Genome-Wide Identification and Characterization of SPL Family Genes in Chenopodium quinoa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Growth Conditions, and Stress Treatments
2.2. Identification of SPL and MIR156/7 Family Members in the Quinoa Genome
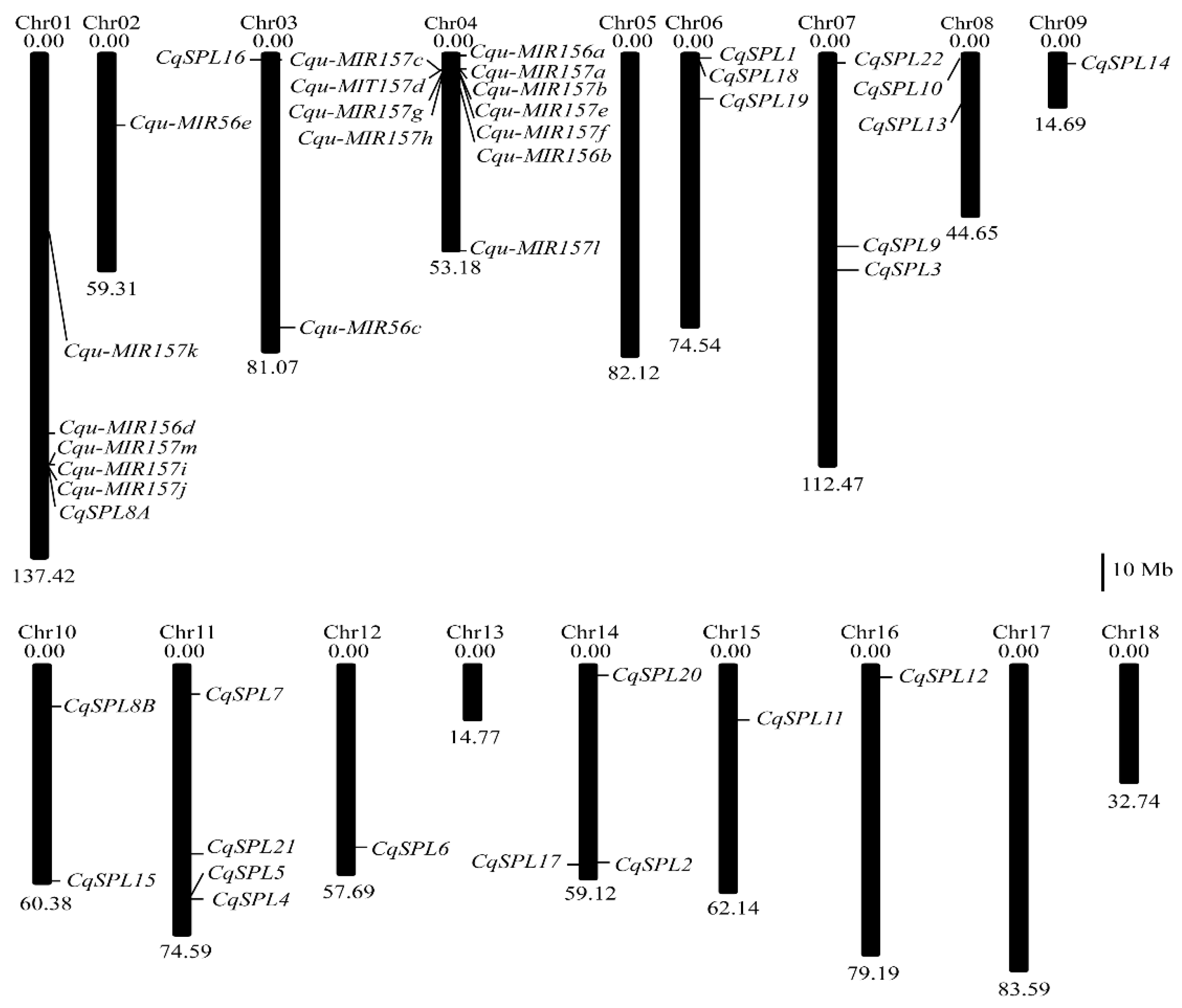
2.3. Location of Genes on Chromosomes
2.4. Prediction of CqSPL Genes Targeted by miR156/7
2.5. Sequence Alignment and Phylogenetic Analysis
2.6. Gene Structure and Conserved Motifs
2.7. Identification and Analysis of Promoters
2.8. RNA Extraction and qRT-PCR Analysis
2.9. Data Analysis
3. Results
3.1. Identification of SPL Family Genes in the Quinoa Genome
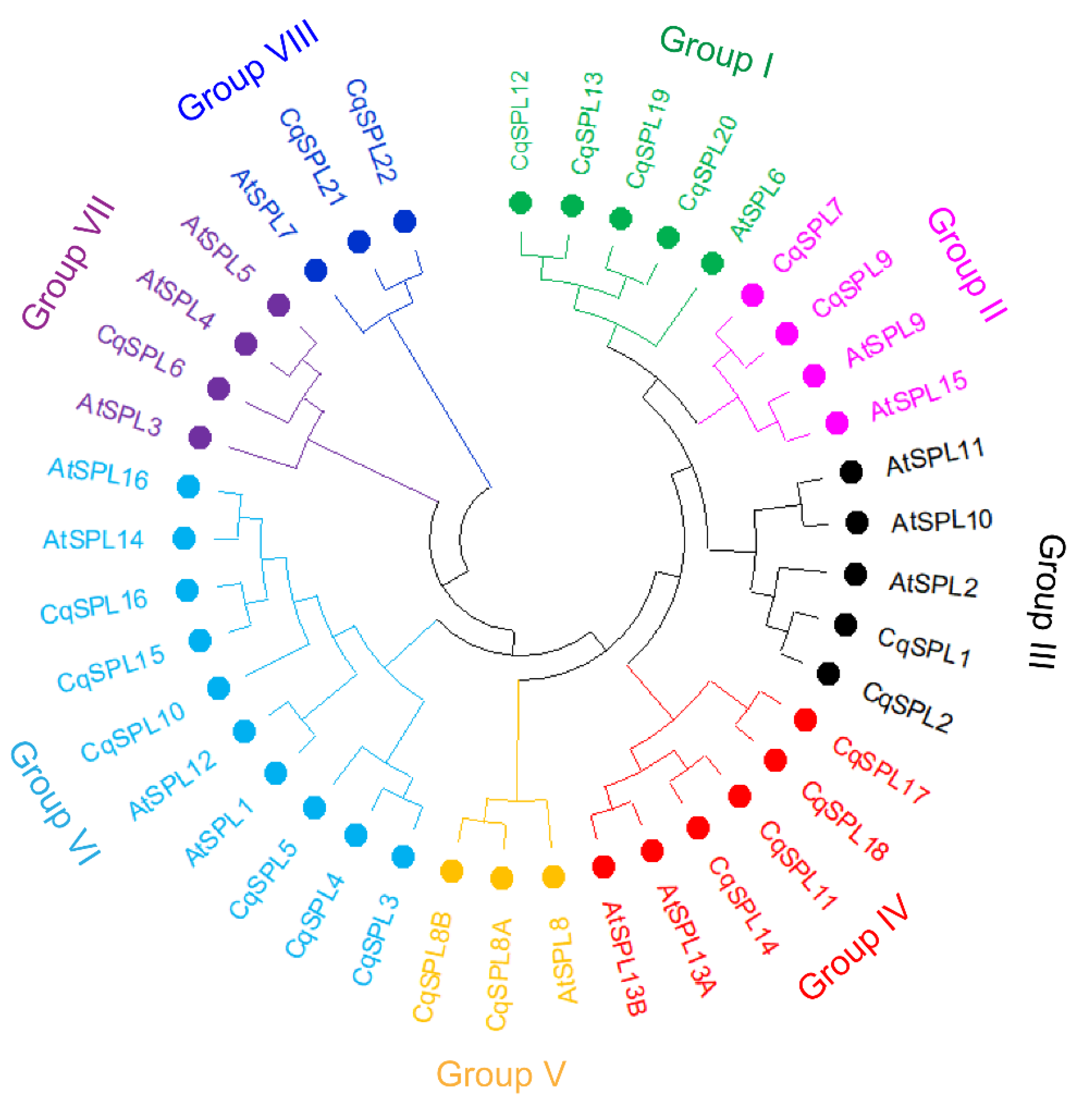
3.2. Phylogeny of CqSPL Genes
3.3. Structure of CqSPL Genes and Domain/Motif Analysis of Their Proteins
3.4. Promoter Analysis of CqSPL Genes
3.5. Tissue Expression Patterns of CqSPL Genes
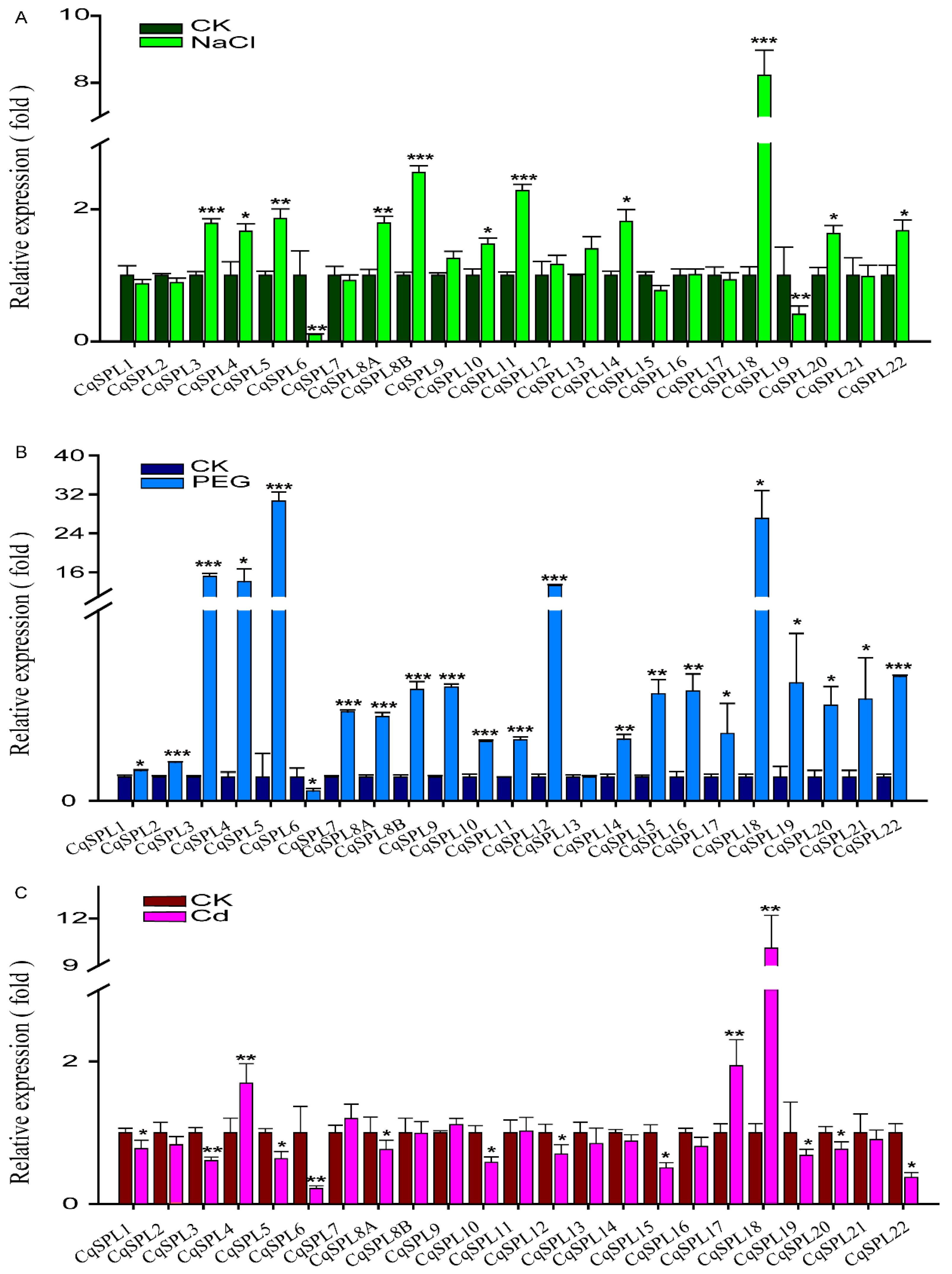
3.6. Expression of CqSPL Genes under Drought, Salt, and Cd Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Birkenbihl, R.P.; Jach, G.; Saedler, H.; Huijser, P. Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 2005, 352, 585–596. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.H.; Hohmann, S.; Nettesheim, K.; Saedler, H.; Huijser, P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 1997, 12, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.A.; Harper, L.C.; Krueger, R.W.; Dellaporta, S.L.; Freeling, M. liguleless1 encodes a nuclear-localized protein required for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 1997, 11, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Cardon, G.; Hohmann, S.; Klein, J.; Nettesheim, K.; Saedler, H.; Huijser, P. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 1999, 237, 91–104. [Google Scholar] [CrossRef]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Hou, H.; Li, J.; Gao, M.; Singer, S.D.; Wang, H.; Mao, L.; Fei, Z.; Wang, X. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 2013, 8, e59358. [Google Scholar] [CrossRef]
- Li, C.; Lu, S. Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 2014, 14, 131. [Google Scholar] [CrossRef]
- Mao, H.-D.; Yu, L.-J.; Li, Z.-J.; Yan, Y.; Han, R.; Liu, H.; Ma, M. Genome-wide analysis of the SPL family transcription factors and their responses to abiotic stresses in maize. Plant Gene 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Zhang, W.; Li, B.; Yu, B. Genome-wide identification, phylogeny and expression analysis of the SBP-box gene family in maize (Zea mays). J. Integr. Agric. 2016, 15, 29–41. [Google Scholar] [CrossRef]
- Kavas, M.; Kizildogan, A.K.; Abanoz, B. Comparative genome-wide phylogenetic and expression analysis of SBP genes from potato (Solanum tuberosum). Comput. Biol. Chem. 2017, 67, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Sun, W.; Ma, Z.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification of the SPL gene family in Tartary Buckwheat (Fagopyrum tataricum) and expression analysis during fruit development stages. BMC Plant Biol. 2019, 19, 299. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Liu, Y.; Ma, L.; Wang, X.; Zhang, D.; Han, Y.; Ding, Q.; Ma, L. Genome-wide identification, phylogeny and expression analysis of the SPL gene family in wheat. BMC Plant Biol. 2020, 20, 420. [Google Scholar] [CrossRef] [PubMed]
- Unte, U.S.; Sorensen, A.M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef]
- Zhang, Y.; Schwarz, S.; Saedler, H.; Huijser, P. SPL8, a local regulator in a subset of gibberellin-mediated developmental processes in Arabidopsis. Plant Mol. Biol. 2007, 63, 429–439. [Google Scholar] [CrossRef]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef]
- Wang, J.W.; Schwab, R.; Czech, B.; Mica, E.; Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 2008, 20, 1231–1243. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.; Cai, W.J.; Wang, S.; Shan, C.M.; Wang, L.J.; Chen, X.Y. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 2010, 22, 2322–2335. [Google Scholar] [CrossRef]
- Xue, X.Y.; Zhao, B.; Chao, L.M.; Chen, D.Y.; Cui, W.R.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet. 2014, 10, e1004266. [Google Scholar] [CrossRef]
- Ioannidi, E.; Rigas, S.; Tsitsekian, D.; Daras, G.; Alatzas, A.; Makris, A.; Tanou, G.; Argiriou, A.; Alexandrou, D.; Poethig, S.; et al. Trichome patterning control involves TTG1 interaction with SPL transcription factors. Plant Mol. Biol. 2016, 92, 675–687. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Höhmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Garcia-Molina, A.; Hohmann, S.; Berndtgen, R.; Huijser, P. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 2013, 75, 566–577. [Google Scholar] [CrossRef]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef]
- Martin, R.C.; Asahina, M.; Liu, P.-P.; Kristof, J.R.; Coppersmith, J.L.; Pluskota, W.E.; Bassel, G.W.; Goloviznina, N.A.; Nguyen, T.T.; Martínez-Andújar, C.; et al. The regulation of post-germinative transition from the cotyledon- to vegetative-leaf stages by microRNA-targeted Squamosa Promoter-Binding Protein LIKE13 in Arabidopsis. Seed Sci. Res. 2010, 20, 89–96. [Google Scholar] [CrossRef]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, S.; Liu, Q.; Wu, K.; Zhang, J.; Wang, S.; Wang, Y.; Chen, X.; Zhang, Y.; Gao, C.; et al. The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 2015, 47, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Q. Boosting Rice Yield by Fine-Tuning SPL Gene Expression. Trends Plant Sci. 2017, 22, 643–646. [Google Scholar] [CrossRef]
- Chuck, G.; Whipple, C.; Jackson, D.; Hake, S. The maize SBP-box transcription factor encoded by tasselsheath4 regulates bract development and the establishment of meristem boundaries. Development 2010, 137, 1243–1250. [Google Scholar] [CrossRef]
- Chuck, G.S.; Brown, P.J.; Meeley, R.; Hake, S. Maize SBP-box transcription factors unbranched2 and unbranched3 affect yield traits by regulating the rate of lateral primordia initiation. Proc. Natl. Acad. Sci. USA 2014, 111, 18775–18780. [Google Scholar] [CrossRef]
- Wang, H.; Nussbaum-Wagler, T.; Li, B.; Zhao, Q.; Vigouroux, Y.; Faller, M.; Bomblies, K.; Lukens, L.; Doebley, J.F. The origin of the naked grains of maize. Nature 2005, 436, 714–719. [Google Scholar] [CrossRef]
- Manning, K.; Tor, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef]
- Ferreira e Silva, G.F.; Silva, E.M.; Azevedo, M.A.S.; Guivin, M.A.; Ramiro, D.A.; Figueiredo, C.R.; Carrer, H.; Peres, L.E.; Nogueira, F.T. microRNA156-targeted SPL/SBP box transcription factors regulate tomato ovary and fruit development. Plant J. 2014, 78, 604–618. [Google Scholar] [CrossRef]
- Cui, L.; Zheng, F.; Wang, J.; Zhang, C.; Xiao, F.; Ye, J.; Li, C.; Ye, Z.; Zhang, J. miR156a-targeted SBP-Box transcription factor SlSPL13 regulates inflorescence morphogenesis by directly activating SFT in tomato. Plant Biotechnol. J. 2020, 18, 1670–1682. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.; Wang, X.; Ye, B.; Jin, M.; Chen, W.; Wang, Y.; Zhou, Y.; Blanks, A.M.; Gu, M.; Zhang, P.; et al. Molecular and functional characterization of the SBP-box transcription factor SPL-CNR in tomato fruit ripening and cell death. J. Exp. Bot. 2020, 71, 2995–3011. [Google Scholar] [CrossRef] [PubMed]
- Yun, J.; Sun, Z.; Jiang, Q.; Wang, Y.; Wang, C.; Luo, Y.; Zhang, F.; Li, X. The miR156b-GmSPL9d module modulates nodulation by targeting multiple core nodulation genes in soybean. New Phytol. 2022, 233, 1881–1899. [Google Scholar] [CrossRef]
- Zhang, L.; He, G.; Li, Y.; Yang, Z.; Liu, T.; Xie, X.; Kong, X.; Sun, J. PIL transcription factors directly interact with SPLs and repress tillering/branching in plants. New Phytol. 2022, 233, 1414–1425. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA Promoter Binding Protein-Like7 Is a Central Regulator for Copper Homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef]
- Kropat, J.; Tottey, S.; Birkenbihl, R.P.; Depege, N.; Huijser, P.; Merchant, S. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 2005, 102, 18730–18735. [Google Scholar] [CrossRef]
- Chen, W.W.; Jin, J.F.; Lou, H.Q.; Liu, L.; Kochian, L.V.; Yang, J.L. LeSPL-CNR negatively regulates Cd acquisition through repressing nitrate reductase-mediated nitric oxide production in tomato. Planta 2018, 248, 893–907. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ding, H.; Jiang, H.; Wang, H.; Chen, K.; Duan, J.; Feng, S.; Wu, G. Regulation of cadmium tolerance and accumulation by miR156 in Arabidopsis. Chemosphere 2020, 242, 125168. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, J.; Jiang, D.; Hong, Y.; Xu, J.; Zheng, S.; Yang, J.; Chen, W. The miR157-SPL-CNR module acts upstream of bHLH101 to negatively regulate iron deficiency responses in tomato. J. Integr. Plant Biol. 2022, 64, 1059–1075. [Google Scholar] [CrossRef]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef]
- Wang, J.; Ye, Y.; Xu, M.; Feng, L.; Xu, L.A. Roles of the SPL gene family and miR156 in the salt stress responses of tamarisk (Tamarix chinensis). BMC Plant Biol. 2019, 19, 370. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Yan, S.; Jing, Y.; Yang, R.; Zhang, Y.; Zhou, Y.; Zhu, Y.; Sun, J. MIR156-Targeted SPL9 Is Phosphorylated by SnRK2s and Interacts With ABI5 to Enhance ABA Responses in. Front. Plant Sci. 2021, 12, 708573. [Google Scholar] [CrossRef] [PubMed]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.R.; Baurle, I. Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef]
- Ma, Y.; Xue, H.; Zhang, F.; Jiang, Q.; Yang, S.; Yue, P.; Wang, F.; Zhang, Y.; Li, L.; He, P.; et al. The miR156/SPL module regulates apple salt stress tolerance by activating MdWRKY100 expression. Plant Biotechnol. J. 2021, 19, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Feyissa, B.A.; Arshad, M.; Gruber, M.Y.; Kohalmi, S.E.; Hannoufa, A. The interplay between miR156/SPL13 and DFR/WD40-1 regulate drought tolerance in alfalfa. BMC Plant Biol. 2019, 19, 434. [Google Scholar] [CrossRef]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, L.J.; Wang, J.W.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2017, 8, 13925. [Google Scholar] [CrossRef]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A Regulatory Network for miR156-SPL Module in. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef]
- Li, F.; Guo, X.; Liu, J.; Zhou, F.; Liu, W.; Wu, J.; Zhang, H.; Cao, H.; Su, H.; Wen, R. Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Transcription Factor in Chenopodium quinoa. Genes 2019, 10, 500. [Google Scholar] [CrossRef]
- Lopez-Marques, R.L.; Norrevang, A.F.; Ache, P.; Moog, M.; Visintainer, D.; Wendt, T.; Osterberg, J.T.; Dockter, C.; Jorgensen, M.E.; Salvador, A.T.; et al. Prospects for the accelerated improvement of the resilient crop quinoa. J. Exp. Bot. 2020, 71, 5333–5347. [Google Scholar] [CrossRef]
- Filho, A.M.; Pirozi, M.R.; Borges, J.T.; Pinheiro Sant’Ana, H.M.; Chaves, J.B.; Coimbra, J.S. Quinoa: Nutritional, functional, and antinutritional aspects. Crit. Rev. Food Sci. Nutr. 2017, 57, 1618–1630. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa, L.; Gonzalez, J.A.; Barrios-Masias, F.H.; Fuentes, F.; Murphy, K.M. Quinoa Abiotic Stress Responses: A Review. Plants 2018, 7, 106. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Su, C.; Dong, C.H. Genome-Wide Transcriptomic and Proteomic Exploration of Molecular Regulations in Quinoa Responses to Ethylene and Salt Stress. Plants 2021, 10, 2281. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.E.; Ho, Y.S.; Lightfoot, D.J.; Schmockel, S.M.; Li, B.; Borm, T.J.; Ohyanagi, H.; Mineta, K.; Michell, C.T.; Saber, N.; et al. The genome of Chenopodium quinoa. Nature 2017, 542, 307–312. [Google Scholar] [CrossRef]
- Golicz, A.A.; Steinfort, U.; Arya, H.; Singh, M.B.; Bhalla, P.L. Analysis of the quinoa genome reveals conservation and divergence of the flowering pathways. Funct. Integr. Genom. 2020, 20, 245–258. [Google Scholar] [CrossRef]
- Wu, Q.; Bai, X.; Zhao, W.; Shi, X.; Xiang, D.; Wan, Y.; Wu, X.; Sun, Y.; Zhao, J.; Peng, L.; et al. Investigation into the underlying regulatory mechanisms shaping inflorescence architecture in Chenopodium quinoa. BMC Genom. 2019, 20, 658. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, B.; Wei, X. Genome wide identification and expression pattern analysis of the GRAS family in quinoa. Funct. Plant Biol. 2021, 48, 948–962. [Google Scholar] [CrossRef]
- Sun, W.; Yu, H.; Ma, Z.; Yuan, Y.; Wang, S.; Yan, J.; Xu, X.; Chen, H. Molecular Evolution and Local Root Heterogeneous Expression of the Chenopodium quinoa ARF Genes Provide Insights into the Adaptive Domestication of Crops in Complex Environments. J. Mol. Evol. 2021, 89, 287–301. [Google Scholar] [CrossRef]
- Li, F.; Liu, J.; Guo, X.; Yin, L.; Zhang, H.; Wen, R. Genome-wide survey, characterization, and expression analysis of bZIP transcription factors in Chenopodium quinoa. BMC Plant Biol. 2020, 20, 405. [Google Scholar] [CrossRef]
- Yue, H.; Chang, X.; Zhi, Y.; Wang, L.; Xing, G.; Song, W.; Nie, X. Evolution and Identification of the WRKY Gene Family in Quinoa (Chenopodium quinoa). Genes 2019, 10, 131. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.D.; Ling, L.Z.; Yi, T.S. Evolution and divergence of SBP-box genes in land plants. BMC Genom. 2015, 16, 787. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhang, S.; Chen, F.; Liu, B.; Wu, L.; Li, F.; Zhang, J.; Bao, M.; Liu, G. Genome-wide identification and characterization of the SBP-box gene family in Petunia. BMC Genom. 2018, 19, 193. [Google Scholar] [CrossRef] [PubMed]
- Salinas, M.; Xing, S.; Hohmann, S.; Berndtgen, R.; Huijser, P. Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato. Planta 2012, 235, 1171–1184. [Google Scholar] [CrossRef]
- Yu, N.; Yang, J.C.; Yin, G.T.; Li, R.S.; Zou, W.T. Genome-wide characterization of the SPL gene family involved in the age development of Jatropha curcas. BMC Genom. 2020, 21, 368. [Google Scholar] [CrossRef]
- Liu, Y.; Aslam, M.; Yao, L.A.; Zhang, M.; Wang, L.; Chen, H.; Huang, Y.; Qin, Y.; Niu, X. Genomic analysis of SBP gene family in Saccharum spontaneum reveals their association with vegetative and reproductive development. BMC Genom. 2021, 22, 767. [Google Scholar] [CrossRef]
- Sommer, F.; Kropat, J.; Malasarn, D.; Grossoehme, N.E.; Chen, X.; Giedroc, D.P.; Merchant, S.S. The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell 2010, 22, 4098–4113. [Google Scholar] [CrossRef]
- Gou, J.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Wen, J.; Wang, Z.Y. From model to crop: Functional characterization of SPL8 in M. truncatula led to genetic improvement of biomass yield and abiotic stress tolerance in alfalfa. Plant Biotechnol. J. 2018, 16, 951–962. [Google Scholar] [CrossRef]
- Gou, J.; Tang, C.; Chen, N.; Wang, H.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Allen, R.D.; et al. SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 2019, 222, 1610–1623. [Google Scholar] [CrossRef]
- Gao, F.; Nan, F.; Feng, J.; Lv, J.; Liu, Q.; Xie, S. Identification and characterization of microRNAs in Eucheuma denticulatum by high-throughput sequencing and bioinformatics analysis. RNA Biol. 2016, 13, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. Functional Evolution in the Plant Squamosa-Promoter Binding Protein-Like (SPL) Gene Family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.M.; Liu, Y.Q.; Chen, D.Y.; Xue, X.Y.; Mao, Y.B.; Chen, X.Y. Arabidopsis Transcription Factors SPL1 and SPL12 Confer Plant Thermotolerance at Reproductive Stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef]
- Riese, M.; Hohmann, S.; Saedler, H.; Munster, T.; Huijser, P. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 2007, 401, 28–37. [Google Scholar] [CrossRef]
- Gao, Z.; Ma, C.; Zheng, C.; Yao, Y.; Du, Y. Advances in the regulation of plant salt-stress tolerance by miRNA. Mol. Biol. Rep. 2022, 49, 5041–5055. [Google Scholar] [CrossRef]
- Lan, T.; Zheng, Y.; Su, Z.; Yu, S.; Song, H.; Zheng, X.; Lin, G.; Wu, W. OsSPL10, an SBP-Box Gene, Plays a Dual Role in Salt Tolerance and Trichome Formation in Rice (Oryza sativa L.). G3 Genes Genomes Genet. 2019, 9, 4107–4114. [Google Scholar] [CrossRef]
- Jerome Jeyakumar, J.M.; Ali, A.; Wang, W.M.; Thiruvengadam, M. Characterizing the Role of the miR156-SPL Network in Plant Development and Stress Response. Plants 2020, 9, 1206. [Google Scholar] [CrossRef]
- Ning, K.; Chen, S.; Huang, H.; Jiang, J.; Yuan, H.; Li, H. Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tissue Organ Cult. 2017, 130, 469–481. [Google Scholar] [CrossRef]
- Li, S.; Cheng, Z.; Li, Z.; Dong, S.; Yu, X.; Zhao, P.; Liao, W.; Yu, X.; Peng, M. MeSPL9 attenuates drought resistance by regulating JA signaling and protectant metabolite contents in cassava. Theor. Appl. Genet. 2022, 135, 817–832. [Google Scholar] [CrossRef]








| Gene Name | Gene ID | CDS (bp) | Amino Acids | MW (kD) | pI | Subcellular Localization |
|---|---|---|---|---|---|---|
| CqSPL1 | AUR62028919 | 1389 | 462 | 50.27 | 8.90 | Nucl |
| CqSPL2 | AUR62005629 | 1389 | 462 | 50.31 | 8.67 | Nucl |
| CqSPL3 | AUR62002563 | 2937 | 978 | 108.45 | 6.44 | Nucl/Cyto |
| CqSPL4 | AUR62029983 | 3570 | 1189 | 132.14 | 7.01 | Nucl/Cyto |
| CqSPL5 | AUR62029984 | 2943 | 980 | 108.87 | 6.30 | Cyto/Nucl |
| CqSPL6 | AUR62019452 | 609 | 202 | 21.12 | 9.81 | Nucl |
| CqSPL7 | AUR62024322 | 1050 | 349 | 37.73 | 7.64 | Nucl |
| CqSPL8A | AUR62004146 | 849 | 282 | 31.95 | 9.41 | Nucl |
| CqSPL8B | AUR62013707 | 861 | 286 | 32.59 | 9.41 | Nucl |
| CqSPL9 | AUR62012061 | 1122 | 373 | 39.56 | 8.45 | Nucl |
| CqSPL10 | AUR62011728 | 2955 | 984 | 110.16 | 5.76 | Nucl |
| CqSPL11 | AUR62029416 | 1044 | 347 | 38.02 | 8.76 | Nucl |
| CqSPL12 | AUR62039662 | 1638 | 545 | 60.40 | 6.26 | Nucl |
| CqSPL13 | AUR62032118 | 1689 | 562 | 62.08 | 6.87 | Nucl |
| CqSPL14 | AUR62003425 | 1050 | 349 | 38.42 | 8.74 | Nucl |
| CqSPL15 | AUR62042534 | 2769 | 922 | 101.71 | 6.75 | Nucl |
| CqSPL16 | AUR62035190 | 3321 | 1106 | 121.76 | 7.06 | Nucl |
| CqSPL17 | AUR62005645 | 1080 | 359 | 39.00 | 9.36 | Nucl |
| CqSPL18 | AUR62028905 | 954 | 317 | 34.78 | 9.05 | Nucl |
| CqSPL19 | AUR62003075 | 1500 | 499 | 55.39 | 5.89 | Nucl |
| CqSPL20 | AUR62007890 | 1272 | 423 | 46.75 | 6.57 | Nucl |
| CqSPL21 | AUR62042853 | 2151 | 716 | 80.13 | 6.32 | Nucl |
| CqSPL22 | AUR62042654 | 2124 | 707 | 79.25 | 5.74 | Nucl |
| Name | Chr. | Precursor Length (bp) | Position | Database | Original Name |
|---|---|---|---|---|---|
| Cqu-MIR156a | 4 | 166 | 1458734-1458899 | PmiREN * | Cqu-MIR156a |
| Cqu-MIR156b | 4 | 85 | 13677430-13677514 | PmiREN | Cqu-MIR156b |
| Cqu-MIR156c | 3 | 197 | 75723156-75723352 | PmiREN | Cqu-MIR156c |
| Cqu-MIR156d | 1 | 86 | 107403506-107403591 | PmiREN | Cqu-MIR156d |
| Cqu-MIR156e | 2 | 87 | 22325967-22326053 | This study | |
| Cqu-MIR157a | 4 | 87 | 6735955-6736041 | PmiREN | Cqu-MIR156e |
| Cqu-MIR157b | 4 | 82 | 6736193-6736274 | PmiREN | Cqu-MIR156f |
| Cqu-MIR157c | 4 | 87 | 6743900-6743986 | PmiREN | Cqu-MIR156g |
| Cqu-MIR157d | 4 | 84 | 6744076-6744159 | PmiREN | Cqu-MIR156h |
| Cqu-MIR157e | 4 | 87 | 6786472-6786558 | PmiREN | Cqu-MIR156i |
| Cqu-MIR157f | 4 | 82 | 6786710-6786791 | PmiREN | Cqu-MIR156j |
| Cqu-MIR157g | 4 | 87 | 6794420-6794506 | PmiREN | Cqu-MIR156k |
| Cqu-IR157h | 4 | 85 | 6794596-6794680 | PmiREN | Cqu-MIR156l |
| Cqu-MIR157i | 1 | 83 | 115037617-115037699 | PmiREN | Cqu-MIR156m |
| Cqu-MIR157j | 1 | 87 | 115046902-115046988 | PmiREN | Cqu-MIR156n |
| Cqu-MIR157k | 1 | 144 | 46037964-46038107 | PmiREN | Cqu-MIR156o |
| Cqu-MIR157l | 4 | 132 | 52526028-52526159 | PmiREN | Cqu-MIR156p |
| Cqu-MIR157m | 1 | 85 | 115031692-115031776 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Cao, H.; Zhang, M.; Deng, S.; Li, T.; Xing, S. Genome-Wide Identification and Characterization of SPL Family Genes in Chenopodium quinoa. Genes 2022, 13, 1455. https://doi.org/10.3390/genes13081455
Zhao H, Cao H, Zhang M, Deng S, Li T, Xing S. Genome-Wide Identification and Characterization of SPL Family Genes in Chenopodium quinoa. Genes. 2022; 13(8):1455. https://doi.org/10.3390/genes13081455
Chicago/Turabian StyleZhao, Hongmei, Huaqi Cao, Mian Zhang, Sufang Deng, Tingting Li, and Shuping Xing. 2022. "Genome-Wide Identification and Characterization of SPL Family Genes in Chenopodium quinoa" Genes 13, no. 8: 1455. https://doi.org/10.3390/genes13081455
APA StyleZhao, H., Cao, H., Zhang, M., Deng, S., Li, T., & Xing, S. (2022). Genome-Wide Identification and Characterization of SPL Family Genes in Chenopodium quinoa. Genes, 13(8), 1455. https://doi.org/10.3390/genes13081455






