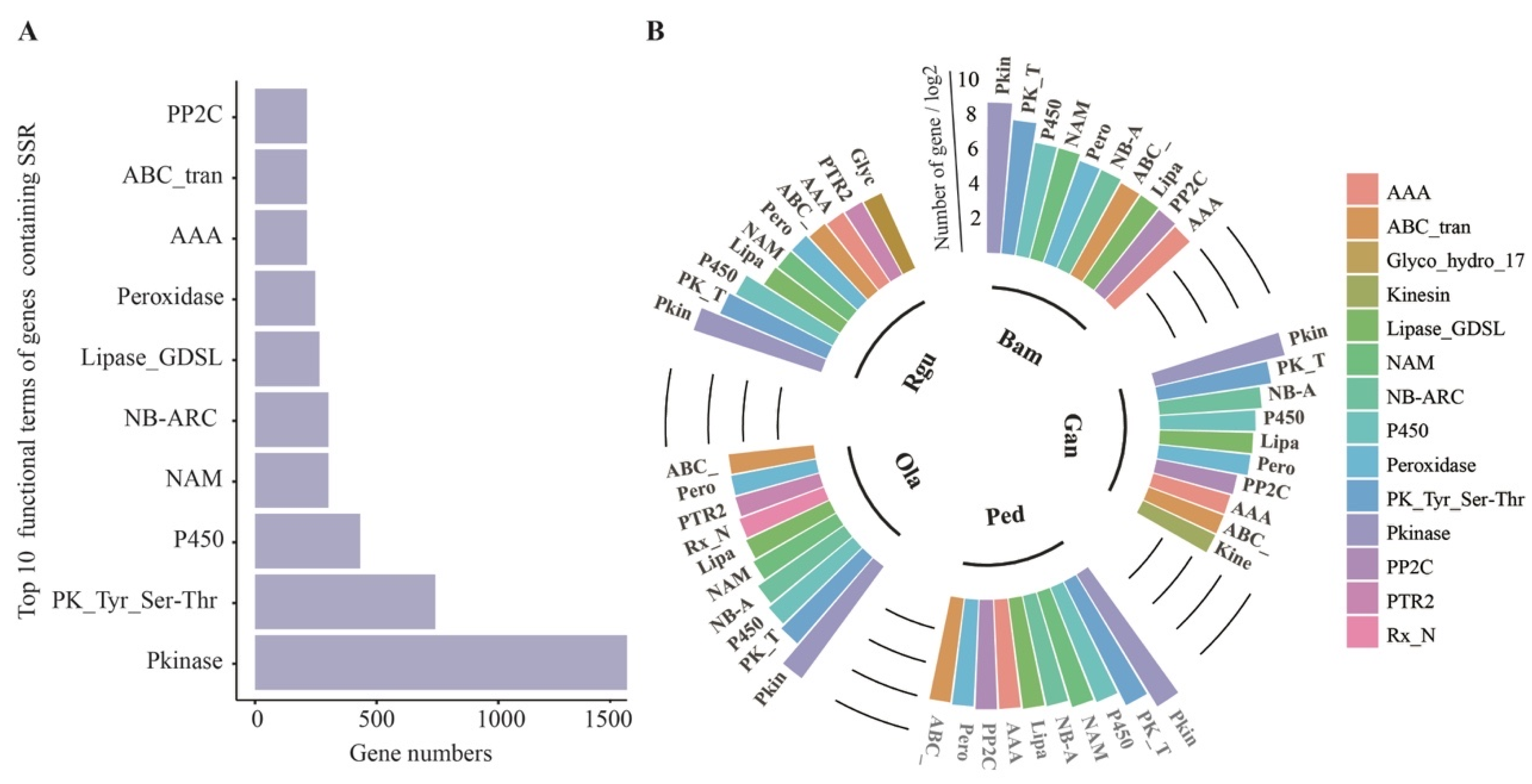
3.3. Statistical Analysis of the SSR-Containing Gene Functions
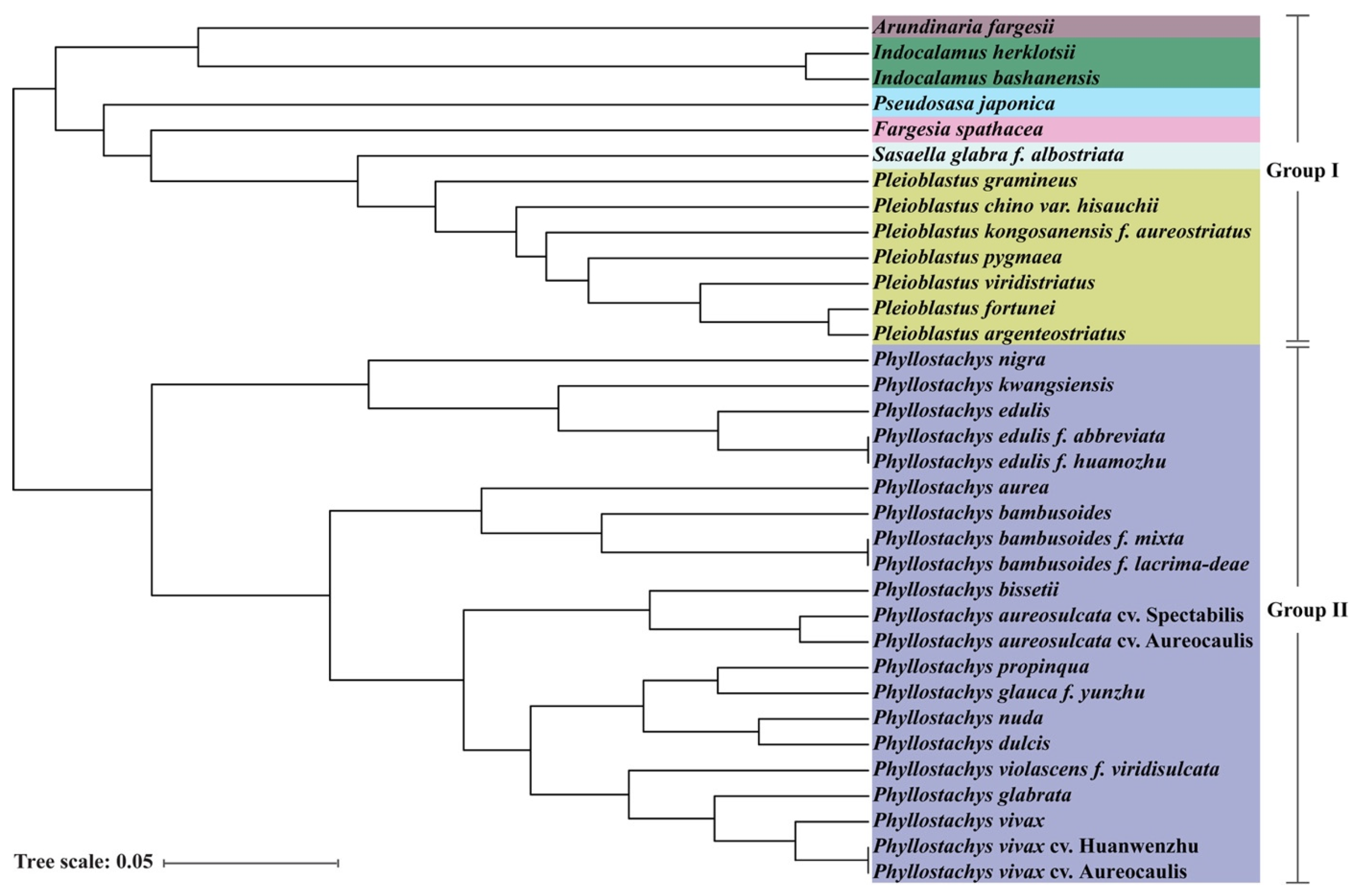
3.4. Analysis of the Peroxidase Gene Family
3.5. Development SSR Primer Pairs in Peroxidase Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Garrido-Cardenas, J.A.; Mesa-Valle, C.; Manzano-Agugliaro, F. Trends in plant research using molecular markers. Planta 2018, 247, 543–557. [Google Scholar] [CrossRef]
- Song, X.; Yang, Q.; Bai, Y.; Gong, K.; Wu, T.; Yu, T.; Pei, Q.; Duan, W.; Huang, Z.; Wang, Z.; et al. Comprehensive analysis of SSRs and database construction using all complete gene-coding sequences in major horticultural and representative plants. Hortic. Res. 2021, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, N.; Guo, Y.; Bai, Y.; Wu, T.; Yu, T.; Feng, S.; Zhang, Y.; Wang, Z.; Liu, Z.; et al. Comprehensive identification and characterization of simple sequence repeats based on the whole-genome sequences of 14 forest and fruit trees. For. Res. 2021, 1, 7. [Google Scholar] [CrossRef]
- Wang, X.T.; Zhang, Y.J.; Qiao, L.; Chen, B. Comparative analyses of simple sequence repeats (SSRs) in 23 mosquito species genomes: Identification, characterization and distribution (Diptera: Culicidae). Insect Sci. 2019, 26, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Zane, L.; Bargenlloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xing, M.; Song, L.; Yan, J.; Lu, W.; Zeng, A. Genome-wide analysis of simple sequence repeats in cabbage (Brassica oleracea L.). Front. Plant Sci. 2021, 12, 726084. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Yadav, I.S.; Jindal, M.; Sharma, P.K.; Dhillon, G.S.; Boora, R.S.; Arora, N.K.; Gill, M.I.S.; Chhuneja, P.; Mittal, A. Development of genome-wide functional markers using draft genome assembly of guava (Psidium guajava L.) cv. Allahabad Safeda to Expedite Molecular Breeding. Front. Plant Sci. 2021, 12, 708332. [Google Scholar] [CrossRef]
- Chen, J.; Guo, Y.; Hu, X.; Zhou, K. Comparison of the chloroplast genome sequences of 13 oil-tea camellia samples and identification of an undetermined oil-tea camellia species from Hainan province. Front. Plant Sci. 2021, 12, 798581. [Google Scholar] [CrossRef]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Nevo, E. Microsatellites within genes: Structure, function, and evolution. Mol. Biol. Evol. 2004, 21, 991–1007. [Google Scholar] [CrossRef]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites genomic distribution putative functions and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef]
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, M.; Yrjälä, K.; Vinod, K.K.; Sharma, A.; Cho, J.; Satheesh, V.; Zhou, M. Genetics and genomics of moso bamboo (Phyllostachys edulis): Current status, future challenges, and biotechnological opportunities toward a sustainable bamboo industry. Food Energy Secur. 2020, 9, e229. [Google Scholar] [CrossRef]
- Clark, L.G.; Londoño, X.; Ruiz-Sanchez, E. Bamboo Taxonomy and Habitat. In Bamboo; Liese, W., Köhl, M., Eds.; Springer: Cham, Switzerland, 2015; pp. 1–30. [Google Scholar] [CrossRef]
- Zhao, H.; Sun, S.; Ding, Y.; Wang, Y.; Yue, X.; Du, X.; Wei, Q.; Fan, G.; Sun, H.; Lou, Y.; et al. Analysis of 427 genomes reveals moso bamboo population structure and genetic basis of property traits. Nat. Commun. 2021, 12, 5466. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Bai, T.; Dai, H.; Wei, Q.; Zhang, W.; Ding, Y. Microsatellite markers revealed moderate genetic diversity and population differentiation of moso bamboo (Phyllostachys edulis)—a primarily asexual reproduction species in China. Tree Genet. Genomes 2017, 13, 130. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Peng, Z.; Sun, H.; Yue, X.; Lou, Y.; Dong, L.; Wang, L.; Gao, Z. Developing genome-wide microsatellite markers of bamboo and their applications on molecular marker assisted taxonomy for accessions in the genus Phyllostachys. Sci. Rep. 2015, 5, 8018. [Google Scholar] [CrossRef]
- Du, Q.; Pan, W.; Xu, B.; Li, B.; Zhang, D. Polymorphic simple sequence repeat (SSR) loci within cellulose synthase (PtoCesA) genes are associated with growth and wood properties in Populus tomentosa. New Phytol. 2013, 197, 763–776. [Google Scholar] [CrossRef]
- Guo, Z.H.; Ma, P.F.; Yang, G.Q.; Hu, J.Y.; Liu, Y.L.; Xia, E.H.; Zhong, M.C.; Zhao, L.; Sun, G.L.; Xu, Y.X.; et al. Genome sequences provide insights into the reticulate origin and unique traits of woody bamboos. Mol. Plant. 2019, 12, 1353–1365. [Google Scholar] [CrossRef]
- Yang, K.; Li, L.; Lou, Y.; Zhu, C.; Li, X.; Gao, Z. A regulatory network driving shoot lignification in rapidly growing bamboo. Plant Physiol. 2021, 187, 900–916. [Google Scholar] [CrossRef]
- Begovic, L.; Abicic, I.; Lalic, A.; Lepedus, H.; Cesar, V.; Leljak-Levanic, D. Lignin synthesis and accumulation in barley cultivars differing in their resistance to lodging. Plant Physiol. Biochem. 2018, 133, 142–148. [Google Scholar] [CrossRef]
- Hou, S.; Ren, X.; Yang, Y.; Wang, D.; Du, W.; Wang, X.; Li, H.; Han, Y.; Liu, L.; Sun, Z. Genome-wide development of polymorphic microsatellite markers and association analysis of major agronomic traits in core germplasm resources of tartary buckwheat. Front. Plant Sci. 2022, 13, 819008. [Google Scholar] [CrossRef]
- Terrett, O.M.; Dupree, P. Covalent interactions between lignin and hemicelluloses in plant secondary cell walls. Curr. Opin. Biotechnol. 2019, 56, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Cui, D.; Ye, Z.H. Secondary cell wall biosynthesis. New Phytol. 2019, 221, 1703–1723. [Google Scholar] [CrossRef]
- Luo, L.; Li, L. Molecular understanding of wood formation in trees. For. Res. 2022, 2, 5. [Google Scholar] [CrossRef]
- Zhao, H.; Gao, Z.; Wang, L.; Wang, J.; Wang, S.; Fei, B.; Chen, C.; Shi, C.; Liu, X.; Zhang, H.; et al. Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience 2018, 7, giy115. [Google Scholar] [CrossRef] [PubMed]
- Thiel, T.; Michalek, W.; Varshney, R.K.; Graner, A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 2003, 106, 411–422. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Munch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—new capabilities and interfaces. Nucleic. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic. Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Boutet, E.; Damien, L.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.J.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: How to use the entry view. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit incorporating γ-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Peng, Z.; Lu, Y.; Li, L.; Zhao, Q.; Feng, Q.; Gao, Z.; Lu, H.; Hu, T.; Yao, N.; Liu, K.; et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat. Genet. 2013, 45, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.M.; Fan, S.H.; Gao, J.; Li, X.P.; Cai, C.J.; Peng, Z.H. Extract genomic DNA from Phyllostachys edulis by CTAB-based method. For. Res. 2006, 19, 725–728. [Google Scholar]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Holland, M.M.; Parson, W. GeneMarker(R) HID: A reliable software tool for the analysis of forensic STR data. J. Forensic Sci. 2011, 56, 29–35. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarer: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Werck-Reichhart, D.; Feyereisen, R. Cytochromes P450: A success story. Genome Biol. 2000, 1, reviews3003. [Google Scholar] [CrossRef]
- Ichino, T.; Yazaki, K. Modes of secretion of plant lipophilic metabolites via ABCG transporter-dependent transport and vesicle-mediated trafficking. Curr. Opin. Plant Biol. 2022, 66, 102184. [Google Scholar] [CrossRef]
- Francoz, E.; Ranocha, P.; Nguyen-Kim, H.; Jamet, E.; Burlat, V.; Dunand, C. Roles of cell wall peroxidases in plant development. Phytochemistry 2015, 112, 15–21. [Google Scholar] [CrossRef]
- Huang, C.J.; Chu, F.H.; Huang, Y.S.; Tu, Y.C.; Hung, Y.M.; Tseng, Y.H.; Pu, C.E.; Hsu, C.T.; Chao, C.H.; Chou, Y.S.; et al. SSR individual identification system construction and population genetics analysis for Chamaecyparis formosensis. Sci. Rep. 2022, 12, 4126. [Google Scholar] [CrossRef]
- Kumar, S.P.J.; Susmita, C.; Sripathy, K.V.; Agarwal, D.K.; Pal, G.; Singh, A.N.; Kumar, S.; Rai, A.K.; Simal-Gandara, J. Molecular characterization and genetic diversity studies of Indian soybean (Glycine max (L.) Merr.) cultivars using SSR markers. Mol. Biol. Rep. 2022, 49, 2129–2140. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, Y.; Wang, Y.; Han, R.; Liang, Z.; He, Q.; Jia, Q. SSR loci analysis in transcriptome and molecular Marker development in Polygonatum sibiricum. Biomed Res. Int. 2022, 2022, 4237913. [Google Scholar] [CrossRef]
- Bonthala, B.; Abdin, M.Z.; Arya, L.; Pandey, C.D.; Sharma, V.; Yadav, P.; Verma, M. Genome-wide SSR markers in bottle gourd: Development, characterization, utilization in assessment of genetic diversity of National Genebank of India and synteny with other related cucurbits. J. Appl. Genet. 2022, 63, 237–263. [Google Scholar] [CrossRef]
- Jiang, W.X.; Zhang, W.J.; Ding, Y.L. Development of polymorphic microsatellite markers for Phyllostachys edulis (Poaceae), an important bamboo species in China. Appl. Plant Sci. 2013, 1, 1200012. [Google Scholar] [CrossRef]
- Portis, E.; Lanteri, S.; Barchi, L.; Portis, F.; Valente, L.; Toppino, L.; Rotino, G.L.; Acquadro, A. Comprehensive characterization of simple sequence repeats in eggplant (Solanum melongena L.) genome and construction of a web resource. Front. Plant Sci. 2018, 9, 401. [Google Scholar] [CrossRef]
- Cho, Y.G.; Ishii, T.; Temnykh, S.; Chen, X.; Lipovich, L.; McCouch, S.R.; Park, W.D.; Ayres, N.; Cartinhour, S. Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza sativa L.). Theor. Appl. Genet. 2000, 100, 713–722. [Google Scholar] [CrossRef]
- Passardi, F.; Theiler, G.; Zamocky, M.; Cosio, C.; Rouhier, N.; Teixera, F.; Margis-Pinheiro, M.; Ioannidis, V.; Penel, C.; Falquet, L.; et al. PeroxiBase: The peroxidase database. Phytochemistry 2007, 68, 1605–1611. [Google Scholar] [CrossRef]
- Koua, D.; Cerutti, L.; Falquet, L.; Sigrist, C.J.; Theiler, G.; Hulo, N.; Dunand, C. PeroxiBase: A database with new tools for peroxidase family classification. Nucleic Acids Res. 2009, 37, D261–D266. [Google Scholar] [CrossRef]
- Fawal, N.; Li, Q.; Savelli, B.; Brette, M.; Passaia, G.; Fabre, M.; Mathe, C.; Dunand, C. PeroxiBase: A database for large-scale evolutionary analysis of peroxidases. Nucleic Acids Res. 2013, 41, D441–D444. [Google Scholar] [CrossRef]
- Fagerstedt, K.V.; Kukkola, E.M.; Koistinen, V.V.; Takahashi, J.; Marjamaa, K. Cell wall lignin is polymerised by class III secretable plant peroxidases in Norway spruce. J. Integr. Plant Biol. 2010, 52, 186–194. [Google Scholar] [CrossRef]
- Shigeto, J.; Tsutsumi, Y. Diverse functions and reactions of class III peroxidases. New Phytol. 2016, 209, 1395–1402. [Google Scholar] [CrossRef]
- Song, X.; Ge, T.; Li, Y.; Hou, X. Genome-wide identification of SSR and SNP markers from the non-heading Chinese cabbage for comparative genomic analyses. BMC Genom. 2015, 16, 328. [Google Scholar] [CrossRef]
- Ma, S.; Han, C.; Zhou, J.; Hu, R.; Jiang, X.; Wu, F.; Tian, K.; Nie, G.; Zhang, X. Fingerprint identification of white clover cultivars based on SSR molecular markers. Mol. Biol. Rep. 2020, 47, 8513–8521. [Google Scholar] [CrossRef]
- Du, Q.; Gong, C.; Pan, W.; Zhang, D. Development and application of microsatellites in candidate genes related to wood properties in the Chinese white poplar (Populus tomentosa Carr.). DNA Res. 2013, 20, 31–44. [Google Scholar] [CrossRef] [Green Version]




| Species | Type | Genome Size (Mb) | SSR Number | Density (SSRs/Mb) |
|---|---|---|---|---|
| B. amplexicaulis | Woody | 848.6 | 107,976 | 127.2 |
| G. angustifolia | Woody | 1708.6 | 159,530 | 93.4 |
| P. edulis | Woody | 1907.6 | 185,102 | 97.0 |
| O. latifolia | Herbaceous | 646.6 | 87,197 | 134.9 |
| R. guianensis | Herbaceous | 626.4 | 98,511 | 157.3 |
| Total | 5737.8 | 638,316 | 111.2 |
| Primer ID | Genes | Motif | Position | Genotype Number | Na | PIC |
|---|---|---|---|---|---|---|
| SSR2 | PH02Gene25200 | (TAT)5 | Intron | 2 | 2 | 0.03 |
| SSR3 | PH02Gene00439 | (TACA)5 | Intron | 12 | 7 | 0.72 |
| SSR6 | PH02Gene02739 | (AG)6 | 5′ UTR | 2 | 3 | 0.40 |
| SSR7 | PH02Gene03921 | (GC)6 | Intron | 9 | 8 | 0.58 |
| SSR8 | PH02Gene05936 | (AGCAG)4 | 5′ UTR | 8 | 7 | 0.63 |
| SSR11 | PH02Gene12228 | (CT)7 | Intron | 12 | 9 | 0.75 |
| SSR13 | PH02Gene18893 | (TG)21 | 3′ UTR | 20 | 16 | 0.87 |
| SSR14 | PH02Gene21597 | (TGAT)4 | Intron | 5 | 3 | 0.37 |
| SSR17 | PH02Gene25407 | (CG)6 | Exon | 7 | 6 | 0.48 |
| SSR19 | PH02Gene33375 | (CT)11 | 5′ UTR | 3 | 3 | 0.16 |
| SSR23 | PH02Gene44535 | (ACC)8 | Exon | 4 | 3 | 0.17 |
| SSR32 | PH02Gene50471 | (CGG)5 | Exon | 2 | 2 | 0.03 |
| SSR37 | PH02Gene27682 | (CT)7 | Intron | 12 | 9 | 0.81 |
| SSR40 | PH02Gene44580 | (TGCA)4 | Intron | 2 | 2 | 0.08 |
| SSR43 | PH02Gene19592 | (CGA)5 | Exon | 13 | 9 | 0.78 |
| Mean | 7.60 | 5.93 | 0.46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Xiao, X.; Li, G.; Zhu, C.; Yang, K.; Feng, X.; Lou, Y.; Gao, Z. Comprehensive Analyses of Simple Sequence Repeat (SSR) in Bamboo Genomes and Development of SSR Markers with Peroxidase Genes. Genes 2022, 13, 1518. https://doi.org/10.3390/genes13091518
Liu Y, Xiao X, Li G, Zhu C, Yang K, Feng X, Lou Y, Gao Z. Comprehensive Analyses of Simple Sequence Repeat (SSR) in Bamboo Genomes and Development of SSR Markers with Peroxidase Genes. Genes. 2022; 13(9):1518. https://doi.org/10.3390/genes13091518
Chicago/Turabian StyleLiu, Yan, Xiaoyan Xiao, Guangzhu Li, Chenglei Zhu, Kebin Yang, Xiaohu Feng, Yongfeng Lou, and Zhimin Gao. 2022. "Comprehensive Analyses of Simple Sequence Repeat (SSR) in Bamboo Genomes and Development of SSR Markers with Peroxidase Genes" Genes 13, no. 9: 1518. https://doi.org/10.3390/genes13091518
APA StyleLiu, Y., Xiao, X., Li, G., Zhu, C., Yang, K., Feng, X., Lou, Y., & Gao, Z. (2022). Comprehensive Analyses of Simple Sequence Repeat (SSR) in Bamboo Genomes and Development of SSR Markers with Peroxidase Genes. Genes, 13(9), 1518. https://doi.org/10.3390/genes13091518






