Antimicrobial Resistance Analysis of Escherichia coli Isolated from Pigeons in Qingdao, Shandong Province, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation and Identification of Escherichia coli
2.3. Antimicrobial Susceptibility Test
2.4. PCR Detection of Antimicrobial Resistance Genes
3. Results
3.1. Isolation and Identification Results of Bacteria
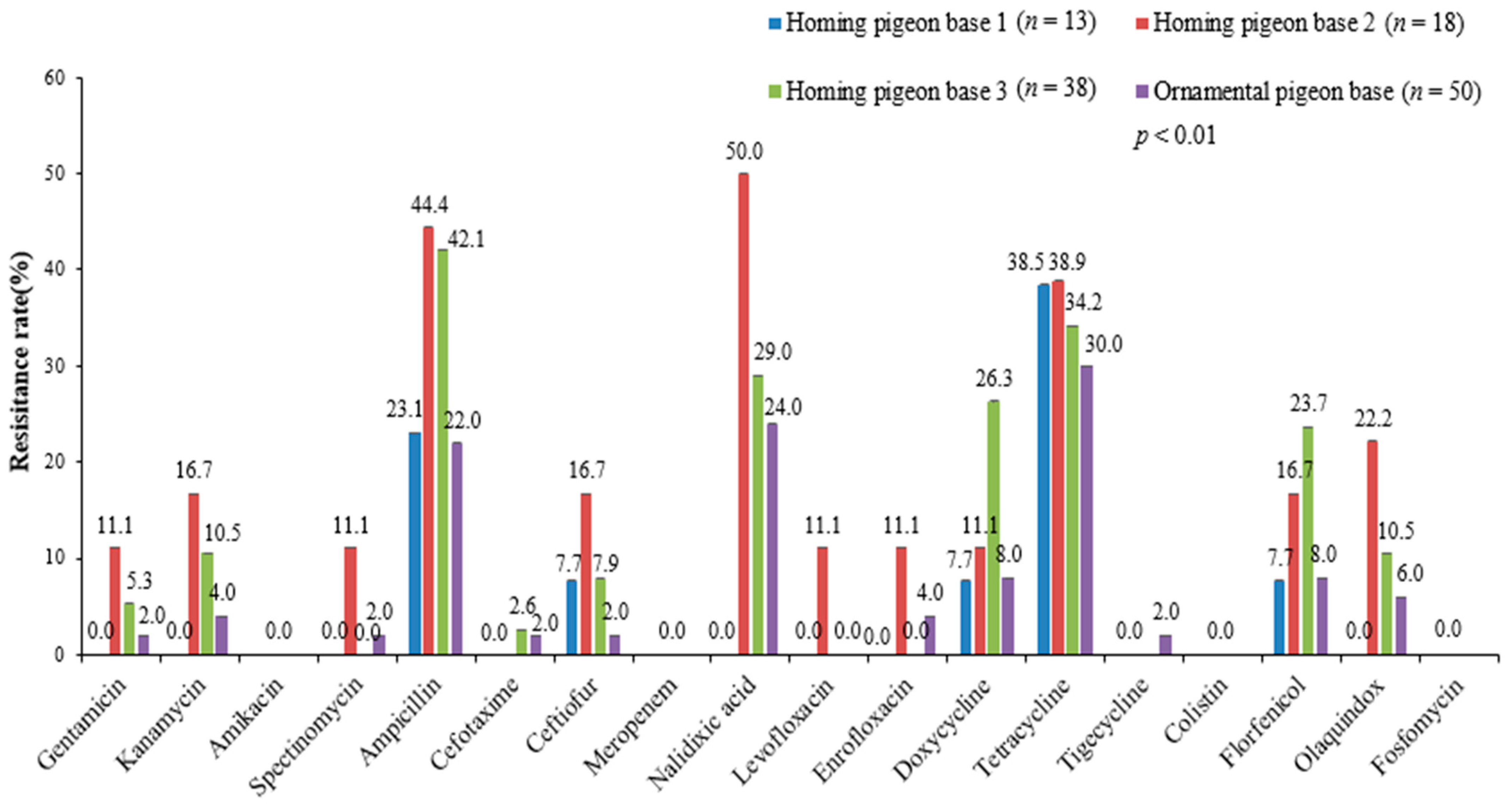
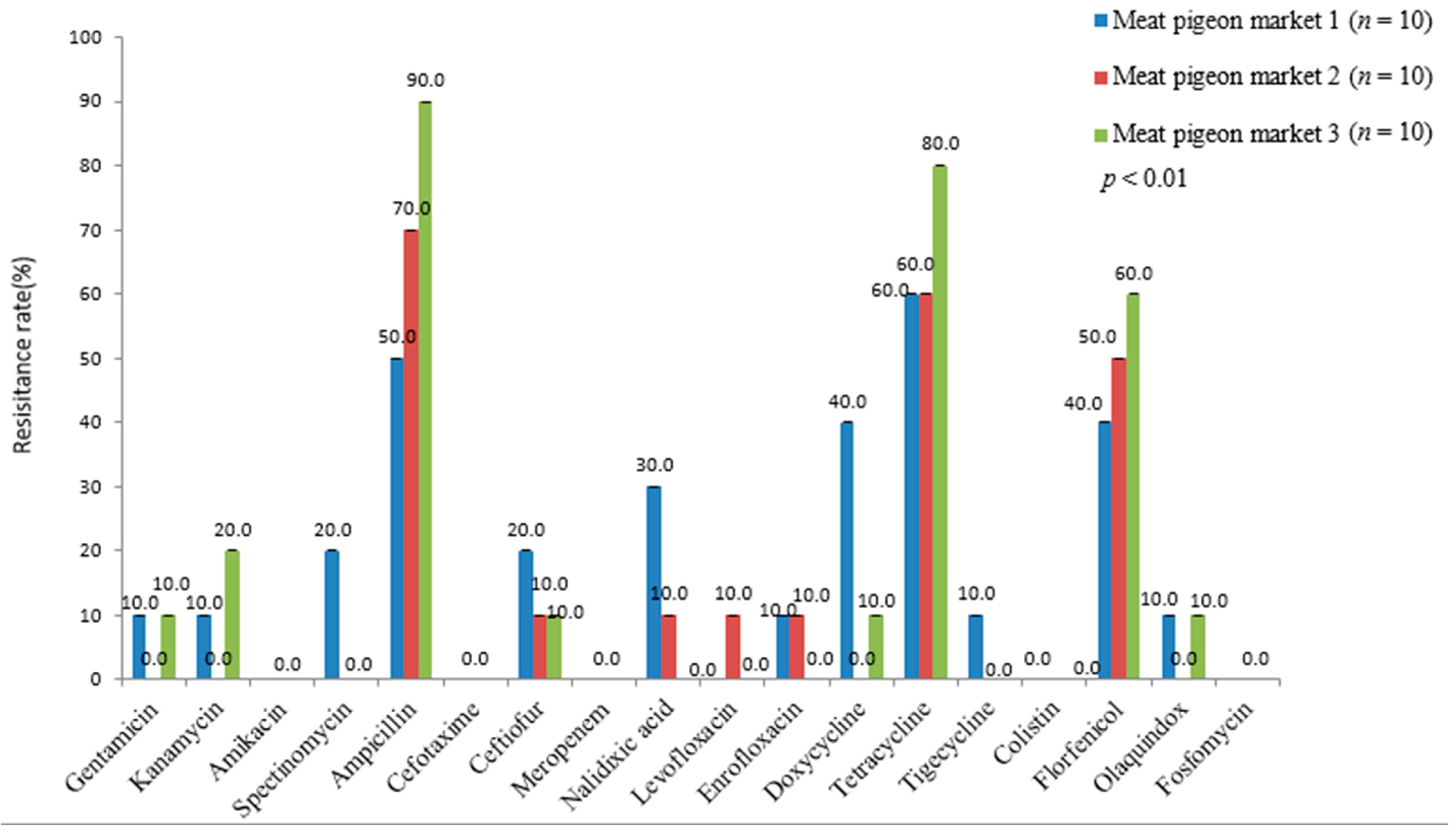
3.2. Antimicrobial Susceptibility Test
3.3. Multi-Drug Resistance of Isolates
3.4. PCR Test Results of Antimicrobial Resistance Genes
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, H.X. Isolation, Identification and Drug Sensitivity Test of Escherichia coli in Meat Pigeons. China Anim. Husb. 2016, 4, 44–45. [Google Scholar]
- Lu, C.P. Veterinary Microbiology, 5th ed.; China Agriculture Press: Beijing, China, 2013. [Google Scholar]
- Qu, Z.H.; Guo, X.Q.; Liu, T.; Li, J. Investigation and analysis of drug resistance of pathogenic Escherichia coli in meat pigeons. Heilongjiang Anim. Sci. Vet. Med. 2013, 3, 104–105. [Google Scholar]
- Xu, B.; Zhang, X.L.; He, Q.; Su, B. Isolation, Identification and Drug Sensitivity Test of Escherichia coli in Squash Pigeon. Anim. Husb. Vet. Med. 2013, 45, 123–124. [Google Scholar]
- Lv, M.N.; Sun, M.F.; Shen, H.D. Drug resistance analysis of Escherichia coli and Salmonella from pigeon origin in Guangdong from 2011 to 2018. Anim. Husb. Sci. Vet. Inf. 2019, 12, 46–48. [Google Scholar]
- Wang, H.B.; Zhu, L.X.; Gao, G.S.; Shi, Q.M. Isolation, Identification, Detection of Virulence Gene and Drug Sensitivity Test of Escherichia coli from Pigeon. J. Hebei Norm. Univ. Sci. Technol. 2019, 33, 48–53. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2015. [Google Scholar]
- Liu, J.H.; Wei, S.Y.; Ma, J.Y.; Zeng, Z.L.; Lü, D.; Yang, G.X.; Chen, Z.L. Detection and characterisation of CTX-M and CMY-2 β-lactamases among Escherichia coli isolates from farm animals in Guangdong Province of China. Int. J. Antimicrob. Agents 2007, 29, 576–581. [Google Scholar] [CrossRef] [PubMed]
- Brinas, L.; Moreno, M.A.; Zarazaga, M.; Porrero, C.; Saenz, Y.; Garcia, M.; Dominguez, L.; Torres, C. Detection of CMY-2, CTX-M-14, and SHV-12 β-Lactamases in Escherichia coli Fecal-Sample Isolates from Healthy Chickens. Antimicrob. Agents Chemother. 2003, 47, 2056. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Arakawa, Y. 16S ribosomal RNA methylation: Emerging resistance mechanism against aminoglycosides. Clin. Infect. Dis. 2007, 45, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Walsh, T.R.; Cuvillier, V. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef]
- Rebelo, A.R.; Bortolaia, V.; Kjeldgaard, J.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Hansen, I.M.; Guerra, B.; Malorny, B.; Borowiak, M.; Hammerl, J.A. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Eurosurveillance 2018, 23, 17-00672. [Google Scholar] [CrossRef]
- Borowiak, M.; Baumann, B.; Fischer, J. Development of a Novel mcr-6 to mcr-9 Multiplex PCR and Assessment of mcr-1 to mcr-9 Occurrence in Colistin-Resistant Salmonella enterica Isolates From Environment, Feed, Animals and Food (2011-2018) in Germany. Front. Microbiol. 2020, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.Y. Investigation and Analysis of Virulence Factors and Antibiotical Resistance of Poultry Originated E. coli Strains in Qingdao Region; Northeast Agricultural University: Harbin, China, 2014. [Google Scholar]
- Wang, K.; Liu, Z.H.; Zhao, L.; Liu, X.D.; Ren, H.Y.; Hu, L.F.; Hao, Z.H.; Wang, J.Q. Study on Epidemicity of Chicken blaNDM and mcr-1 Resistance Genes in Qingdao. Chin. J. Vet. Med. 2020, 56, 64–68. [Google Scholar]
- Bai, W.F.; Qing-Lin, W.U.; Jin, L.M.; Chen, Y.H. The Separation of E. coli in Pigeons and Its Sensitivity Test to Drugs. Acta Ecol. Anim. Domastici 2013, 34, 58–61. [Google Scholar]
- Xie, G.D.; Luo, H.P.; Ren, X.; Chen, Y.W.; Wen, Y.U.; Cui, S.H. Isolation, identification and drug resistance analysis of Escherichia coli and Salmonella in Beijing pigeon meat. J. Food Saf. Qual. 2019, 10, 48–53. [Google Scholar]
- McMillan, E.A.; Gupta, S.K.; Williams, L.E.; Jové, T.; Hiott, L.M.; Woodley, T.A.; Barrett, J.B.; Jackson, C.R.; Wasilenko, J.L.; Simmons, M.; et al. Antimicrobial Resistance Genes, Cassettes, and Plasmids Present in Salmonella Enterica Associated with United States Food Animals. Front. Microbiol. 2019, 10, 832. [Google Scholar] [CrossRef]
- Lu, T.W.; Liu, B.T.; Zhang, Q.D.; Liu, H.Q.; Zou, M. Epidemiological Analysis of Extended—spectrum β—lactamas—producing Escherichia coli from Cattle in Qingdao. Chin. J. Vet. Med. 2020, 56, 25–28. [Google Scholar]
- Singer, A.C.; Shaw, H.; Rhodes, V.; Hart, A. Review of Antimicrobial Resistance in the Environment and Its Relevance to Environmental Regulators. Front. Microbiol. 2016, 7, 1728. [Google Scholar] [CrossRef]
- Huijbers, P.M.C.; Blaak, H.; de Jong, M.C.M.; Graat, E.A.M.; Vandenbroucke-Grauls, C.M.J.E.; de Roda Husman, A.M.E. Role of the environment in the transmission of antimicrobial resistance to humans: A review. Environ. Sci. Technol. 2015, 49, 11993–12004. [Google Scholar] [CrossRef]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 0026-2017. [Google Scholar] [CrossRef]
- Hasan, B.; Laurell, K.; Rakib, M.M.; Ahlstedt, E.; Hernandez, J.; Caceres, M.; Jarhult, J.D. Fecal carriage of Extended-Spectrum β-Lactamases in healthy humans, poultry, and wild birds in Leon, Nicaragua-a shared pool of bla(CTX-M) genes and possible interspecies clonal spread of Extended-Spectrum β-Lactamases-producing Escherichia coli. Microb. Drug Resist. 2016, 22, 682–687. [Google Scholar] [CrossRef]
- Borges, C.A.; Cardozo, M.V.; Beraldo, L.G.; Oliveira, E.S.; Maluta, R.P.; Barboza, K.B.; Werther, K.; Ãvila, F.A. Wild birds and urban pigeons as reservoirs for diarrheagenic Escherichia coli with zoonotic potential. J. Microbiol. 2017, 55, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Aljeldah, M.M. Antimicrobial Resistance and Its Spread Is a Global Threat. Antibiotics 2022, 11, 1082. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]

| Species, Gene, Primer | Sequence (5′–3′) | Amplicon Size, Base Pairs | |
|---|---|---|---|
| Extended-Spectrum β-Lactamases (ESBLs) gene | |||
| blaCTX-M-9G | blaCTX-M-9G-F | TGA CCG TAT TGG GAG TTT G | 615 |
| blaCTX-M-9G-R | ACC AGT TAC AGC CCT TCG | ||
| blaCTX-M-1G | blaCTX-M-1G-F | CTT CCA GAA TAA GGA ATC CC | 765 |
| blaCTX-M-1G-R | CGT CTA AGG CGA TAA ACA AA | ||
| 16S rRNA methylase gene | |||
| armA | armA-F | ATT CTG CCT ATC CTA ATT GG | 315 |
| armA-R | ACC TAT ACT TTA TCG TCG TC | ||
| rmtA | rmtA-F | CTA GCG TCC ATC CTT TCC TC | 635 |
| rmtA-R | TTG CTT CCA TGC CCT TGC C | ||
| rmtB | rmtB-F | GCT TTC TGC GGG CGA TGT AA | 173 |
| rmtB-R | ATG CAA TGC CGC GCT CGT AT | ||
| rmtC | rmtC-F | CGA AGA AGT AAC AGC CAA AG | 711 |
| rmtC-R | ATC CCA ACA TCT CTC CCA CT | ||
| rmtD | rmtD-F | CGG CAC GCG ATT GGG AAG C | 401 |
| rmtD-R | CGG AAA CGA TGC GAC GAT | ||
| carbapenemase-encoding gene | |||
| blaNDM | blaNDM-F | GGT TTG GCG ATC TGG TTT TC | 621 |
| blaNDM-R | CGG AAT GGC TCA TCA CGA TC | ||
| blaKPC | blaKPC-Fm | CGT CTA GTT CTG CTG TCT TG | 798 |
| blaKPC-Rm | CTT GTC ATC CTT GTT AGG CG | 232 | |
| plasmid-mediated colistin resistance (mcr) gene | |||
| mcr-1 | mcr-1-F | AGT CCG TTT GTT CTT GTG GC | 320 |
| mcr-1-R | AGA TCC TTG GTC TCG GCT TG | ||
| mcr-2 | mcr-2-F | CAA GTG TGT TGG TCG CAG TT | 715 |
| mcr-2-R | TCT AGC CCG ACA AGC ATA CC | ||
| mcr-3 | mcr-3-F | AAA TAA AAA TTG TTC CGC TTA TG | 929 |
| mcr-3-R | AAT GGA GAT CCC CGT TTT T | ||
| mcr-4 | mcr-4-F | TCA CTT TCA TCA CTG CGT TG | 1116 |
| mcr-4-R | TTG GTC CAT GAC TAC CAA TG | ||
| mcr-5 | mcr-5-F | ATG CGG TTG TCT GCA TTT ATC | 1644 |
| mcr-5-R | TCA TTG TGG TTG TCC TTT TCT G | ||
| Species | Multiple Resistant Isolates (%) | |||
|---|---|---|---|---|
| Triple-Drug Resistance | Quadruple-Drug Resistance | Quintuple-Drug Resistance | Sixfold-Drug Resistance | |
| Homing pigeon (n = 69) | 8 (11.6) | 5 (7.2) | 3 (4.3) | 3 (4.3) |
| Ornamental pigeon (n = 50) | 1 (2.0) | 1 (2.0) | 1 (2.0) | 0 (0.0) |
| Meat pigeon (n = 30) | 9 (30.0) | 4 (13.3) | 1 (3.3) | 1 (3.3) |
| The total (n = 149) | 18 (12.1) | 10 (6.7) | 5 (3.4) | 4 (2.7) |
| Antimicrobial Resistance Gene | Number of Isolates (%) |
|---|---|
| ESBLs genes | |
| blaCTX-M-9G | 0 (0.0) |
| blaCTX-M-1G | 2 (1.3) |
| 16S rRNA methylase genes | |
| armA | 0 (0.0) |
| rmtA | 0 (0.0) |
| rmtB | 0 (0.0) |
| rmtC | 0 (0.0) |
| rmtD | 0 (0.0) |
| carbapenemase-encoding genes | |
| blaNDM | 0 (0.0) |
| blaKPC | 0 (0.0) |
| mcr genes | |
| mcr-1~mcr-5 | 0 (0.0) |
| mcr-6~mcr-9 | 0 (0.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Hu, C. Antimicrobial Resistance Analysis of Escherichia coli Isolated from Pigeons in Qingdao, Shandong Province, China. Genes 2022, 13, 1510. https://doi.org/10.3390/genes13091510
Wang A, Hu C. Antimicrobial Resistance Analysis of Escherichia coli Isolated from Pigeons in Qingdao, Shandong Province, China. Genes. 2022; 13(9):1510. https://doi.org/10.3390/genes13091510
Chicago/Turabian StyleWang, Anqi, and Changmin Hu. 2022. "Antimicrobial Resistance Analysis of Escherichia coli Isolated from Pigeons in Qingdao, Shandong Province, China" Genes 13, no. 9: 1510. https://doi.org/10.3390/genes13091510
APA StyleWang, A., & Hu, C. (2022). Antimicrobial Resistance Analysis of Escherichia coli Isolated from Pigeons in Qingdao, Shandong Province, China. Genes, 13(9), 1510. https://doi.org/10.3390/genes13091510





