Japanese Flounder HMGB1: A DAMP Molecule That Promotes Antimicrobial Immunity by Interacting with Immune Cells and Bacterial Pathogen
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Ethics
2.2. Sequence Analysis
2.3. Quantitative Real-Time Reverse Transcription-PCR (qRT-PCR)
2.4. Recombinant Protein Purification and Polyclonal Antibody Preparation
2.5. Detection of Extracellular HMGB1 and Lactate Dehydrogenase (LDH)
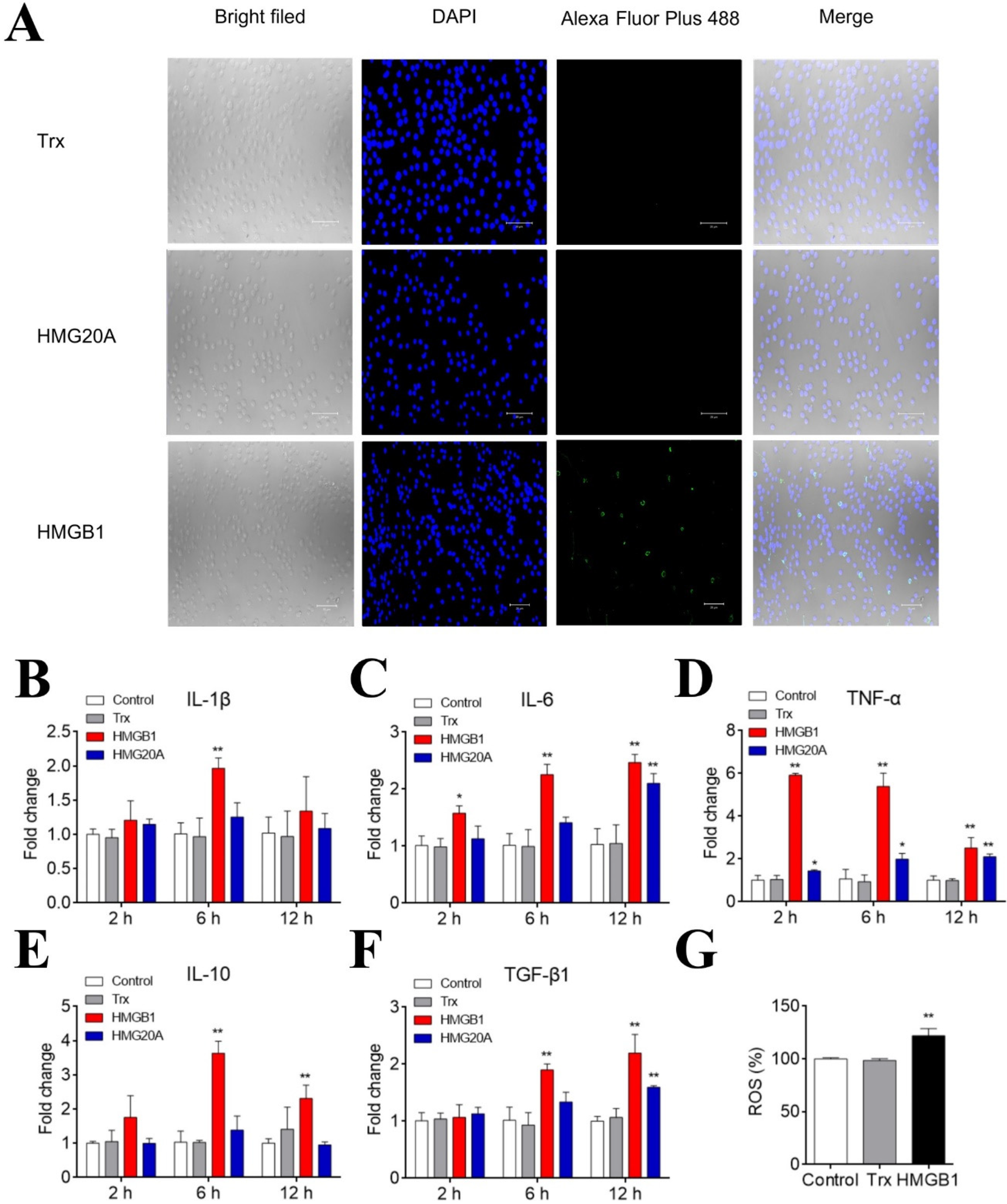
2.6. Interaction of HMGB1 and HMG20A with PBLs
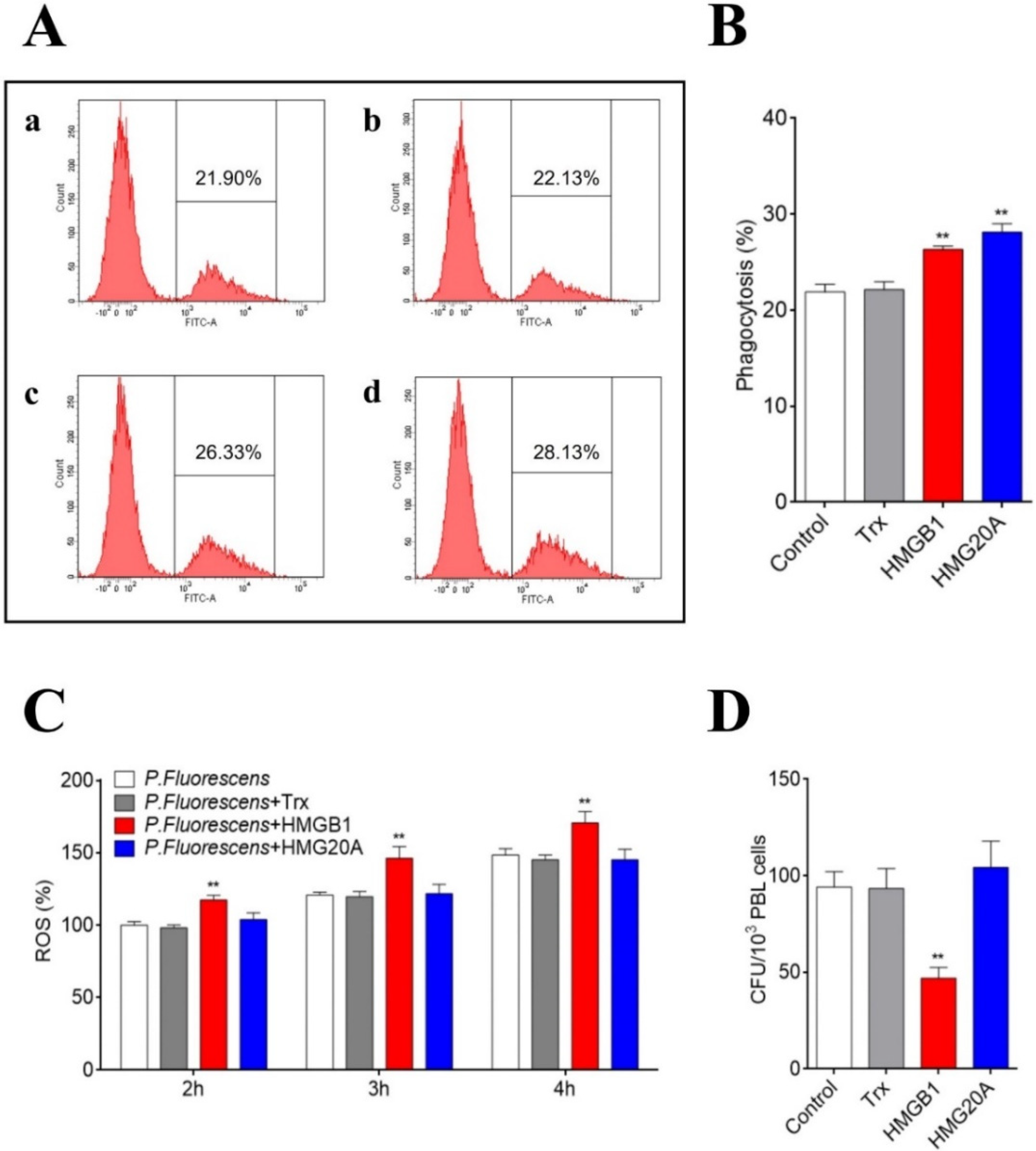
2.7. Effects of HMGB1 and HMG20A on Immune Gene Expression, ROS Production, and Phagocytosis
2.8. Interaction of HMGB1 and HMG20A with Bacteria
2.9. Effects of HMGB1 and HMG20A on Bacterial Agglutination
2.10. Effects of HMGB1/HMG20A on Bacterial Infection and ROS Production in PBLs
2.11. Statistical Analysis
3. Results
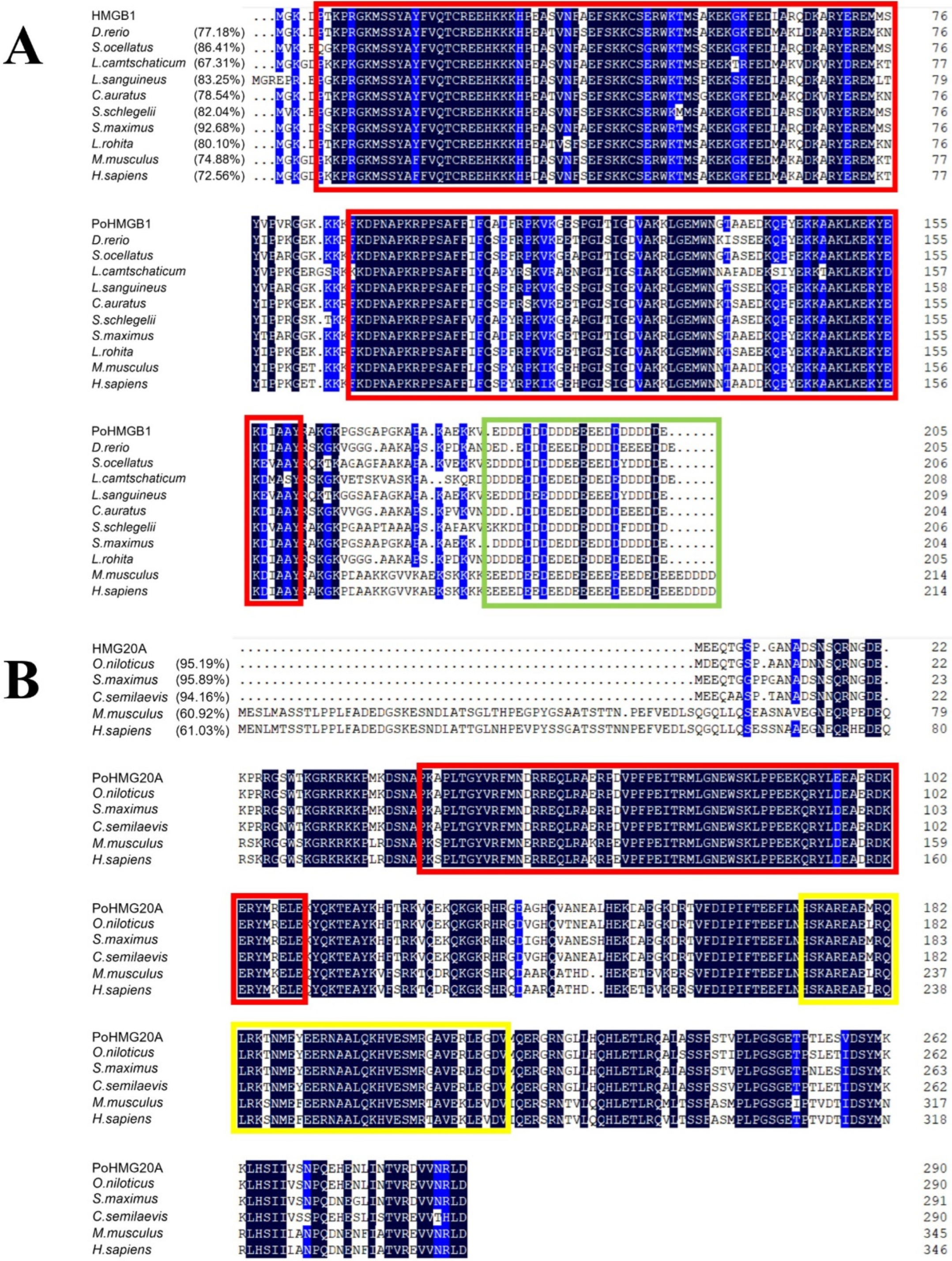
3.1. Sequence Characteristics of Flounder HMGB1 and HMG20A
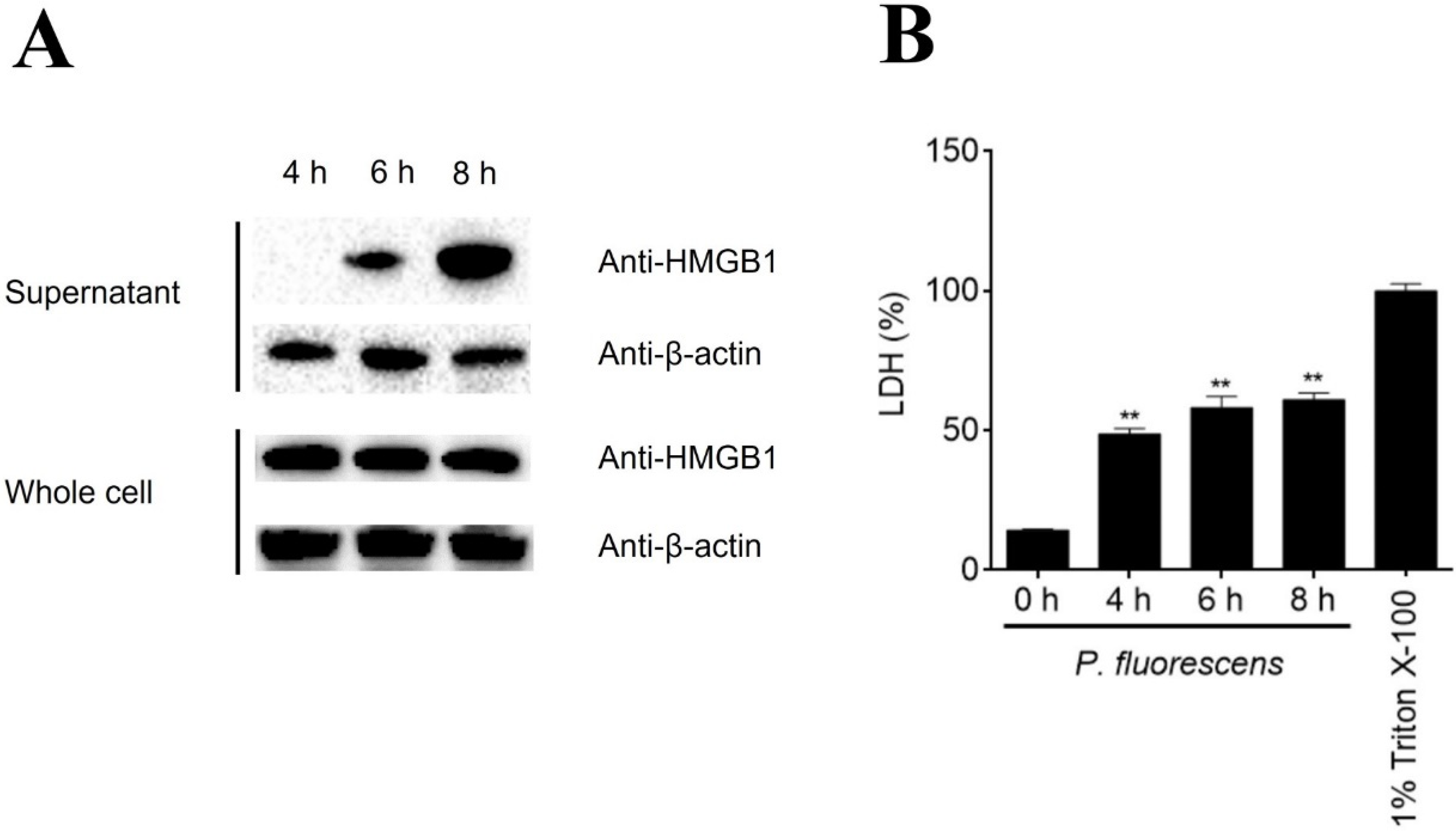
3.2. HMGB1 Is Released from PBLs during Bacterial Infection and Induces PBL Activation
3.3. HMGB1 Binds to Various Bacteria in a HMG20A-Competitive Manner and Causes Bacterial Agglutination
3.4. HMGB1 Promotes the Antibacterial Activity of PBLs and Impairs Bacterial Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lotze, M.T.; Tracey, K.J. High-mobility group box 1 protein (HMGB1): Nuclear weapon in the immune arsenal. Nat. Rev. Immunol. 2005, 5, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, G.H.; Johns, E.W. Isolation and characterisation of two calf-thymus chromatin non-histone proteins with high contents of acidic and basic amino acids. Eur. J. Biochem. 1973, 40, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, G.H.; Sanders, C.; Johns, E.W. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem. 1973, 38, 14–19. [Google Scholar] [CrossRef]
- Sumoy, L.; Carim, L.; Escarceller, M.; Nadal, M.; Gratacos, M.; Pujana, M.; Estivill, X.; Peral, B. HMG20A and HMG20B map to human chromosomes 15q24 and 19p13.3 and constitute a distinct class of HMG-box genes with ubiquitous expression. Cytogenet. Genome Res. 2000, 88, 62–67. [Google Scholar] [CrossRef]
- Thomas, J.O.; Travers, A.A. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001, 26, 167–174. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Czura, C.J.; Tracey, K.J. The cytokine activity of HMGB1. J. Leukoc. Biol. 2005, 78, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kwak, M.S.; Kim, H.S.; Lee, B.; Kim, Y.H.; Son, M.; Shin, J.-S. Immunological significance of HMGB1 post-translational modification and redox biology. Front. Immunol. 2020, 11, 1189. [Google Scholar] [CrossRef]
- Lee, S.-A.; Kwak, M.S.; Kim, S.; Shin, J.-S. The role of high mobility group box 1 in innate immunity. Yonsei Med. J. 2014, 55, 1165–1176. [Google Scholar] [CrossRef]
- Dupont, N.; Jiang, S.; Pilli, M.; Ornatowski, W.; Bhattacharya, D.; Deretic, V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011, 30, 4701–4711. [Google Scholar] [CrossRef]
- Lu, B.; Nakamura, T.; Inouye, K.; Li, J.; Tang, Y.; Lundbäck, P.; Valdes-Ferrer, S.I.; Olofsson, P.S.; Kalb, T.; Roth, J. Novel role of PKR in inflammasome activation and HMGB1 release. Nature 2012, 488, 670–674. [Google Scholar] [CrossRef]
- Qin, S.; Wang, H.; Yuan, R.; Li, H.; Ochani, M.; Ochani, K.; Rosas-Ballina, M.; Czura, C.J.; Huston, J.M.; Miller, E. Role of HMGB1 in apoptosis-mediated sepsis lethality. J. Exp. Med. 2006, 203, 1637–1642. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lundbäck, P.; Ottosson, L.; Erlandsson-Harris, H.; Venereau, E.; Bianchi, M.E.; Al-Abed, Y.; Andersson, U.; Tracey, K.J. Redox modifications of cysteine residues regulate the cytokine activity of HMGB1. Mol. Med. 2021, 27, 58. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.H.; Shin, J.-S. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 2006, 177, 7889–7897. [Google Scholar] [CrossRef] [PubMed]
- Nogueira-Machado, J.A.; Volpe, C.M.d.O.; Veloso, C.A.; Chaves, M.M. HMGB1, TLR and RAGE: A functional tripod that leads to diabetic inflammation. Expert Opin. Ther. Targets 2011, 15, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Sims, G.P.; Rowe, D.C.; Rietdijk, S.T.; Herbst, R.; Coyle, A.J. HMGB1 and RAGE in inflammation and cancer. Annu. Rev. Immunol. 2009, 28, 367–388. [Google Scholar] [CrossRef]
- van Beijnum, J.R.; Buurman, W.A.; Griffioen, A.W. Convergence and amplification of toll-like receptor (TLR) and receptor for advanced glycation end products (RAGE) signaling pathways via high mobility group B1 (HMGB1). Angiogenesis 2008, 11, 91–99. [Google Scholar] [CrossRef]
- Yu, M.; Wang, H.; Ding, A.; Golenbock, D.T.; Latz, E.; Czura, C.J.; Fenton, M.J.; Tracey, K.J.; Yang, H. HMGB1 signals through toll-like receptor (TLR) 4 and TLR2. Shock 2006, 26, 174–179. [Google Scholar] [CrossRef]
- Gong, W.; Li, Y.; Chao, F.; Huang, G.; He, F. Amino acid residues 201–205 in C-terminal acidic tail region plays a crucial role in antibacterial activity of HMGB1. J. Biomed. Sci. 2009, 16, 83. [Google Scholar] [CrossRef]
- Zetterström, C.K.; Strand, M.-L.; Söder, O. The high mobility group box chromosomal protein 1 is expressed in the human and rat testis where it may function as an antibacterial factor. Hum. Reprod. 2006, 21, 2801–2809. [Google Scholar] [CrossRef]
- Zetterström, C.K.; Bergman, T.; Rynnel-Dagöö, B.; Erlandsson Harris, H.; Söder, O.; Andersson, U.; Boman, H.G. High mobility group box chromosomal protein 1 (HMGB1) is an antibacterial factor produced by the human adenoid. Pediatric Res. 2002, 52, 148–154. [Google Scholar] [CrossRef]
- Lorenzo, P.I.; Vazquez, E.M.; López-Noriega, L.; Fuente-Martín, E.; Mellado-Gil, J.M.; Franco, J.M.; Cobo-Vuilleumier, N.; Martínez, J.A.G.; Romero-Zerbo, S.Y.; Perez-Cabello, J.A. The metabesity factor HMG20A potentiates astrocyte survival and reactive astrogliosis preserving neuronal integrity. Theranostics 2021, 11, 6983. [Google Scholar] [CrossRef] [PubMed]
- Rivero, S.; Ceballos-Chavez, M.; Bhattacharya, S.; Reyes, J. HMG20A is required for SNAI1-mediated epithelial to mesenchymal transition. Oncogene 2015, 34, 5264–5276. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Pan, H.-C.; Lin, S.L.; Zhang, W.-Q.; Rauvala, H.; Schachner, M.; Shen, Y.-Q. HMGB1 contributes to regeneration after spinal cord injury in adult zebrafish. Mol. Neurobiol. 2014, 49, 472–483. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Kuja-Panula, J.; Rouhiainen, A.; Chen, Y.-C.; Panula, P.; Rauvala, H. High mobility group box-1 (HMGB1; amphoterin) is required for zebrafish brain development. J. Biol. Chem. 2011, 286, 23200–23213. [Google Scholar] [CrossRef]
- Yang, C.; Peng, L.; Su, J. Two HMGB1 genes from grass carp Ctenopharyngodon idella mediate immune responses to viral/bacterial PAMPs and GCRV challenge. Dev. Comp. Immunol. 2013, 39, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Hodgkinson, J.W.; Li, C.; Kovacevic, N.; Belosevic, M. Identification and functional characterization of the goldfish (Carassius auratus L.) high mobility group box 1 (HMGB1) chromatin-binding protein. Dev. Comp. Immunol. 2014, 44, 245–253. [Google Scholar] [CrossRef]
- Pang, Y.; Xiao, R.; Liu, X.; Li, Q. Identification and characterization of the lamprey high-mobility group box 1 gene. PLoS ONE 2012, 7, e35755. [Google Scholar]
- Zhao, L.; Hu, Y.-H.; Sun, J.-S.; Sun, L. The high mobility group box 1 protein of Sciaenops ocellatus is a secreted cytokine that stimulates macrophage activation. Dev. Comp. Immunol. 2011, 35, 1052–1058. [Google Scholar] [CrossRef]
- Zhang, Z.; Niu, J.; Li, Q.; Huang, Y.; Jiang, B.; Wu, Y.; Huang, Y.; Jian, J. HMG20A from Nile tilapia (Oreochromis niloticus) involved in the immune response to bacterial infection. Fish Shellfish Immunol. 2021, 119, 499–507. [Google Scholar] [CrossRef]
- Li, H.; Sun, Y.; Sun, L. A Teleost CXCL10 Is Both an Immunoregulator and an Antimicrobial. Front. Immunol. 2022, 13, 917697. [Google Scholar] [CrossRef]
- Yu, C.; Zhang, P.; Zhang, T.-F.; Sun, L. IL-34 regulates the inflammatory response and anti-bacterial immune defense of Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2020, 104, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Zhang, P.; Li, X.-P.; Sun, L. Japanese flounder Paralichthys olivaceus interleukin 21 induces inflammatory response and plays a vital role in the immune defense against bacterial pathogen. Fish Shellfish Immunol. 2020, 98, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.-J.; Sun, L. Evaluation of housekeeping genes as references for quantitative real time RT-PCR analysis of gene expression in Japanese flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2011, 30, 638–645. [Google Scholar] [CrossRef]
- Zhou, Z.-X.; Sun, L. Immune effects of R848: Evidences that suggest an essential role of TLR7/8-induced, Myd88-and NF-κB-dependent signaling in the antiviral immunity of Japanese flounder (Paralichthys olivaceus). Dev. Comp. Immunol. 2015, 49, 113–120. [Google Scholar] [CrossRef]
- Li, W.; Guan, X.; Sun, L. Phosphatase and Tensin Homolog (PTEN) of Japanese Flounder—Its Regulation by miRNA and Role in Autophagy, Apoptosis and Pathogen Infection. Int. J. Mol. Sci. 2020, 21, 7725. [Google Scholar] [CrossRef]
- Li, M.-F.; Li, Y.-X.; Sun, L. CD83 is required for the induction of protective immunity by a DNA vaccine in a teleost model. Dev. Comp. Immunol. 2015, 51, 141–147. [Google Scholar] [CrossRef]
- Long, H.; Chen, C.; Zhang, J.; Sun, L. Antibacterial and antiviral properties of tongue sole (Cynoglossus semilaevis) high mobility group B2 protein are largely independent on the acidic C-terminal domain. Fish Shellfish Immunol. 2014, 37, 66–74. [Google Scholar] [CrossRef]
- Cato, L.; Stott, K.; Watson, M.; Thomas, J.O. The interaction of HMGB1 and linker histones occurs through their acidic and basic tails. J. Mol. Biol. 2008, 384, 1262–1272. [Google Scholar] [CrossRef]
- Lenka, S.S.; Paichha, M.; Basu, M.; Samanta, M. LrHMGB1 Shares Structural Similarities with Human HMGB1, and Its Expression Is Induced in Bacterial Infection, Antiviral Vaccination, and Pathogen-Associated Molecular Patterns Stimulation. DNA Cell Biol. 2018, 37, 708–723. [Google Scholar] [CrossRef]
- Erlandsson Harris, H.; Andersson, U. Mini-review: The nuclear protein HMGB1 as a proinflammatory mediator. Eur. J. Immunol. 2004, 34, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Thorburn, J.; Horita, H.; Redzic, J.; Hansen, K.; Frankel, A.E.; Thorburn, A. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die. Cell Death Differ. 2009, 16, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Dumitriu, I.E.; Baruah, P.; Valentinis, B.; Voll, R.E.; Herrmann, M.; Nawroth, P.P.; Arnold, B.; Bianchi, M.E.; Manfredi, A.A.; Rovere-Querini, P. Release of high mobility group box 1 by dendritic cells controls T cell activation via the receptor for advanced glycation end products. J. Immunol. 2005, 174, 7506–7515. [Google Scholar] [CrossRef] [PubMed]
- Faraco, G.; Fossati, S.; Bianchi, M.E.; Patrone, M.; Pedrazzi, M.; Sparatore, B.; Moroni, F.; Chiarugi, A. High mobility group box 1 protein is released by neural cells upon different stresses and worsens ischemic neurodegeneration in vitro and in vivo. J. Neurochem. 2007, 103, 590–603. [Google Scholar] [CrossRef]
- Qu, Y.; Zhan, Y.; Yang, S.; Ren, S.; Qiu, X.; Rehamn, Z.U.; Tan, L.; Sun, Y.; Meng, C.; Song, C. Newcastle disease virus infection triggers HMGB1 release to promote the inflammatory response. Virology 2018, 525, 19–31. [Google Scholar] [CrossRef]
- Vande Walle, L.; Kanneganti, T.-D.; Lamkanfi, M. HMGB1 release by inflammasomes. Virulence 2011, 2, 162–165. [Google Scholar] [CrossRef]
- Cai, J.; Xia, H.; Huang, Y.; Lu, Y.; Wu, Z.; Jian, J. Molecular cloning and characterization of high mobility group box1 (Ls-HMGB1) from humphead snapper, Lutjanus sanguineus. Fish Shellfish Immunol. 2014, 40, 539–544. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Xiang, Z.; Xiao, S.; Yu, F.; Yu, Z. High mobility group box 1 can enhance NF-κB activation and act as a pro-inflammatory molecule in the Pacific oyster, Crassostrea gigas. Fish Shellfish Immunol. 2013, 35, 63–70. [Google Scholar] [CrossRef]
- Wang, M.; Wang, L.; Guo, Y.; Zhou, Z.; Yi, Q.; Zhang, D.; Zhang, H.; Liu, R.; Song, L. A high mobility group box 1 (HMGB1) gene from Chlamys farreri and the DNA-binding ability and pro-inflammatory activity of its recombinant protein. Fish Shellfish Immunol. 2014, 36, 393–400. [Google Scholar] [CrossRef]
- He, Q.; You, H.; Li, X.-M.; Liu, T.-H.; Wang, P.; Wang, B.-E. HMGB1 promotes the synthesis of pro-IL-1β and pro-IL-18 by activation of p38 MAPK and NF-κB through receptors for advanced glycation end-products in macrophages. Asian Pac. J. Cancer Prev. 2012, 13, 1365–1370. [Google Scholar] [CrossRef]
- Ivanov, S.; Dragoi, A.-M.; Wang, X.; Dallacosta, C.; Louten, J.; Musco, G.; Sitia, G.; Yap, G.S.; Wan, Y.; Biron, C.A. A novel role for HMGB1 in TLR9-mediated inflammatory responses to CpG-DNA. Blood J. Am. Soc. Hematol. 2007, 110, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kokkola, R.; Tabibzadeh, S.; Yang, R.; Ochani, M.; Qiang, X.; Harris, H.E.; Czura, C.J.; Wang, H.; Ulloa, L. Structural basis for the proinflammatory cytokine activity of high mobility group box 1. Mol. Med. 2003, 9, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Schiraldi, M.; Raucci, A.; Muñoz, L.M.; Livoti, E.; Celona, B.; Venereau, E.; Apuzzo, T.; De Marchis, F.; Pedotti, M.; Bachi, A. HMGB1 promotes recruitment of inflammatory cells to damaged tissues by forming a complex with CXCL12 and signaling via CXCR4. J. Exp. Med. 2012, 209, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.-P.; Hu, Y.-H.; Yong, L.; Wang, J.-J.; Wang, G.-H.; Wang, R.-J.; Zhang, M. A high-mobility group box 1 that binds to DNA, enhances pro-inflammatory activity, and acts as an anti-infection molecule in black rockfish, Sebastes schlegelii. Fish Shellfish Immunol. 2016, 56, 402–409. [Google Scholar]
- Guo, D.-Y.; Cao, C.; Zhang, X.-Y.; Xiang, L.-X.; Shao, J.-Z. Scavenger receptor SCARA5 acts as an HMGB1 recognition molecule negatively involved in HMGB1-mediated inflammation in fish models. J. Immunol. 2016, 197, 3198–3213. [Google Scholar] [CrossRef]
- Janko, C.; Filipović, M.; Munoz, L.E.; Schorn, C.; Schett, G.; Ivanović-Burmazović, I.; Herrmann, M. Redox modulation of HMGB1-related signaling. Antioxid. Redox Signal. 2014, 20, 1075–1085. [Google Scholar] [CrossRef]
- Andersson, U.; Rauvala, H. Introduction: HMGB1 in Inflammation and Innate Immunity; Wiley Online Library: Hoboken, NJ, USA, 2011; Volume 270, pp. 296–300. [Google Scholar]
- Yanai, H.; Ban, T.; Taniguchi, T. High-mobility group box family of proteins: Ligand and sensor for innate immunity. Trends Immunol. 2012, 33, 633–640. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Antoine, D.J.; Andersson, U.; Tracey, K.J. The many faces of HMGB1: Molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J. Leukoc. Biol. 2013, 93, 865–873. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Gao, C.; Su, B.; Tan, F.; Yang, N.; Wang, G. Expression profiling and microbial ligand binding analysis of high-mobility group box-1 (HMGB1) in turbot (Scophthalmus maximus L.). Fish Shellfish Immunol. 2018, 78, 100–108. [Google Scholar] [CrossRef]
- Miao, J.; Ye, S.; Lan, J.; Ye, P.; Wen, Q.; Mei, L.; Liu, X.; Lin, J.; Zhou, X.; Du, S. Nuclear HMGB1 promotes the phagocytic ability of macrophages. Exp. Cell Res. 2020, 393, 112037. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Yu, C.; Jiang, S.; Sun, L. Japanese Flounder HMGB1: A DAMP Molecule That Promotes Antimicrobial Immunity by Interacting with Immune Cells and Bacterial Pathogen. Genes 2022, 13, 1509. https://doi.org/10.3390/genes13091509
Chen Y, Yu C, Jiang S, Sun L. Japanese Flounder HMGB1: A DAMP Molecule That Promotes Antimicrobial Immunity by Interacting with Immune Cells and Bacterial Pathogen. Genes. 2022; 13(9):1509. https://doi.org/10.3390/genes13091509
Chicago/Turabian StyleChen, Yuan, Chao Yu, Shuai Jiang, and Li Sun. 2022. "Japanese Flounder HMGB1: A DAMP Molecule That Promotes Antimicrobial Immunity by Interacting with Immune Cells and Bacterial Pathogen" Genes 13, no. 9: 1509. https://doi.org/10.3390/genes13091509
APA StyleChen, Y., Yu, C., Jiang, S., & Sun, L. (2022). Japanese Flounder HMGB1: A DAMP Molecule That Promotes Antimicrobial Immunity by Interacting with Immune Cells and Bacterial Pathogen. Genes, 13(9), 1509. https://doi.org/10.3390/genes13091509





