Selection of Suitable Reference Genes for Gene Expression Normalization Studies in Dendrobium huoshanense
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Selection of Candidate Reference Genes and Primers Design
2.4. qPCR Analysis
2.5. Gene Expression Stability Analysis
2.6. Comprehensive Analysis and Validation of Selected Reference Genes
3. Results
3.1. Primer Specificity Verification and PCR efficiency
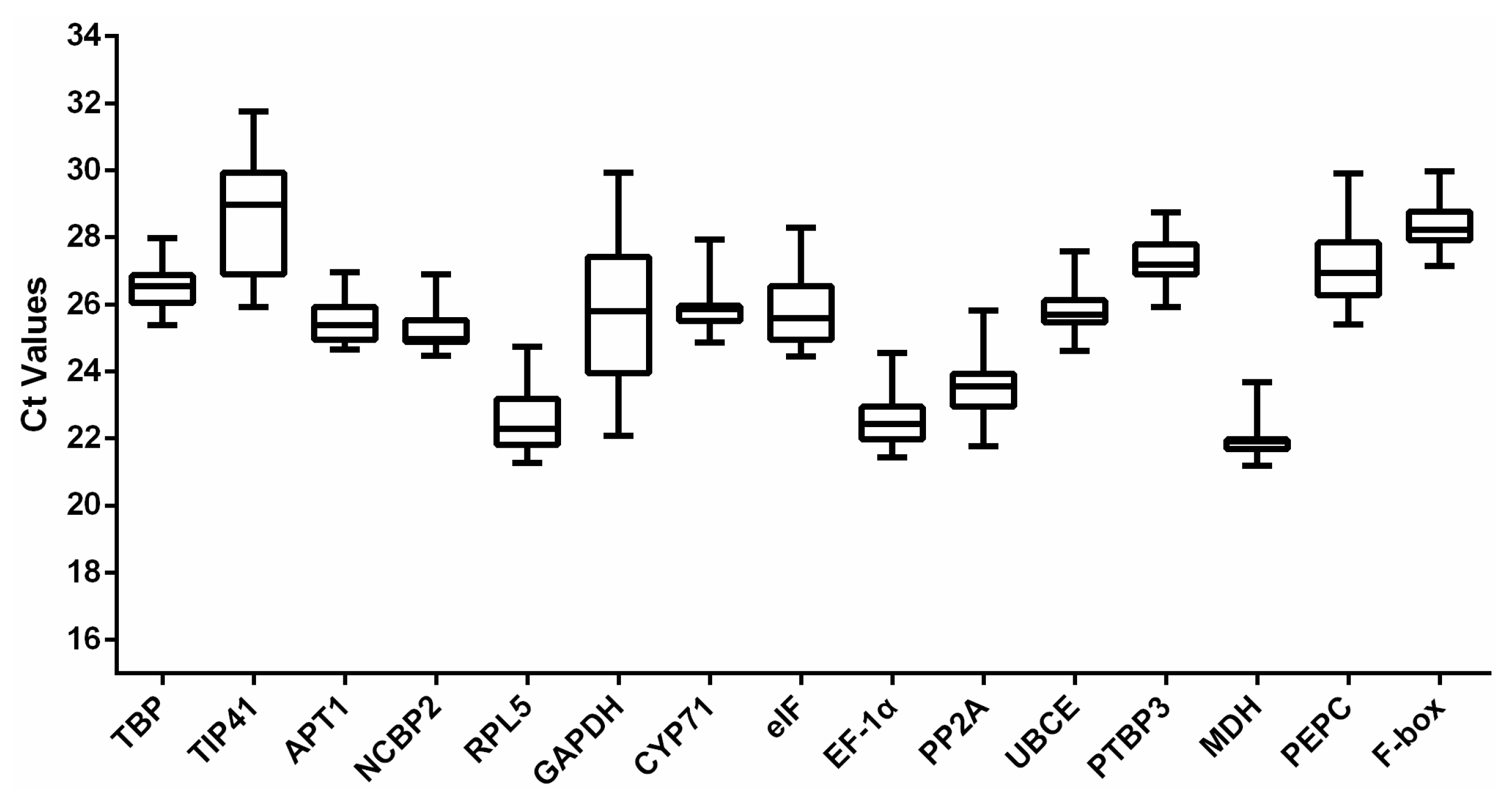
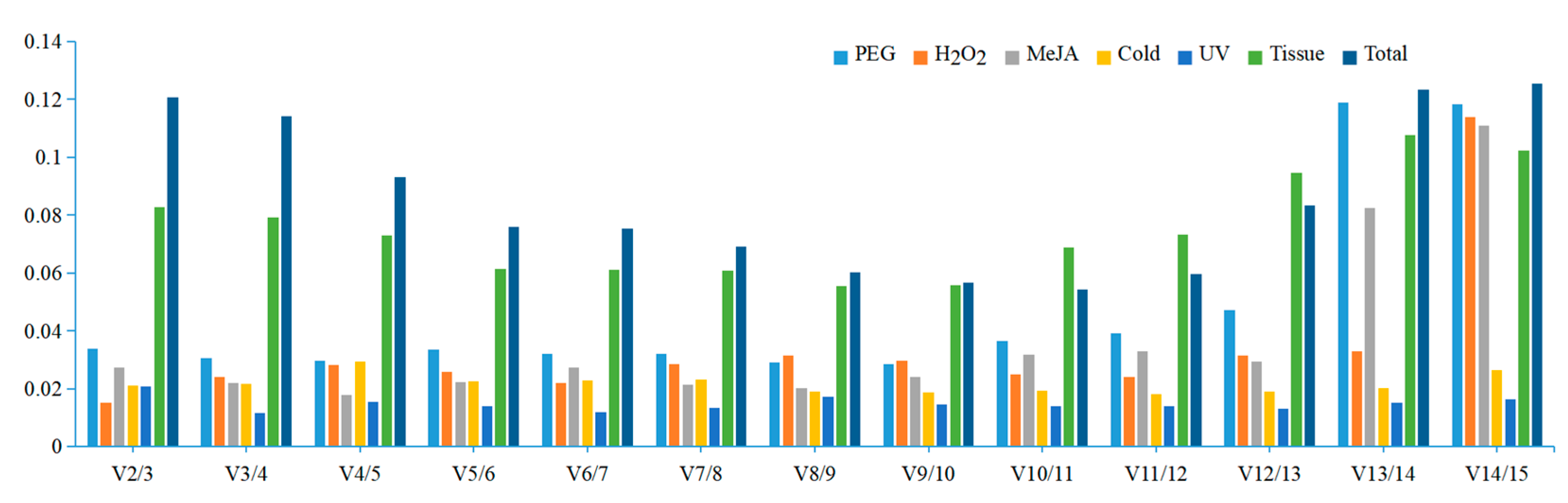
3.2. Expression Profile of the Reference Genes
3.3. The Analysis of Expression Stability of Candidate Reference Genes
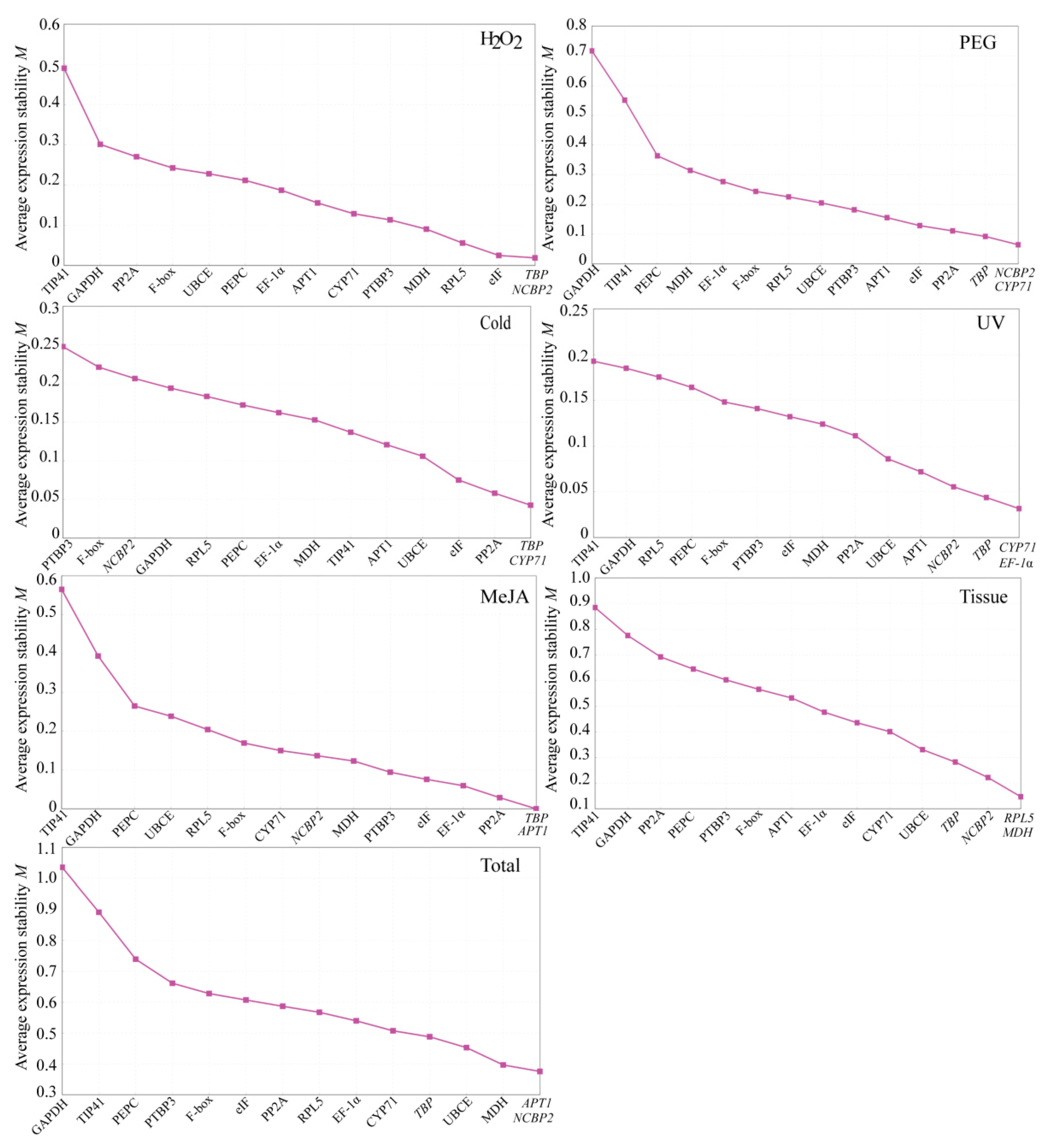
3.3.1. geNorm Analysis
3.3.2. NormFinder Analysis
3.3.3. BestKeeper Analysis
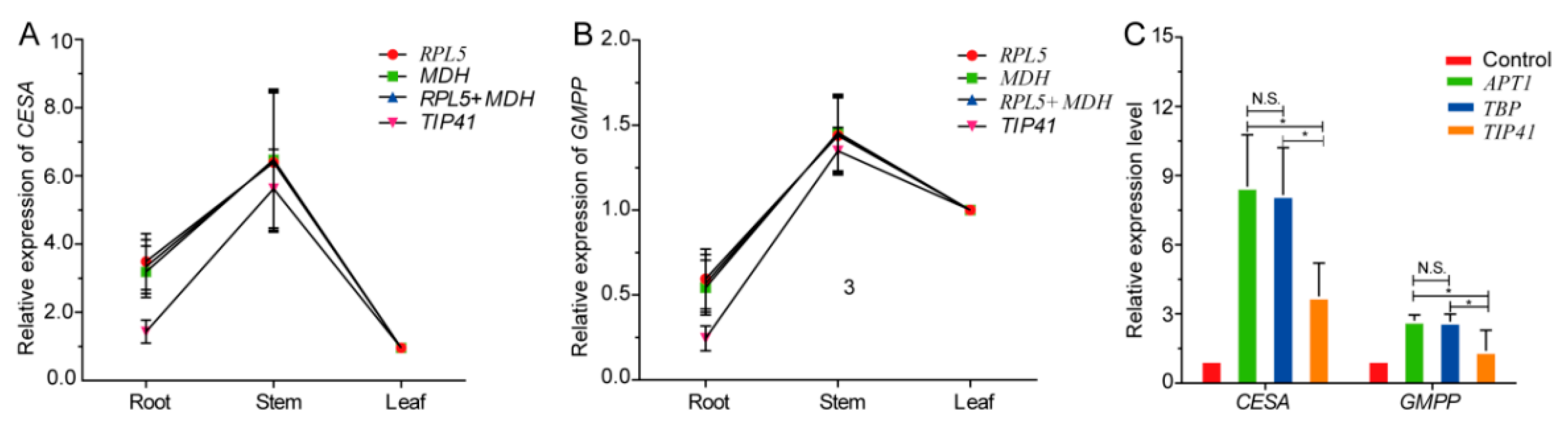
3.4. Comprehensive Analysis and Validation of Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, X.M. Supplement to Compendium of Materia Medica; China Press of Chinese Traditional Medicine: Beijing, China, 2007. [Google Scholar]
- Wu, C.F.; Gui, S.H.; Huang, Y.C.; Dai, Y.F.; Shun, Q.S.; Huang, K.W.; Tao, S.C.; Wei, G. Characteristic fingerprint analysis of Dendrobium huoshanense by ultra-high performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Anal. Methods 2016, 8, 3802–3808. [Google Scholar] [CrossRef]
- Yi, S.Y.; Kang, C.Z.; Wang, W.; Song, X.W.; Xu, T.; Lu, H.B.; Luo, S.L.; Liu, D.; Guo, L.P.; Han, B.X. Comparison of planting modes of Dendrobium huoshanense and analysis of advantages of simulated cultivation. Zhongguo Zhong Yao Za Zhi 2021, 46, 1864–1868. [Google Scholar]
- Chang, C.C.; Ku, A.F.; Tseng, Y.Y.; Yang, W.B.; Fang, J.M.; Wong, C.H. 6,8-Di-C-glycosyl flavonoids from Dendrobium huoshanense. J. Nat. Prod. 2010, 73, 229–232. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Han, B.X.; Peng, H.S.; Wang, X.; Chu, S.S.; Dai, J.; Peng, D.Y. Identification of “Huoshan shihu” Fengdou: Comparative authentication of the Daodi herb Dendrobium huoshanense and its related species by macroscopic and microscopic features. Microsc. Res. Tech. 2017, 80, 712–721. [Google Scholar] [CrossRef]
- Xing, X.H.; Cui, S.W.; Nie, S.P.; Phillips, G.O.; Goff, H.D.; Wang, Q. A review of isolation process, structural characteristics, and bioactivities of water-soluble polysaccharides from Dendrobium plants. Bioact. Carbohydr. Diet. Fibre 2013, 1, 131–147. [Google Scholar] [CrossRef]
- Ng, T.B.; Liu, J.; Wong, J.H.; Ye, X.; Wing, S.S.C.; Tong, Y.; Zhang, K.Y. Review of research o1n Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef]
- Mudiam, M.K.R.; Jin, Q.; Jiao, C.; Sun, S.; Song, C.; Cai, Y.; Lin, Y.; Fan, H.; Zhu, Y. Metabolic analysis of medicinal Dendrobium officinale and Dendrobium huoshanense during different growth years. PLoS ONE 2016, 11, e0146607. [Google Scholar]
- Gao, L.L.; Wang, F.; Hou, T.T.; Geng, C.Y.; Xu, T.; Han, B.X.; Liu, D. Dendrobium huoshanense C.Z.Tang et S.J.Cheng: A review of its traditional uses, phytochemistry, and pharmacology. Front. Pharmacol. 2022, 13, 920823. [Google Scholar] [CrossRef]
- Gu, F.L.; Huang, R.S.; He, X.M.; Chen, N.F.; Han, B.X.; Deng, H. Dendrobium huoshanense polysaccharides prevent inflammatory response of ulcerative colitis rat through inhibiting the NF-κB signaling pathway. Chem. Biodivers. 2021, 18, e2100130. [Google Scholar] [CrossRef]
- Liu, B.; Shang, Z.Z.; Li, Q.M.; Zha, X.Q.; Wu, D.L.; Yu, N.J.; Han, L.; Peng, D.Y.; Luo, J.P. Structural features and anti-gastric cancer activity of polysaccharides from stem, root, leaf and flower of cultivated Dendrobium huoshanense. Int. J. Biol. Macromol. 2020, 143, 651–664. [Google Scholar] [CrossRef]
- Wang, Y.H. Traditional uses, chemical constituents, pharmacological activities, and toxicological effects of Dendrobium leaves: A review. J. Ethnopharmacol. 2021, 270, 113851. [Google Scholar] [CrossRef] [PubMed]
- Han, R.C.; Xie, D.M.; Tong, X.H.; Zhang, W.; Liu, G.; Peng, D.Y.; Yu, N.J. Transcriptomic landscape of Dendrobium huoshanense and its genes related to polysaccharide biosynthesis. Acta Soc. Bot. Pol. 2018, 87, 1–11. [Google Scholar] [CrossRef]
- Yuan, Y.D.; Yu, M.Y.; Jia, Z.H.; Song, X.E.; Liang, Y.Q.; Zhang, J.C. Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genom. 2018, 19, 978. [Google Scholar] [CrossRef] [PubMed]
- Han, R.C.; Liu, L.L.; Liu, J.L.; Xie, D.M.; Peng, D.Y.; Yu, N.J. Cloning and quantitative expression analysis of GMPP gene from Dendrobium huoshanense. Zhongguo Zhong Yao Za Zhi 2019, 44, 1552–1557. [Google Scholar]
- Zhou, P.N.; Pu, T.Z.; Gui, C.; Zhang, X.Q.; Gong, L. Transcriptome analysis reveals biosynthesis of important bioactive constituents and mechanism of stem formation of Dendrobium huoshanense. Sci. Rep. 2020, 10, 2857. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.D.; Zuo, J.J.; Zhang, H.Y.; Li, R.Z.; Yu, M.Y.; Liu, S. Integration of transcriptome and metabolome provides new insights to flavonoids biosynthesis in Dendrobium huoshanense. Front. Plant Sci. 2022, 13, 850090. [Google Scholar] [CrossRef]
- Valoroso, M.C.; De Paolo, S.; Iazzetti, G.; Aceto, S. Transcriptome-wide identification and expression analysis of DIVARICATA-and RADIALIS-like genes of the Mediterranean Orchid Orchis italica. Genome Biol. Evol. 2017, 9, 1418–1431. [Google Scholar] [CrossRef][Green Version]
- An, H.; Zhu, Q.; Pei, W.; Fan, J.; Liang, Y.; Cui, Y.; Lv, N.; Wang, W. Whole-transcriptome selection and evaluation of internal reference genes for expression analysis in protocorm development of Dendrobium officinale Kimura et Migo. PLoS ONE 2016, 11, e0163478. [Google Scholar] [CrossRef]
- Yi, Y.Q.; Liu, L.L.; Zhou, W.Y.; Peng, D.Y.; Han, R.C.; Yu, N.J. Characterization of GMPP from Dendrobium huoshanense yielding GDP-D-mannose. Open Life Sci. 2021, 16, 102–107. [Google Scholar] [CrossRef]
- Wu, J.; Meng, X.X.; Jiang, W.M.; Wang, Z.J.; Zhang, J.; Meng, F.; Yao, X.Y.; Ye, M.J.; Yao, L.; Wang, L.H.; et al. Qualitative proteome-wide analysis reveals the diverse functions of lysine crotonylation in Dendrobium huoshanense. Front. Plant Sci. 2022, 13, 822374. [Google Scholar] [CrossRef]
- De Magalhães, J.P.; Finch, C.E.; Janssens, G. Next-generation sequencing in aging research: Emerging applications, problems, pitfalls and possible solutions. Ageing Res. Rev. 2010, 9, 315–323. [Google Scholar] [CrossRef]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Tan, B.; Guo, W.W. Estimating transgene copy number in precocious trifoliate orange by TaqMan real-time PCR. Plant Cell Tissue Organ Cult. 2012, 109, 363–371. [Google Scholar] [CrossRef]
- Dheda, K.; Huggett, J.F.; Chang, J.S.; Kim, L.U.; Bustin, S.A.; Johnson, M.A.; Rook, G.A.; Zumla, A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 2005, 344, 141–143. [Google Scholar] [CrossRef]
- Liang, W.; Zou, X.; Carballar-Lejarazú, R.; Wu, L.; Sun, W.; Yuan, X.; Wu, S.; Li, P.; Ding, H.; Ni, L.; et al. Selection and evaluation of reference genes for qRT-PCR analysis in Euscaphis konishii Hayata based on transcriptome data. Plant Methods 2018, 14, 42. [Google Scholar] [CrossRef]
- Hou, S.; Zhao, T.; Yang, D.; Li, Q.; Liang, L.; Wang, G.; Ma, Q. Selection and validation of reference genes for quantitative RT-PCR analysis in Corylus heterophylla Fisch × Corylus avellana L. Plants 2021, 10, 159. [Google Scholar] [CrossRef]
- Li, Y.; Liang, X.; Zhou, X.; Wu, Z.; Yuan, L.; Wang, Y.; Li, Y. Selection of reference genes for qRT-PCR analysis in medicinal plant Glycyrrhiza under abiotic stresses and hormonal treatments. Plants 2020, 9, 1441. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, C.; Yang, H.; Lyu, L.; Li, W.; Wu, W. Selection and validation of candidate reference genes for gene expression analysis by RT-qPCR in Rubus. Int. J. Mol. Sci. 2021, 22, 10533. [Google Scholar] [CrossRef]
- Dong, X.M.; Zhang, W.; Zhang, S.B. Selection and validation of reference genes for quantitative real-time PCR analysis of development and tissue-dependent flower color formation in Cymbidium lowianum. Int. J. Mol. Sci. 2022, 23, 738. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, R.; Zhou, Z. Identification and validation of reference genes for gene expression analysis in Schima superba. Genes 2021, 12, 732. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; Xue, J.; Yang, J.; Hu, H.; Cui, J.; Xu, J. Selection and verification of appropriate reference genes for expression normalization in Cryptomeria fortunei under abiotic stress and hormone treatments. Genes 2021, 12, 791. [Google Scholar] [CrossRef]
- He, Y.; Zhong, Y.; Bao, Z.; Wang, W.; Xu, X.; Gai, Y.; Wu, J. Evaluation of Angelica decursiva reference genes under various stimuli for RT-qPCR data normalization. Sci. Rep. 2021, 11, 18993. [Google Scholar] [CrossRef]
- Wang, B.; Duan, H.; Chong, P.; Su, S.; Shan, L.; Yi, D.; Wang, L.; Li, Y. Systematic selection and validation of suitable reference genes for quantitative real-time PCR normalization studies of gene expression in Nitraria tangutorum. Sci. Rep. 2020, 10, 15891. [Google Scholar] [CrossRef]
- Yi, S.Y.; Lin, Q.W.; Zhang, X.J.; Wang, J.; Miao, Y.Y.; Tan, N.H. Selection and validation of appropriate reference genes for quantitative RT-PCR Analysis in Rubia yunnanensis Diels based on transcriptome data. Biomed Res. Int. 2020, 2020, 5824841. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, C.; Zhang, J.; Chen, F.; Gong, Y.; Li, Y.; Su, Y.; Wei, Y.; Zhao, Y. Selection of the reference gene for expression normalization in Papaver somniferum L. under abiotic stress and hormone treatment. Genes 2020, 11, 124. [Google Scholar] [CrossRef]
- Song, H.; Mao, W.; Duan, Z.; Que, Q.; Zhou, W.; Chen, X.; Li, P. Selection and validation of reference genes for measuring gene expression in Toona ciliata under different experimental conditions by quantitative real-time PCR analysis. BMC Plant Biol. 2020, 20, 450. [Google Scholar] [CrossRef]
- Song, C.; Ma, J.B.; Li, G.H.; Pan, H.Y.; Zhu, Y.F.; Jin, Q.; Cai, Y.P.; Han, B.X. Natural composition and biosynthetic pathways of alkaloids in medicinal Dendrobium species. Front. Plant Sci. 2022, 13, 850949. [Google Scholar] [CrossRef]
- Ulrich, M.N.; Muñiz-Padilla, E.; Corach, A.; Hopp, E.; Tosto, D. Validation of reference genes for quantitative PCR in Johnsongrass (Sorghum halepense L.) under glyphosate stress. Plants 2021, 10, 1555. [Google Scholar] [CrossRef]
- Duan, M.; Wang, J.; Zhang, X.; Yang, H.; Wang, H.; Qiu, Y.; Song, J.; Guo, Y.; Li, X. Identification of optimal reference genes for expression analysis in Radish (Raphanus sativus L.) and its relatives based on expression stability. Front. Plant Sci. 2017, 8, 1605. [Google Scholar] [CrossRef]
- Song, Y.; Wang, Y.; Guo, D.; Jing, L. Selection of reference genes for quantitative real-time PCR normalization in the plant pathogen Puccinia helianthi Schw. BMC Plant Biol. 2019, 19, 20. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntof, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaf, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, diferentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Zhang, J.; He, C.; Wu, K.; da Silva, J.A.T.; Zeng, S.; Zhang, X.; Yu, Z.; Xia, H.; Duan, J. Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 2016, 7, 5. [Google Scholar] [CrossRef]
- Sun, H.P.; Li, F.; Ruan, Q.M.; Zhong, X.H. Identification and validation of reference genes for quantitative real-time PCR studies in Hedera helix L. Plant Physiol. Biochem. 2016, 108, 286–294. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Zhuang, H.H.; Fu, Y.P.; He, W.; Wang, L.; Wei, Y.H. Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 2015, 6, 475. [Google Scholar] [CrossRef]
- Deng, Y.; Chen, L.X.; Han, B.X.; Wu, D.T.; Cheong, K.; Chen, N.F.; Zhao, J.; Li, S.P. Qualitative and quantitative analysis of specific polysaccharides in Dendrobium huoshanense by using saccharide mapping and chromatographic methods. J. Pharm. Biomed. Anal. 2016, 129, 163–171. [Google Scholar] [CrossRef]
- Chen, S.T.; Dai, J.; Song, X.W.; Jiang, X.P.; Zhao, Q.; Sun, C.B.; Chen, C.W.; Chen, N.D.; Han, B.X. Endophytic microbiota comparison of Dendrobium huoshanense root and stem in different growth years. Planta Med. 2020, 86, 967–975. [Google Scholar] [CrossRef]
- Li, Z.Q.; Zhou, H.Q.; Ouyang, Z.; Dai, J.; Yue, Q.; Wei, Y.; Han, B.X. Comparison of active ingredients and protective effects of Dendrobium huoshanense of different growth years on acute liver injury. Zhongguo Zhong Yao Za Zhi 2021, 46, 298–305. [Google Scholar] [PubMed]
- Wan, J.Q.; Gong, X.H.; Wang, F.X.; Wen, C.W.; Wei, Y.; Han, B.X.; Ouyang, Z. Comparative analysis of chemical constituents by HPLC-ESI-MSn and antioxidant activities of Dendrobium huoshanense and Dendrobium officinale. Biomed. Chromatogr. 2022, 36, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Han, B.X.; Jing, Y.; Dai, J.; Zheng, T.; Gu, F.L.; Zhao, Q.; Zhu, F.C.; Song, X.W.; Deng, H.; Wei, P.P.; et al. A chromosome-level genome assembly of Dendrobium huoshanense using long reads and Hi-C data. Genome Biol. Evol. 2020, 12, 2486–2490. [Google Scholar] [CrossRef] [PubMed]
- Riseh, R.S.; Skorik, Y.A.; Thakur, V.K.; Pour, M.M.; Tamanadar, E.; Noghabi, S.S. Encapsulation of plant biocontrol bacteria with alginate as a main polymer material. Int. J. Mol. Sci. 2021, 22, 11165. [Google Scholar] [CrossRef]
- Yuan, Y.D.; Tang, X.G.; Jia, Z.H.; Li, C.; Ma, J.Y.; Zhang, J.C. The effects of ecological factors on the main medicinal components of Dendrobium officinale under different cultivation modes. Forests 2020, 11, 94. [Google Scholar] [CrossRef]
- Jiang, M.; Xu, S.Z.; Wen, G.S.; Zhao, C.L. Validation of seven housekeeping genes as reference ones for qRT-PCR normalization in Dendrobium catenatum. Russ. J. Plant Physiol. 2017, 64, 497–508. [Google Scholar] [CrossRef]
- Basu, S.; Pereira, A.; Pinheiro, D.; Wang, H.; Valencia-Jiménez, A.; Siegfried, B.; Louis, J.; Zhou, X.; Velez, A. Evaluation of reference genes for real-time quantitative PCR analysis in southern corn rootworm, Diabrotica undecimpunctata howardi (Barber). Sci. Rep. 2019, 9, 10703. [Google Scholar] [CrossRef]
- Bao, Z.Z.; Zhang, K.D.; Lin, H.F.; Li, C.J.; Zhao, X.R.; Wu, J.; Nian, S.H. Identification and selection of reference genes for quantitative transcript analysis in Corydalis yanhusuo. Genes 2020, 11, 130. [Google Scholar] [CrossRef]
- Wang, Z.L.; Xu, J.Y.; Liu, Y.H.; Chen, J.Y.; Lin, H.F.; Huang, Y.L.; Bian, X.H.; Zhao, Y.C. Selection and validation of appropriate reference genes for real-time quantitative PCR analysis in Momordica charantia. Phytochemistry 2019, 164, 1–11. [Google Scholar] [CrossRef]
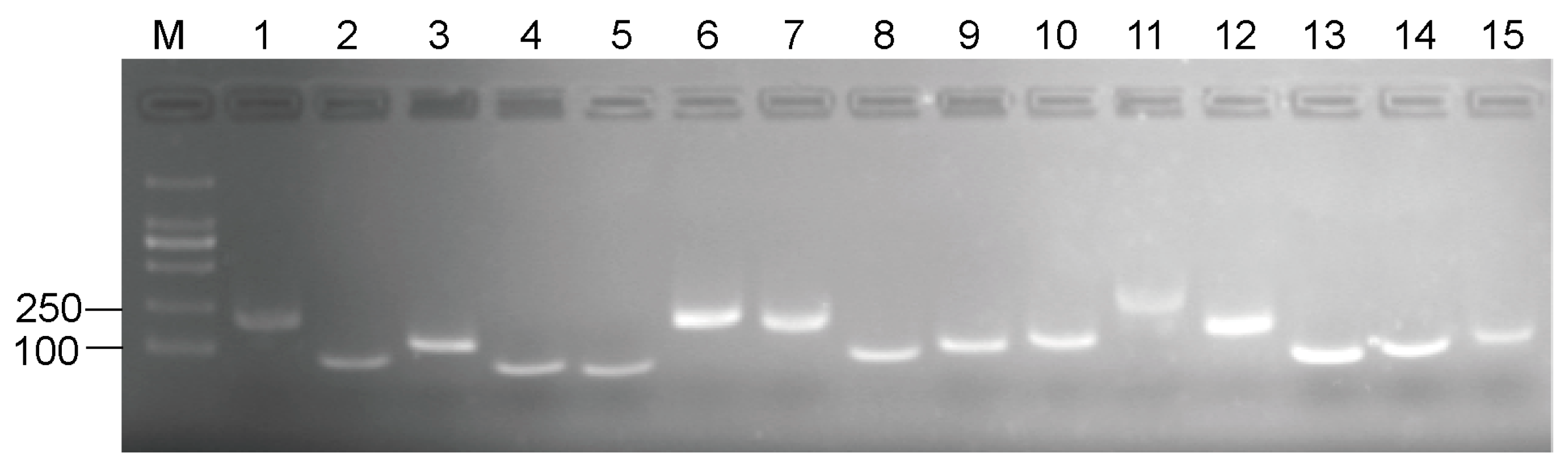
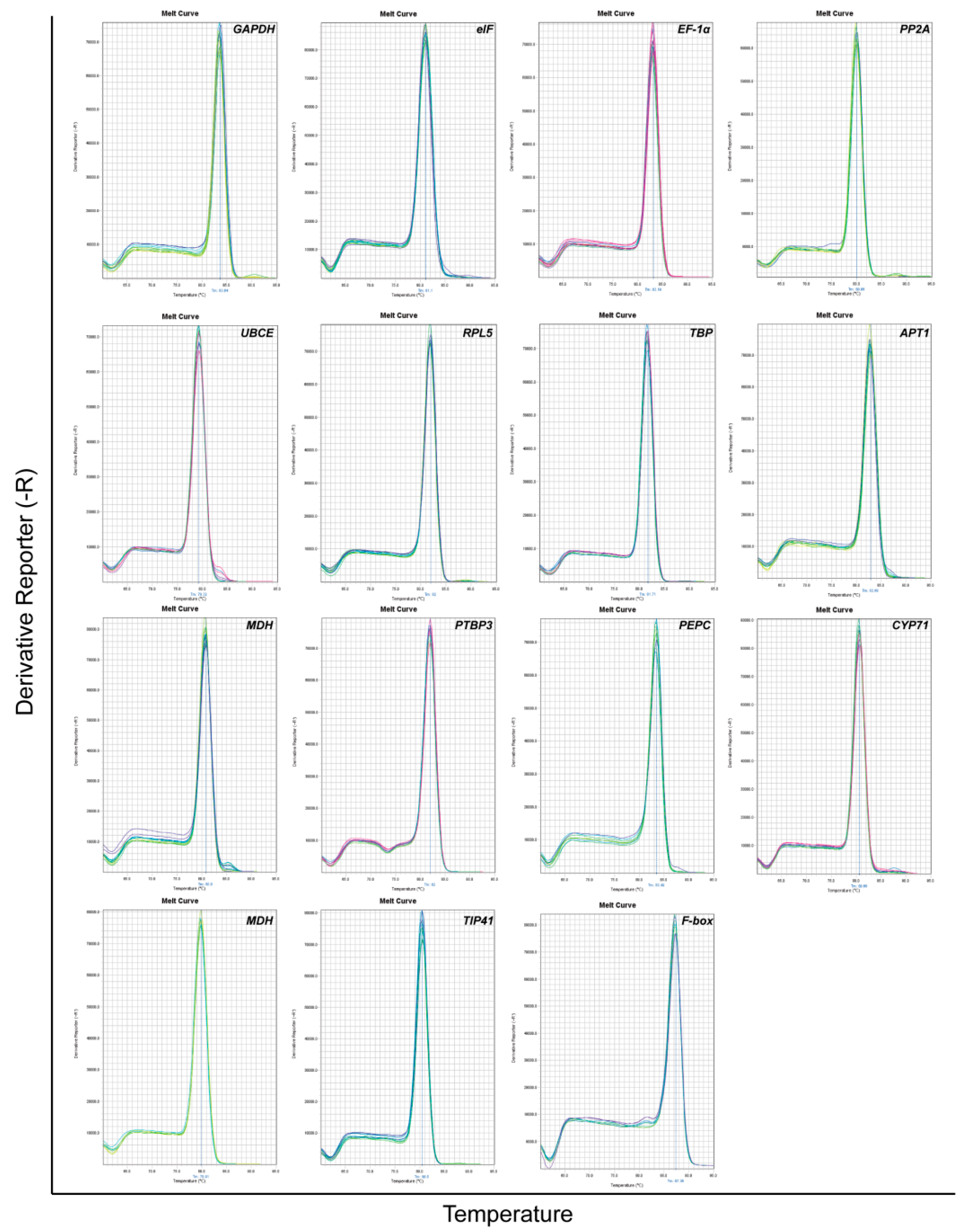
| Gene Symbol | Gene Name | Arabidopsis Homolog Locus | Primer Sequence (5′–3′) | Amplicon Length (bp *) | Primers Tm * (°C) | E * (%) | R2 * |
|---|---|---|---|---|---|---|---|
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | AT1G16300.1 | F: TGCTTACGGCGATTACTTCC | 279 | 58.2/58.6 | 96.979 | 0.994 |
| R: CTTCCAATACGACCAAAACCAT | |||||||
| eIF | Eukaryotic translation initiation factor | AT3G60240.2 | F: CCATTCCATCTGTCCCCCC | 121 | 61.8/60.1 | 97.101 | 0.998 |
| R: GGACCCCTAAACGGAAAACATA | |||||||
| EF-1α | Elongation factor 1-α-like | AT1G07920.1 | F: GCCCAACCGATAAGCCACT | 138 | 59.8/59.1 | 92.403 | 0.997 |
| R: TGAGTCCAGTAGGTCCGAAGG | |||||||
| PP2A | Serine/threonine protein phosphatase 2A | AT3G2580.1 | F: CGCTGCTCTTGGAAAACTGT | 210 | 58.0/59.0 | 96.771 | 0.998 |
| R: ACTTCTGCCTCATTATCACGGA | |||||||
| UBCE | Ubiquitin conjugating enzyme E2 | AT1G50490.1 | F: AATAGATGGAGGCAAGGGAACT | 123 | 59.0/59.0 | 102.92 | 0.997 |
| R: TTGGGATGGAAACAGGAGGT | |||||||
| RPL5 | Ribosomal protein L5 | AT5G39740.1 | F: CGATTTGTTGTGCGATTTACG | 270 | 59.5/59.4 | 97.704 | 0.996 |
| R: CCTCCTGCTTTCTGCTGGTT | |||||||
| TBP | TATA box binding protein like | AT1G55520.1 | F: GGCATCCTTCTGGTATTGTCC | 252 | 58.5/58.4 | 104.77 | 0.998 |
| R: TACGAGCATACTTCCGAGCAG | |||||||
| APT1 | Adenine phosphoribosyltransferase 1 | AT1G27450.1 | F: ATGGCGTCCGTGGATGAA | 114 | 59.8/58.8 | 93.636 | 0.999 |
| R: CAGCAGCAGCGTCGTGATA | |||||||
| MDH | Malate dehydrogenase | AT5G43330.1 | F: TTGCTGATGATGAGTGGCTGAG | 142 | 61.0/59.5 | 98.403 | 0.991 |
| R: CCAAGGACCCAATCACGAAT | |||||||
| PTBP3 | Polypyrimidine tract binding protein homolog 3 | AT1G43190.1 | F: CACCTGACACCCGTGAGTTTG | 209 | 60.9/61.2 | 93.938 | 0.998 |
| R: TTGCCTCTTACCATTCACCTCTATC | |||||||
| PEPC | Phosphoenolpyruvate carboxykinase | AT4G37870.1 | F: TGACATCATCCACAAGCATAGACA | 283 | 60.5/59.4 | 90.502 | 0.995 |
| R: GAAACACCATTATCACTCCAGCA | |||||||
| CYP71 | Cyclophilin 71 | AT3G44600.1 | F: TGAATGGGTCTACAAACAAGGAG | 227 | 58.7/59.6 | 92.806 | 0.993 |
| R: GCAATGTTGTAGGGCTCCAGTA | |||||||
| NCBP2 | nuclear cap binding protein subunit 2 | AT5G44200.1 | F: GACTCCCTGTGGCTTTTGCT | 146 | 59.2/59.4 | 100.38 | 0.999 |
| R: GACCCCATTGTCTTCCTTCTTC | |||||||
| TIP41 | TIP41-like protein | AT4G34270.1 | F: TGGCAGCGAAGCAGTAGAAC | 200 | 60.2/60.8 | 101.45 | 0.996 |
| R: GAACTTTTACGGTCAAGAGGGAT | |||||||
| F-box | F-box protein | AT5G39450.1 | F: TTTCCCCGCAGTTTTCACG | 169 | 58.9/58.8 | 102.65 | 0.990 |
| R: TTGGAACCTTCAGGCGGACT | |||||||
| GMPP | GDP-mannose pyrophosphorylase | AT1G74910.1 | F: GAATGTTCCGAAGCCGTTGT | 130 | 58.9/58.6 | 94.465 | 1 |
| R: AGCAAACTCCCGTTCCTCAT | |||||||
| CESA | cellulose synthase A | AT3G03050.1 | F: CTTTGTTTCAACTGCTGACCCT | 193 | 58.8/58.2 | 94.540 | 0.999 |
| R: ACGACAGAAAGGAACCCATAGA |
| Rank | H2O2 | PEG | Cold | UV | MeJA | Tissue | Total |
|---|---|---|---|---|---|---|---|
| 1 | TBP(0.006) | NCBP2(0.022) | TBP(0.013) | EF-1α(0.012) | TBP(0.015) | MDH(0.145) | NCBP2(0.143) |
| 2 | NCBP2(0.006) | CYP71(0.022) | CYP71(0.029) | CYP71(0.023) | APT1(0.015) | NCBP2(0.147) | MDH(0.205) |
| 3 | eIF(0.009) | TBP(0.029) | PP2A(0.043) | NCBP2(0.037) | PP2A(0.015) | RPL5(0.162) | APT1(0.258) |
| 4 | RPL5(0.027) | PP2A(0.029) | APT1(0.055) | TBP(0.046) | EF-1α(0.027) | UBCE(0.236) | UBCE(0.280) |
| 5 | PTBP3(0.038) | eIF(0.033) | UBCE(0.068) | UBCE(0.050) | eIF(0.032) | TBP(0.283) | TBP(0.293) |
| 6 | MDH(0.047) | UBCE(0.067) | eIF(0.089) | APT1(0.052) | PTBP3(0.041) | CYP71(0.289) | CYP71(0.307) |
| 7 | CYP71(0.091) | APT1(0.099) | TIP41(0.114) | PTBP3(0.089) | MDH(0.070) | F-box(0.336) | PP2A(0.349) |
| 8 | APT1(0.175) | PTBP3(0.130) | EF-1α(0.114) | F-box(0.119) | NCBP2(0.089) | APT1(0.336) | EF-1α(0.355) |
| 9 | EF-1α(0.216) | RPL5(0.190) | GAPDH(0.126) | PEPC(0.122) | CYP71(0.144) | eIF(0.339) | RPL5(0.381) |
| 10 | UBCE(0.218) | MDH(0.231) | PEPC(0.129) | MDH(0.127) | F-box(0.194) | PTBP3(0.440) | eIF(0.381) |
| 11 | F-box(0.238) | F-box(0.239) | MDH(0.131) | RPL5(0.130) | RPL5(0.235) | EF-1α(0.447) | F-box(0.394) |
| 12 | PEPC(0.250) | EF-1α(0.310) | NCBP2(0.163) | PP2A(0.131) | PEPC(0.238) | PEPC(0.575) | PTBP3(0.507) |
| 13 | PP2A(0.264) | PEPC(0.387) | RPL5(0.170) | eIF(0.142) | UBCE(0.262) | PP2A(0.593) | PEPC(0.724) |
| 14 | GAPDH(0.300) | TIP41(1.214) | F-box(0.202) | TIP41(0.146) | GAPDH(0.844) | GAPDH(0.803) | TIP41(1.22) |
| 15 | TIP41(1.18) | GAPDH(1.23) | PTBP3(0.274) | GAPDH(0.149) | TIP41(1.15) | TIP41(1.04) | GAPDH(1.31) |
| Rank | H2O2 | PEG | Cold | UV | MeJA | Tissue | Total |
|---|---|---|---|---|---|---|---|
| 1 | TBP | NCBP2 | EF-1α | EF-1α | eIF | PTBP3 | MDH |
| 0.13 ± 0.03 | 0.10 ± 0.02 | 0.47 ± 0.10 | 0.21 ± 0.05 | 0.37 ± 0.09 | 1.42 ± 0.39 | 1.62 ± 0.36 | |
| 2 | RPL5 | CYP71 | UBCE | CYP71 | EF-1α | PEPC | TBP |
| 0.15 ± 0.03 | 0.20 ± 0.05 | 0.53 ± 0.14 | 0.34 ± 0.08 | 0.40 ± 0.09 | 2.02 ± 0.56 | 1.65 ± 0.44 | |
| 3 | NCBP2 | TBP | CYP71 | TBP | TBP | UBCE | PTBP3 |
| 0.17 ± 0.04 | 0.21 ± 0.06 | 0.53 ± 0.14 | 0.36 ± 0.10 | 0.55 ± 0.14 | 2.06 ± 0.54 | 1.73 ± 0.47 | |
| 4 | eIF | eIF | APT1 | UBCE | APT1 | TBP | UBCE |
| 0.21 ± 0.05 | 0.40 ± 0.11 | 0.59 ± 0.15 | 0.40 ± 0.09 | 0.57 ± 0.14 | 2.09 ± 0.56 | 1.90 ± 0.49 | |
| 5 | PTBP3 | PP2A | TBP | NCBP2 | NCBP2 | EF-1α | CYP71 |
| 0.39 ± 0.10 | 0.47 ± 0.11 | 0.77 ± 0.20 | 0.42 ± 0.10 | 0.62 ± 0.15 | 2.11 ± 0.49 | 1.92 ± 0.50 | |
| 6 | MDH | APT1 | NCBP2 | PTBP3 | MDH | F-box | NCBP2 |
| 0.59 ± 0.13 | 0.62 ± 0.16 | 0.82 ± 0.21 | 0.42 ± 0.12 | 0.64 ± 0.14 | 2.32 ± 0.66 | 2.03 ± 0.51 | |
| 7 | CYP71 | UBCE | GAPDH | MDH | PTBP3 | NCBP2 | F-box |
| 0.60 ± 0.15 | 0.67 ± 0.17 | 0.84 ± 0.21 | 0.44 ± 0.11 | 0.64 ± 0.17 | 2.69 ± 0.69 | 2.18 ± 0.62 | |
| 8 | APT1 | PTBP3 | PTBP3 | PEPC | PP2A | CYP71 | APT1 |
| 0.94 ± 0.24 | 0.73 ± 0.20 | 0.84 ± 0.22 | 0.45 ± 0.11 | 0.67 ± 0.15 | 2.96 ± 0.78 | 2.27 ± 0.58 | |
| 9 | UBCE | RPL5 | PP2A | APT1 | CYP71 | APT1 | EF-1α |
| 0.97 ± 0.25 | 0.97 ± 0.23 | 0.91 ± 0.21 | 0.45 ± 0.12 | 0.85 ± 0.21 | 3.09 ± 0.80 | 2.68 ± 0.61 | |
| 10 | F-box | F-box | TIP41 | RPL5 | F-box | MDH | eIF |
| 0.98 ± 0.27 | 1.06 ± 0.30 | 0.93 ± 0.28 | 0.52 ± 0.11 | 0.94 ± 0.26 | 3.12 ± 0.70 | 2.76 ± 0.71 | |
| 11 | PEPC | MDH | RPL5 | PP2A | PEPC | RPL5 | PP2A |
| 1.06 ± 0.29 | 1.54 ± 0.34 | 0.98 ± 0.21 | 0.52 ± 0.14 | 1.21 ± 0.32 | 3.27 ± 0.76 | 2.84 ± 0.67 | |
| 12 | EF-1α | EF-1α | MDH | F-box | UBCE | eIF | PEPC |
| 1.15 ± 0.25 | 1.77 ± 0.40 | 1.06 ± 0.23 | 0.57 ± 0.16 | 1.47 ± 0.39 | 3.34 ± 0.88 | 3.12 ± 0.84 | |
| 13 | GAPDH | PEPC | eIF | eIF | RPL5 | GAPDH | RPL5 |
| 1.19 ± 0.32 | 1.81 ± 0.48 | 1.09 ± 0.27 | 1.05 ± 0.26 | 1.61 ± 0.35 | 3.92 ± 1.09 | 3.27 ± 0.74 | |
| 14 | PP2A | TIP41 | F-box | TIP41 | GAPDH | TIP41 | TIP41 |
| 1.45 ± 0.34 | 5.01 ± 1.40 | 1.27 ± 0.36 | 1.10 ± 0.25 | 3.89 ± 0.99 | 4.97 ± 1.40 | 5.40 ± 1.54 | |
| 15 | TIP41 | GAPDH | PEPC | GAPDH | TIP41 | PP2A | GAPDH |
| 5.01 ± 1.44 | 5.49 ± 1.45 | 1.96 ± 0.28 | 1.13 ± 0.25 | 4.72 ± 1.31 | 5.28 ± 1.26 | 6.99 ± 1.81 |
| Rank | H2O2 | PEG | Cold | UV | MeJA | Tissue | Total |
|---|---|---|---|---|---|---|---|
| 1 | TBP(1.41) | CYP71(1.41) | TBP(1.97) | EF-1α(1.63) | APT1(1.41) | MDH(1.68) | MDH(1.41) |
| 2 | RPL5(2.00) | NCBP2(1.57) | CYP71(2.45) | CYP71(2.45) | TBP(2.34) | RPL5(2.71) | NCBP2(1.57) |
| 3 | NCBP2(2.21) | TBP(2.45) | PP2A(2.99) | NCBP2(3.83) | EF-1α(2.63) | NCBP2(3.35) | UBCE(3.72) |
| 4 | eIF(3.22) | PP2A(3.94) | UBCE(3.13) | APT1(3.98) | PP2A(3.46) | UBCE(3.94) | TBP(3.76) |
| 5 | PTBP3(5.00) | eIF(4.73) | APT1(5.05) | TBP(4.79) | eIF(3.98) | TBP(4.47) | CYP71(4.82) |
| 6 | MDH(6.24) | APT1(6.70) | EF-1α(5.48) | PEPC(5.42) | MDH(5.66) | PTBP3(5.90) | APT1(5.01) |
| 7 | CYP71(6.74) | UBCE(6.70) | eIF(6.24) | UBCE(6.45) | PTBP3(6.45) | EF-1α(6.48) | PP2A(7.65) |
| 8 | APT1(8.24) | PTBP3(7.48) | GAPDH(7.65) | PTBP3(8.37) | NCBP2(7.74) | CYP71(6.82) | EF-1α(8.00) |
| 9 | EF-1α(8.97) | RPL5(9.00) | TIP41(8.57) | RPL5(8.66) | CYP71(9.00) | F-box(7.61) | PTBP3(8.49) |
| 10 | UBCE(9.97) | F-box(10.2) | MDH(9.06) | MDH(9.84) | F-box(10.0) | eIF(8.53) | RPL5(9.67) |
| 11 | PEPC(11.2) | MDH(11.5) | NCBP2(9.16) | F-box(10.1) | RPL5(11.2) | APT1(9.19) | eIF(10.2) |
| 12 | F-box(11.5) | EF-1α(11.7) | PEPC(11.5) | PP2A(10.4) | PEPC(12.2) | PEPC(9.64) | F-box(10.5) |
| 13 | PP2A(13.2) | PEPC(13.0) | RPL5(12.5) | TIP41(10.6) | UBCE(12.5) | PP2A(13.2) | PEPC(13.0) |
| 14 | GAPDH(13.7) | TIP41(14.0) | PTBP3(12.7) | GAPDH(11.0) | GAPDH(14.0) | GAPDH(13.7) | TIP41(14.0) |
| 15 | TIP41(15.0) | GAPDH(15.0) | F-box(14.2) | eIF(12.3) | TIP41(15.0) | TIP41(15.0) | GAPDH(15.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, S.; Lu, H.; Tian, C.; Xu, T.; Song, C.; Wang, W.; Wei, P.; Gu, F.; Liu, D.; Cai, Y.; et al. Selection of Suitable Reference Genes for Gene Expression Normalization Studies in Dendrobium huoshanense. Genes 2022, 13, 1486. https://doi.org/10.3390/genes13081486
Yi S, Lu H, Tian C, Xu T, Song C, Wang W, Wei P, Gu F, Liu D, Cai Y, et al. Selection of Suitable Reference Genes for Gene Expression Normalization Studies in Dendrobium huoshanense. Genes. 2022; 13(8):1486. https://doi.org/10.3390/genes13081486
Chicago/Turabian StyleYi, Shanyong, Haibo Lu, Chuanjun Tian, Tao Xu, Cheng Song, Wei Wang, Peipei Wei, Fangli Gu, Dong Liu, Yongping Cai, and et al. 2022. "Selection of Suitable Reference Genes for Gene Expression Normalization Studies in Dendrobium huoshanense" Genes 13, no. 8: 1486. https://doi.org/10.3390/genes13081486
APA StyleYi, S., Lu, H., Tian, C., Xu, T., Song, C., Wang, W., Wei, P., Gu, F., Liu, D., Cai, Y., & Han, B. (2022). Selection of Suitable Reference Genes for Gene Expression Normalization Studies in Dendrobium huoshanense. Genes, 13(8), 1486. https://doi.org/10.3390/genes13081486






