Abstract
Several syndromic forms of digestive cancers are known to predispose to early-onset gastric tumors such as Hereditary Diffuse Gastric Cancer (HDGC) and Lynch Syndrome (LS). LSII is an extracolonic cancer syndrome characterized by a tumor spectrum including gastric cancer (GC). In the current work, our main aim was to identify the mutational spectrum underlying the genetic predisposition to diffuse gastric tumors occurring in a Tunisian family suspected of both HDGC and LS II syndromes. We selected the index case “JI-021”, which was a woman diagnosed with a Diffuse Gastric Carcinoma and fulfilling the international guidelines for both HDGC and LSII syndromes. For DNA repair, a custom panel targeting 87 candidate genes recovering the four DNA repair pathways was used. Structural bioinformatics analysis was conducted to predict the effect of the revealed variants on the functional properties of the proteins. DNA repair genes panel screening identified two variants: a rare MSH2 c.728G>A classified as a variant with uncertain significance (VUS) and a novel FANCD2 variant c.1879G>T. The structural prediction model of the MSH2 variant and electrostatic potential calculation showed for the first time that MSH2 c.728G>A is likely pathogenic and is involved in the MSH2-MLH1 complex stability. It appears to affect the MSH2-MLH1 complex as well as DNA-complex stability. The c.1879G>T FANCD2 variant was predicted to destabilize the protein structure. Our results showed that the MSH2 p.R243Q variant is likely pathogenic and is involved in the MSH2-MLH1 complex stability, and molecular modeling analysis highlights a putative impact on the binding with MLH1 by disrupting the electrostatic potential, suggesting the revision of its status from VUS to likely pathogenic. This variant seems to be a shared variant in the Mediterranean region. These findings emphasize the importance of testing DNA repair genes for patients diagnosed with diffuse GC with suspicion of LSII and colorectal cancer allowing better clinical surveillance for more personalized medicine.
1. Introduction
Gastric carcinoma (GC) is the fifth most common cancer worldwide with approximately one million new cases registered in 2018 (5.7%) with significant geographical distribution variations. It represents the third largest cause of cancer-related death with 783,000 deaths worldwide in 2018, which represents 8.2% of all cancer deaths [1,2]. In Tunisia, as in other countries, the sporadic form of GC is the most frequent, estimated at 637 new cases per year and ranks the 7th among all diagnosed cancers [1]. Inherited syndromic forms of digestive cancers with age occurrence before the age of 50 (gastric and colorectal) seem to be in relatively higher proportions in the Tunisian population, suggesting a genetic susceptibility [3]. However, no epidemiological data are available, and incidence rates of hereditary forms are lacking. Several syndromic forms are known to predispose to gastric tumors such as hereditary diffuse gastric cancer (OMIM: 137215), lynch syndrome (OMIM: 120435), Peutz Jegher Syndrome (OMIM: 175200) and Li Fraumeni syndrome (OMIM: 151623) [4].
Indeed, 3–5% of GCs are caused by autosomal dominant inherited mutations and familial aggregation occurs in approximately 10% of the cases [5]. Hereditary diffuse gastric cancer (HDGC) syndrome is known as the most frequent autosomal dominant form of GC and is a poorly differentiated morphologic type [6]. Approximately 40% of HDGC cases have germline mutations in CDH1 (E-Cadherin, OMIM: 192090), and more than 100 CDH1 mutations have been reported in HDGC cases. Lynch Syndrome (LS) (OMIM: 120435), or HNPCC (Hereditary Non Polyposis Colorectal Cancer), is the most prevalent inherited CCR (colorectal cancer) susceptibility syndrome, responsible for up to 6% of all CRCs. It is characterized by point mutations and/or large rearrangements in DNA mismatch repair (MMR) genes resulting in a loss of MMR complex function and microsatellite instability (MSI). The MMR genes associated with this syndrome are: MLH1 (OMIM: 609310), MSH2 (OMIM: 609309), MSH6 (OMIM: 600678) and PMS2 (OMIM: 600259) [7,8].
In addition, germline deletions in the 3′end of EPCAM gene (OMIM: 613244), directly upstream to MSH2, have been found to underlie a small proportion of LS cases (<5%) through methylation induced transcriptional silencing of MSH2 [9].
LS has been subdivided into two subtypes: LSI known as site-specific colonic cancer syndrome, and LSII or extracolonic cancer syndrome (OMIM: 120435) [10]. The former subtype includes 2–3% of all CCRs and is characterized by a susceptibility to colorectal tumors in the absence of diffuse polyposis. The latest subtype is associated with a high risk of various tumors types such as endometrium [11,12,13], gastric [5,14,15,16], breast [17], biliary tract, urinary tract [18,19], small bowel [10], ovarian [20,21,22], brain and skin cancers [23,24,25].
In the context of LS II, MMR genes germline mutation carriers represent 50 to 80% lifetime risk to develop CCR, 40 to 60% risk of developing endometrial cancer (OMIM: 608089) and 13 to 19% risk to develop gastric tumors [26]. Moreover, GC is, among LS carriers, far less common than colorectal at an average older age in western countries [14]. It is well-established that MMR genes dysfunction is linked to LS susceptibility. Indeed, current data assume that MSH2, MLH1, and EpCAM mutation carriers have 13% of cumulative risk incidence of developing gastric tumors at an age of 75 years; however, MSH6 carriers have only 3% of the risk [27]. Even though GC is part of the LS tumors spectrum the risk of developing GC in LS families is unknown and surveillance strategies for GC in LS are still controversial [14]. To our best knowledge, few studies have investigated the molecular basis and the genetic mutational profile underlying this form of GC. In Tunisia, some molecular and epidemiological studies have been reported in colorectal cases in LS families [3,28,29]. However, no study has investigated the molecular basis of GC cases in LS. Hence, it is of crucial importance to set up oncogenetic counseling based on specific genetic tests adapted to the Tunisian population. This will help in early detection of individuals and families at high risk of developing LSII and will consequently reduce the mortality and morbidity due to the disease.
Our main objective in the current work was to identify the genetic mutational profile underlying the DGC occurring in a Tunisian family suspected with both HDGC and LSII by using a classical sequencing and target NGS approaches. We used a custom panel targeting 87 candidate genes recovering the four DNA mismatch repair pathway (MMR, BER, NER and HR) known to be involved in DNA repair disorders. The proband diagnosed with a DGC was screened for a repair genes panel after HDGC syndrome exclusion.
2. Material and Methods
2.1. Patient
This study was conducted according to the declaration of Helsinki and with the approval of the Institutional review board (IRB) of Institut Pasteur de Tunis. Four individuals, belonging to the same large Tunisian family suspected with LSII according to revised Bethesda Guidelines, were investigated after participants wrote informed consent (Figure 1). The index case has been investigated first. She was a woman, “JI-021”, from a consanguineous marriage, fulfilling the first and fifth criteria of the Revised Bethesda Guidelines and the third criteria of IGCLC with no alcohol, smoking consumption habits, spicy foods with low consumption of meats. She was referred first for CDH1 germline genetic testing to the Gastroenterology Department of Mohamed Tahar Maamouri Medical Hospital in Nabeul, Tunisia for HDGC suspicion since she was diagnosed with diffuse gastric carcinoma (DGC) (T3NxM1) at the antrum at an age of 52. This index case had an aggressive phenotype since diffuse gastric tumors are known to be an aggressive gastric tumor histotype. This patient had peritoneal carcinosis, and she was therefore not operable and the only treatment she had was a palliative chemotherapy. This index case has a healthy 23-year-old daughter, three unaffected brothers (47, 53 and 55 years old) and two affected sisters (29 and 47 years old). Her youngest sister, “JI-021-E”, suffered from primary sterility, developed successively ovarian and gastric carcinomas and was treated by hysterectomy and a total gastrectomy and died at the same age. The other sister, “JI-021-D”, developed a BC, and she died at the same age. The index case’s father, “JI-021-H”, suffered from renal failure and diabetes and died at the age of 65. In addition, this family seems to have a strong history of CCR from the paternal side (five cases) without clear information regarding their relationship with the index case. He had a brother and a sister who died, respectively, at the ages of 50 (Probably from CCR), “JI-021-J”, and 32 years old from unknown causes, and a third sister, “JI-021-F”, who developed a CCR before the age of 50 and died at the age of 65. Her daughter, “JI-021-G”, also developed a CCR at the age of 50. Table 1 shows all the clinicopathological criteria of the index case.
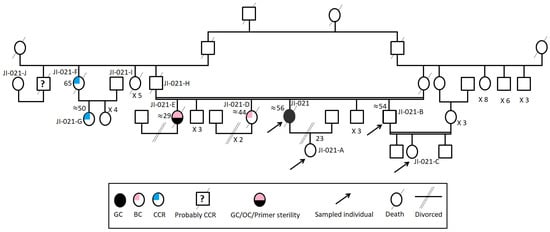
Figure 1.
The familial pedigree of the index case “JI-021”. It included gastric, colon, breast and ovarian cancers. It showed a tumor spectrum typical of LS II form.

Table 1.
Description of clinicopathological features and family history of the index case.
2.2. Sanger Sequencing
2.2.1. CDH1 Coding Regions Sanger Sequencing
Referring to 2015 IGCLC [30] for the search of CDH1 and CTNNA1 [31] germline mutations, a complete sequencing of these two genes was carried out. Primers were designed using Primer Express™ Software version 2.0 Applied Biosystems Thermo Fisher Scientific [32] as shown in Table S1, used in our previous study [33]. Forward and reverse primers incorporated the extensions 18F tail (ACCGTTAGTTAGCGATTT) and 18R tail (CGGATAGCAAGCTCGT), respectively, at their 5′ ends. Then, generated data were analyzed using SeqScape Version 3.2 (Thermo Fisher, Multiple Life Technologies Corporation, Carlsbad, CA, USA) and BioEdit Sequence Alignment Editor Version 7.2.5.
2.2.2. MSH2 Sanger Sequencing
We designed two pairs of primers (Forward primer sequence 5′AGAGGAGGAATTCTGATCAC3′, and reverse primer sequence 5′CTGATTCTCCATTTCTGGC3′) to amplify the following region: MSH2: exon 4 (NM_000251.2) (Table S1). PCR reactions were performed on genomic DNAs (gDNAs), following standard protocols, pursued by Sanger sequencing using an automated sequencer (ABI 3500; Applied Biosystems, Foster City, CA, USA) using a cycle sequencing reaction kit (Big Dye Terminator kit, Applied Biosystems, CA, USA). Data were analyzed by BioEdit Sequence Alignment Editor Version 7.2.5.
2.3. Large CDH1 Deletions/Duplications Analysis by Multiplex Ligation-Dependent Probe Amplification (MLPA) Assay
The index case’s genomic DNA was tested for copy number changes using Multiplex Ligation Dependent Probe Amplification (MLPA). It was performed using the SALSA P083-D2 CDH1 MLPA kit (MRC-Holland, Amsterdam, Holland), following manufacturer’s instructions. The kit contains 35 probes for the CDH1 gene, one upstream flanking probe and one downstream flanking probe. MLPA products were run on the ABI Prism 3730 xl Genetic Analyzer (Applied Biosystems Thermo Fisher, CA, USA) and analyzed with the Peak Scanner™ Software v1.0 (Applied Biosystems Thermo Fisher, CA, USA).
2.4. Next Generation Sequencing
Blood samples have been collected from the index case and her consent relatives and have been sampled in the gastroenterology department. Genomic DNA was isolated from peripheral blood using Qiagen Kit and salting-out, and it was used for the library preparation for NGS. HaloPlexHS assay incorporating molecular barcodes for high-sensitivity sequencing was used as a custom design (HaloPlexHS). Using SureDesign (Agilent Technologies Inc., Santa Clara, CA, USA), probes were generated to cover the exons and 15 bp of the surrounding intronic sequences of a total of 87 candidate genes known to be involved in DNA repair disorders [34].
Amplicon libraries were prepared, from genomic DNA of “JI-021”, using the HaloPlexHS PCR target enrichment system dedicated to Ion Torrent PGM according to the manufacturer’s recommendations. Massively parallel sequencing was performed on an Ion Torrent PGM (Thermo Fisher Scientific). Raw data generated by the PGM sequencer were analyzed using the VarAFTsoftware version 2.5, which is freely available online (https://varaft.eu/, accessed on 20 July 2022). Genome assembly and nucleotide coordinates were referenced to the GRCh37 version of the human genome.
2.5. Variants Filtering Strategy
We aimed herein to select pathogenic or potentially pathogenic variants associated with the development of LSII by removing neutral variants and sequencing errors. We prioritized rare functional variants (missense, nonsense, splice site variants, and indels) and excluded variants with a Minor Allele Frequency (MAF) > 0.01 in dbSNP137, and 138, in the Exome Variant Server (http://evs.gs.washington.edu/EVS/), accessed on 16 April 2022, 1000 Genomes Project (http://www.1000genomes.org/, accessed on 16 April 2022), or Exome Aggregation Consortium database (ExAC), Cambridge, MA (http://exac.broadinstitute.org, accessed on 16 April 2022), and the genome Aggregation Database (gnomAD) (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA398795, accessed on 16 April 2022).
We also removed all synonymous and homozygous variants considering that LS is an autosomal dominant syndrome. All variants with a Depth ≤ 15 have been removed as well as variants classified as Benign and Likely Benign in ClinVar. Variants predicted with tolerated effect by more than two in silico prediction tools have also been removed.
Indeed, various in silico prediction tools have been used to assess the functional effect and pathogenicity of the selected variants such as UMD predictor (http://umd-predictor.eu/, accessed on 16 April 2022), Sorting Intolerant From Tolerant (SIFT) (http://sift.jcvi.org/, accessed on 16 April 2022), used to examine the degree of conservation for amino acid residues across species and to find changes in protein structure and function, PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/, accessed on 16 April 2022), Protein Variation Effect Analyzer (PROVEAN) (http://provean.jcvi.org/, accessed on 16 April 2022), to filter sequence variants to identify nonsynonymous or indel variants that are predicted to be functionally important. Mutation Taster (http://www. mutationtaster.org/, accessed on 16 April 2022), has been used to assess the impact of mutations on protein function and to look at effects on splicing sites, mRNA expression, MAPP-MMR (http://mappmmr.blueankh.com/, accessed on 16 April 2022), to accurate classification of missense variants in MMR genes, FATHMM (http://fathmm.biocompute.org.uk/, accessed on 16 April 2022), to predict the functional consequences of both coding variants (Non-Synonymous single nucleotide variants) and non-coding variants and LRT. Variants not previously reported in healthy controls and classified as pathogenic were evaluated for sequencing depth and visually inspected using the Integrative Genomic Viewer (IGV) before validation by Sanger sequencing.
2.6. Immunohistochemical Study
The index case “JI-021” tumor gastric tissue was tested for E-cadherin, MSH2 and MLH1 expression profiles by immunohistochemistry. Briefly, 3–4 µm tissue sections were obtained from FFPE gastric tissue and all immunohistochemistry steps were conducted as preconized in the Novolink MPolymer Detection Systems Kit (Leica Biosystems United States/Biopole, Tunisia). Antigen retrieval was performed by incubation of tissue sections in a 10 mM Sodium Citrate buffer (pH 6.0) (RE7113, Leica Biosystems United States/Biopole, Tunisia) for 20 min in 95 °C heated water.
2.6.1. E-Cadherin
Gastric tissue was incubated forward overnight with anti-E-Cadherin primary mouse monoclonal antibody (NCL-L-E-Cad, clone 36B5, Novocastra TM, Biopole) recognizing the Nt external domain of E-cadherin. The immunostaining reaction was visualized by adding the Diaminobenzidine DAB as a chromogenic substrate using Novolink MPolymer Detection Systems Kit (Biopole).
2.6.2. MSH2 and MLH1
Primary antibodies anti-MLH1 (ES05) and anti-MSH2 (25D12) Leica Biosystems were diluted to 1:50 and were added and incubated for 60 min. The immunostaining reaction was visualized by adding the AEC as a chromogenic substrate (Novolink MPolymer Detection Systems Kit (Biopole)).
2.7. Preparing the Structures
2.7.1. The Structure of Two Complexes
MSH2/MSH6 [35] and MSH2/MSH3 [36] were used in our computational study to analyze the effect of the variant R243Q on the MSH2 protein. Missing segments and atoms were added using MODELLER [37]. We also used the structure of the N-terminus domain of the DNA mismatch repair protein MLH1 [38]. The variant at position 243 of MSH2 in both complexes was performed in silico using FoldX [39].
2.7.2. Protein–Protein Docking
We conducted a protein–protein docking experiment using ZDOCK version 3.0 [40] using the structure of MLH1 as a ligand (the smallest partner) and the complex MSH2/MSH6 as a receptor (the largest partner). Input options were kept to their default values. We have, however, restrained the docking interface on MLH1 only to solve exposed residues of the large beta-sheet from the N-terminal domain. We generated 2000 complexes from which we took the best ten according to the Zdock scoring function. We then refined the complexes using sander from AMBER 19 [41] molecular dynamics package by running a restrained minimization consisting of 1000 steps of steepest descent (250 steps) and conjugate gradient (750 steps) algorithms. Restraints were applied to backbone atoms of the complex. A non-bonded cutoff of 12 Angstroms was applied for the refinement.
2.7.3. Electrostatic Potential Calculation
The electrostatic potential calculation [42] has been conducted using the Adaptive Poisson–Boltzmann Solver (APBS version 1.4.1) software package.
The PDB structure of a protein is first converted to the PQR format using the PDB2PQR server [43]. The PARSE force field was used to assign the partial charges to each atom and the ionization state of charged residues was assigned according to the calculation by PROPKA. The electrostatic potential calculation was effected using an implicit solvent model. The internal dielectric constants of the solute and the solvent are fixed to 2.0 and 78.45, respectively, and the temperature is set to 298.15 K. The cutoff for non-bonded interactions was set to 15 Angstroms. Visualization of the electrostatic potential was generated using the Chimera molecular viewer.
2.7.4. Stability Analysis
We estimated the free energy of folding between the wild type and the mutant structure (ΔΔGWt–Mut) using DynaMut [44].
3. Results
3.1. Sanger Sequencing
CDH1 Sanger Sequencing
As a result, we have found no deleterious or potentially deleterious variants in the coding and flanking regions of both CDH1 and CTNNA1 genes as it was mentioned in our previous study [33]. This finding suggests other candidate genes/variants predisposing to DGC.
3.2. Large CDH1 Gene Deletions/Duplications Analysis by MLPA Assay
Since heterozygous large genes rearrangements are hard to detect by conventional PCR-based sequencing of gDNA, we searched for possible rearrangements of the CDH1 locus using the MLPA assay [45]. Results were processed by the Coffalyser.Net software (MRC-Holland, Amsterdam, Holland). Our results showed no large rearrangements in the CDH1 gene among this index case, as it is described in our previous study [33].
3.3. E-Cadherin Immunohistochemistry Expression Profile
In order to assess the protein expression level of E-cadherin (CDH1 gene product) in the index case gastric tumor tissue, we have performed an immunohistochemistry by using a monoclonal antibody directed against the Nt domain of E-cadherin. Our result showed a negative immunostaining expression in tumor cells compared to normal adjacent glandular cells as control (Figure 2A). The adjacent normal gland showed a membranous normal E-cadherin expression pattern.
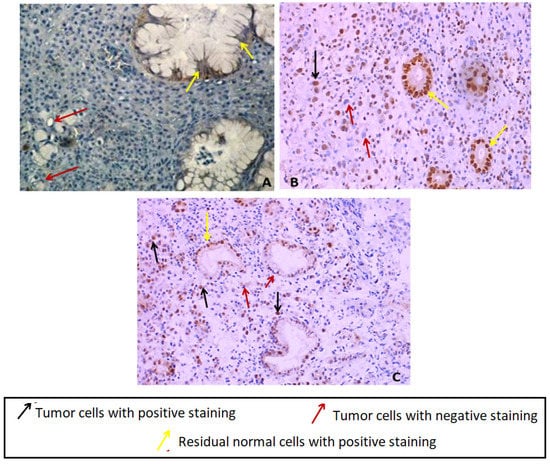
Figure 2.
(A) Immunostaining of E-Cadherin in index case GC tissues showing a loss of E-cadherin in tumor cells compared to normal glandular adjacent cells showing positive membranous staining (×400). (B) Nuclear positive immunostaining of MLH1 in index case GC tissue (×400). (C) Uncomplete nuclear positive immunostaining of MSH2 in index case GC tissue (×200) original magnification.
3.4. Selection of Variants of Interest Detected by Targeted DNA Repair Genes Panel
Besides IGCLC inclusion criteria, this index case also fulfilled Bethesda Guidelines inclusion criteria for testing LS (First, Fourth and Fifth criteria). Hence, this case was a candidate for the screening of a panel of 87 candidate genes. Using SureDesign (Agilent Technologies Inc.), probes were generated to cover exons and 15 bp flanking sequences. The size of the final target region was 251.689 kpb with 33,828 amplicons and the mean coverage was 99.74% of the target region. As a result, we identified 33 variants. Statistical distribution of variants is illustrated in (Figure 3). Exonic variants represented 23 out of 33 variants. Among the 23 exonic variants 20 were novel (Table S2).
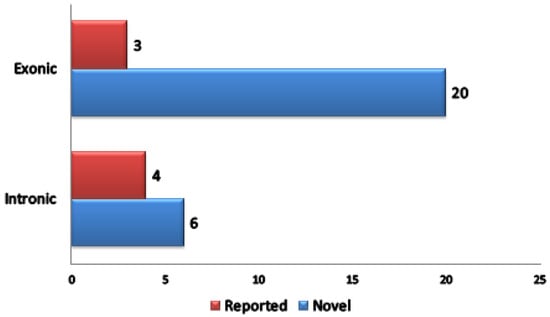
Figure 3.
Statistical distribution of variants identified after filtering strategy.
Among the exonic variants, only the rare MSH2 (rs63751455) variant (with a MAF less than 0.01 in the general population) corresponding to substitutions c.728G>A p.R243Q, classified as a VUS in ClinVar, has been selected for Sanger sequencing validation and for structural model effect prediction analysis. Different in silico prediction tools (15) were used to evaluate the potential functional effect of this variant (Table S2). The variant is located in a conserved protein domain of the MSH2 protein among several species. Using Mutation taster, this variant has been described as “disease causing” and seems to affect the protein structure. Both Polyphen and SIFT described it as “damaging”. UMD predictor classified it as “pathogenic” and Provean as “deleterious”. In addition, according to the Human Splice Finder tool this variant results in a creation of a new acceptor splicing site and a new silent splicing site “Exonic Splicing Silencers ” (ESS) (wild: 0.55/mutated: 0.62). The MAPP-MMR tool classified it as neutral with a score equal to 2.130. In brief, by using other online supplementary prediction tools (DANN, FATHMM v2.3, Mutation Assessor, LRT…), this variant has been described as “deleterious or pathogenic” by 13 out of 15 tested tools.
This variant has been confirmed for the index case “JI-021” in an heterozygous state and not found in the index case’s relatives (index case’s daughter, brother and her niece). This variant has been forward searched in three other gastric cancer cases, “JI-002”, “JI-030” and “JI-036”, belonging to unrelated families suspected with LS II and fulfilling the first criterion of Bethesda guidelines for the first case “JI-002” and the fifth criterion for both “JI-030” and “JI-036” cases based on their family history of cancer. As a result, it has been found in none of the three screened index cases suspected with LSII. A search of potential linkage disequilibrium of this variant with known pathogenic variants has been performed, and currently no information is available in LDproxy Tool [46].
Additionally, based on structural analysis and DynaMut score on the theoretical functional effect, a novel identified variant on the FANCD2 gene has been selected for in silico prediction analysis as well.
3.5. MSH2 and MLH1 Immunohistochemistry Staining in the Index Case Tumor Gastric Tissue
The immunohistochemistry analysis of MSH2 and MLH1 proteins in the index case gastric tissue has shown a positive diffuse nuclear immunostaining for MLH1 and nuclear immunostaining of MSH2 in tumor cells and normal adjacent residual glandular cells with loss of MSH2 staining in some tumor cells (incomplete immunostaining) (Figure 2B,C). Since no significant difference was noticed between mutated and wildtype models regarding the putative impact on the collective motion of the residues using MSH2/MSH6, MSH2/MSH3 and MSH2 (monomeric protein) structures, and based on MSH2/MLH1 effect, only MSH2 and MLH1 proteins have been investigated by IHC. Moreover, we no longer have FFPE gastric tissue available for this case since it was a small biopsy.
3.6. Molecular Modeling of MSH2 Variant on the MSH2/MSH6/MLH1 Complex
The variant R243Q is located on the connector domain of MSH2 at a loop connecting two alpha-helices (Figure 4A). Most of the atoms of the side chain of arginine are highly packed against other residues forming the core of the domain while the polar guanidine group is exposed and forms a hydrogen bond with the D240 main chain. With such a configuration of R243, we thought first that the variant could impact the conformational function of the connector domain to other parts of the protein. We then run a calculation of the normal modes for the Wild Type (WT) form (R243) and the mutant form (Q243) to investigate the putative impact on the collective motion of the residues using MSH2/MSH6, MSH2/MSH3 and MSH2 (monomeric protein) structures, but no significant difference was noticed We then investigated the possibility that the connector domain is involved in protein–protein interaction with other partners. Indeed, there have been different arguments showing that the connector domain of MSH2 is involved in the interaction with a heterodimer of MLH1/PMS1 [47]. We chose to run a protein–protein docking experiment to predict the ternary complex MSH2/MSH6/MLH1 (Figure 4B). Noting that the docking analysis was conducted only using the wild type structure of MSH2 since the docking score for solving protein–protein complexes are not reliable to determine the stability of a binary association. In other words, the docking was conducted to predict surface patches on MSH2 that are likely to bind MLH1. Of the 10 complexes with the best ZDOCK energy score, six show a binding mode where the connector domain interacts with MLH1, while in the docking solution with the best energy score, R243 is part of the interface.
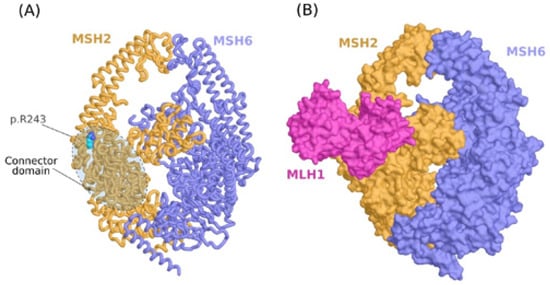
Figure 4.
Molecular modeling analysis of p.R243Q variant. (A) Molecular model of the MSH2/MSH6 complex showing the location of the variant on the connector domain of MSH2. (B) Ternary complex of MSH2/MSH6/MLH1 predicted by protein-protein docking.
As the p.R243Q variant involves a substitution of a positively charged residue with a neutral amino acid, we proceeded by calculating the electrostatic potential of MSH2 in both WT and mutant forms. At the site of the variant, the electrostatic potential is majorly electronegative (Figure 5A). We noticed the occurrence of some neutral surface patches and a slightly electropositive region at the exact position of R243 in the wild type form. In the mutant form, however, the electronegativity increases. We verified that the electrostatic potential at the site of the variant is compatible with the interaction interface of MLH1 by calculating the electrostatic potential of the latter protein. Indeed, we found that the protein–protein interface is exclusively electropositive in MLH1 (Figure 5B).
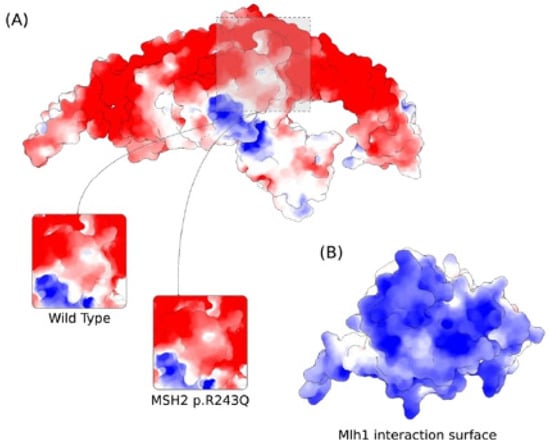
Figure 5.
Predicted effect of MSH2 p.R243Q variant on the electrostatic properties of MSH2 calculated by the Adaptive Poisson–Boltzmann Solver. Red and Blue colors correspond to potentials of electronegative and electropositive intensities, respectively. The maximum intensity values are −5 and +5 kb T e−1. (A) Electrostatic potential of MSH2 showing the variation at the site of the mutation for the wild type and p.R243Q forms. (B) Electrostatic potential of MLH1 at the interaction surface with MSH2.
3.7. Molecular Modeling of the FANCD2 Novel Variant
Among the other putative deleterious variants, we identified a novel p.A627S variant on FANCD2. FANCD2 forms a protein–protein semi-symmetrical complex with FANCI, which is involved in the physio-pathological mechanism of Fanconi anemia cancer. The substitution is located at the N-terminal end of an alpha helix of the FANCD2 NTD domain. The variant p.A627S is located near a monoubiquitination site on FANCD2 K561 residue (Figure S1). Ubiquitination can activate the Fanconi anemia DNA repair pathway [48]. The variant induces a significant destabilization of the folding energy calculated using DynaMut as well as a perturbation of the local flexibility of the structure [49]. The ΔΔG calculated by DynaMut shows an unfavorable energy of −1 kcal/mol. Moreover, four other tools including EnCoM, mCSM, SDM and DUET also show an unfavorable ΔΔG of −1.8, −2.9 and −2.1, respectively. Therefore, all the tools are in concordance about the destabilizing effect of the p.A627S variant.
4. Discussion
Currently, screening for serious diseases such as cancer represents one of the priorities of public health, and therefore of medical research. Indeed, thanks to the development of highly throughput technologies, particularly in the field of genomics, it became easier to elucidate the molecular mechanisms underlying the development and progression of hereditary cancers. In addition, such sequencing approaches contributed significantly to the identification of various genetic actionable variants allowing the development of molecular screening for early diagnosis of complex syndromic cancers, particularly in under-investigated populations such as the North African population.
GC is the second most common extracolonic tumor in LS (HNPCC) [50,51]. Although the risk of extracolonic tumors including gastric seems to be higher in MSH2 compared to MLH1 mutation carriers [52], the association between germline mutational profile and clinical phenotype is generally weak in LS [15]. Few studies have performed molecular comprehensive investigation on patients with gastric tumors even if they are LS mutation carriers or not.
In the current study, we investigated a DGC patient “JI-021” belonging to a Tunisian family from the north-east of Tunisia with suspicion of both HDGC and LSII (according to the tumor’s cluster family history) shedding light on the molecular basis of GC in this region known to have relatively high proportion of digestive cancer syndromes (no epidemiological data available). Based on the International Gastric Cancer Linkage Consortium (IGCLC), this case meets the third criterion for CDH1 germline screening. As a result, we found no pathogenic variant in the CDH1 gene coding and exons flanking regions [33]. In addition, no large rearrangements have been identified by MLPA in this gene for this patient [33]. This result is not surprising since the lack of CDH1 pathogenic variant DGC patients meeting the IGCLC testing criteria has already been reported. Indeed, Jakubowska and colleagues have sequenced the entire coding region of CDH1 gene in 86 Polish cancer patients from families fulfilling the criteria of HDGC and have found no deleterious mutations in the CDH1 gene [53].
They have concluded that CDH1 mutations do not contribute to DGC in Poland. They have, however, some limitations, namely less restrictive criteria of GC patients selection and lack of analysis for large genomic deletions. In fact, our study has covered the CDH1 entire coding region (16 exons), splice junctions as well as large rearrangements analysis. Nevertheless, we need to investigate the non-coding regions particularly regulatory transcriptional regions and promoter to confirm the definitive HDGC exclusion in this case since its corresponding FFPE tumor tissue harbored a loss of E-cadherin expression in tumor cells comparing to normal adjacent gland (Figure 2A) suggesting a putative CDH1 gene transcription defect at germinal or somatic level.
In addition to the IGCLC inclusion criteria, the index case “JI-021” also fulfilled Bethesda Guidelines inclusion criteria for LS testing (1st and 5th criteria). In the current study, our index case gDNA was screened via a custom panel targeting 87 DNA repair genes. Although several studies have used panels covering additional genes other than DNA repair as LS known associated genes or candidate genes, we aimed to focus on a panel covering the four DNA repair pathways in order to identify new candidate DNA repair genes/variants predisposing to such cancer syndrome. As a result, we have identified the MSH2 (c.728G>A p.R243Q) variant in the index case “JI-021”, but not in her asymptomatic relatives (with available DNAs). According to our best knowledge, this is the first study investigating the genetic predisposition of GC case in the context of LSII in Tunisia, and very few studies have reported such an investigation worldwide [14,15].
Indeed, Capelle and colleagues have reported 2014 mutations identified in the MMR genes as part of LS in 236 Dutch families, and among them GC was diagnosed in 32 subjects (1.6%), including 22 (69%) with family history of GC. The risk of developing GC was rated at 4.8% for carriers of the MLH1 gene mutation and at 9% for carriers of the MSH2 mutation. However, among 378 MSH6 identified mutations, no carrier had GC [14].
Nevertheless, with the emergence of precision oncology and development of germline panels, several studies have investigated LS susceptibility genes in individuals with cancers and individuals at risk to develop hereditary cancers.
It is well known that LS is histologically and genetically a complex disease with heterogeneous molecular background. Indeed, it seems that different families with the same LS inclusion criteria could have different mutational profiles in newly identified susceptibility genes not initially known to be associated with LS. These identified mutations/variants appear to have low frequencies among screened cases and families. This reported result is in concordance with our finding since our MSH2 c.728G>A identified variant has been found only in one case among the three screened GC cases belonging to LSII suspected families. While several other works have investigated the molecular background of CCR in the context of LS worldwide [54,55,56,57,58,59,60,61], only three studies have been conducted in Tunisia in this field. Indeed, Moussa and colleagues have reported germline mutations in MMR genes in 11/31 families suspected for LS (six MSH2 and five MLH1 mutations). Among the six MSH2 identified variants, our MSH2 c.728G>A variant has been found in only two probands (52 and 38 years old) belonging to two families sharing the fifth Bethesda guidelines selection criterion and the same mutational profile as well as a common newly identified MSH2 c.1413dupA mutation. These authors reported that MSH2 c.728G>A (p.R243Q) variant was predicted as probably non-pathogenic since the substituted Arginine residue was relatively little conserved across phylogeny with weak physicochemical difference between arginine and glutamine. However, this claim does not consider the complexity of domain–domain cooperatively, the protein–protein interactions and the fact that Arg residues are positively charged and Gln is neutral. They have found this variant in two families, the first one with five cases of CCR, the second with two cases of CCR. Both families shared the fifth Bethesda Guidelines criterion and two mutations “MSH2 c.728G>A” and “c.1413dupA”. Compared to our results, the proband “JI-021” carrying the same MSH2 c.728G>A (p.R243Q) variant shared the same age of GC onset (52) and the same Bethesda criteria (1st and 5th) with the two lastly reported Tunisian families. These homologies suggest that the MSH2 c.728G>A (p.R243Q) variant might be a LS/LSII predisposing variant in Tunisian population; however, supplementary molecular investigation among a larger cohort is needed to confirm this hypothesis. Intriguingly, Moussa and colleagues noted that the age of cancer onset as well as the tumors spectrum were similar in families with and without MMR germline mutations, which is in contradiction with other studies conducted in other populations [3]. More recently, next generation sequencing of MMR genes, POLE and POLD1 was performed, by Ben Sghaier and colleagues, to identify the genetic mechanisms underlying CCR in 24 Tunisian probands. As results, they identified, in six cases, five germline variants in MLH1, a somatic pathogenic variant in MSH2 (c.2557G>T) and a germline variant in MSH2 (c.1413 dupA), which was previously reported in the study of Ben Sghaier and colleagues [28]. In addition, a team in our lab have identified in a LS family with discordant twins, an MSH2 pathogenic mutation (c.1552C>T;p.Q518X) previously reported only in a Portuguese LS family and shared with all investigated family members suggesting that this variant is a causal mutation [29]. In addition, an Algerian recent study have screened MLH1, MSH2 and MSH6 genes among 21 families from East of Algeria and have identified the MSH2 (c.728G>A) mutation in two cases (44 and 38 years old) with CCR belonging to the same family with colon cancer family history [62]. The MSH2 (c.728G>A) variant seems to be identified only in Mediterranean countries. These findings support the fact that this variant might be an LSII rather than LSI predisposing variants in Tunisian population and appears to be a shared variant in the Mediterranean region. Indeed, previous work in our lab has reported that the Tunisian population shares founder variants with other North African and Middle Eastern populations for 43 inherited conditions [63]. Additionally, other variants appeared to be specific to the Tunisian population and shared by other populations as the case for 11-β hydroxylase deficiency as well as breast and ovarian hereditary cancer [64,65,66,67]. Such a phenomenon could certainly be explained by all migratory waves that occurred with the colonial period resulting in the occurrence of new mutations with an important impact on the genetic diversity of the Tunisian population. All these data highlight the importance of founder and shared mutations in decision making tools for diagnosis and prevention of diseases in North Africa, Middle East and migrant populations living in Europe or Mediterranean region [63].
Moreover, our index case harboring the MSH2 variant (c.728G>A) has an aggressive phenotype since DG tumors are known to be an aggressive gastric tumor histotype. This patient had peritoneal carcinosis, and she was therefore not operable and the only treatment she had was a palliative chemotherapy. The MSH2 and MLH1 immunohistochemistry analysis in index case gastric tumor tissue has shown a positive nuclear immunostaining in tumor cells for MLH1 and an incomplete nuclear immunostaining for MSH2 protein (Figure 2B,C). This result is in good agreement with in silico prediction analyses of MSH2 variant since the predicted effect involves the MSH2/MLH1 complex destabilization, which has no effect on the protein tissue expression and detection. According to our best knowledge, this study is one of the rare investigations of the MMR proteins expression profile in DGC tissue in a LSII suspected family. The MMR protein expression analysis by IHC is usually performed in CRC tissues according to international recommendation for LS diagnosis in routine. However, few studies have investigated MMR status in tumor tissues corresponding to other LS-associated cancers such as GC. Indeed, it has been reported that MMR deficiency seems to be much more frequent in CRC tumors compared to gastric tumors belonging to LS families [68]. These authors have reported that among 45 LS confirmed families 31 cases had GC and only four gastric cases were MMR deficient versus 27 MMR proficient cases.
Shedding light on the molecular pathogenesis of GC in LS settings, gastric tumors have been included in the modified Amsterdam criteria II, however the clinical consensus criteria for gastric tumors diagnosis in LS suspected cases/families still remain unclear and germline mutations search as well as MSI status are not yet clearly indicated for gastric tumors. Actually there is growing evidence that such tumors from MMR gene germline mutation carriers are part of LS tumor spectrum and relative risk of GC in LS mutation carriers is reported to be higher by 4–19 compared to the general population in western countries [50,51,69] at least by 2-fold in endemic areas in Asia [16] as our investigated region. In fact, taking into account the gene–environment interactions, our investigated family came from an endemic region of Tunisia with relatively high incidence of digestive cancers known with significant exposure of residents to pesticides and characterized by the spread of water with high levels of toxic chemicals; however, no epidemiological or environmental studies have been carried out in this region. These findings highlight the crucial importance of early screening of individuals at high risk of developing such tumors, particularly for these under-investigated in Tunisia.
In the context of LS, this variant is classified in class 3 (Variant with uncertain significance ‘VUS’) by the LOVD database according to the InSiGHT classifications (http://insight-database.org/ accessed on 20 July 2022) and no functional studies on its effect on MSH2 protein structure, function or its interactions with other partners are available. Hence, we performed several in silico prediction analyses using online prediction tools and databases (15 prediction tools), as well as structural modeling of MSH2 known complexes.
The variant p.R243Q as part of the interface between MSH2 and MLH1 appears to be involved in the destabilization of the protein–protein interaction. In their work, Groothuizen et al. [70] suggest the proximity of the connector domain to the MutL binding surface with MutS. Nevertheless, the structure of MutS deviates significantly from that of MSH2 to be able to infer any homology-based conclusions about the contribution of the residue at the 243rd position, hence the utility of our protein–protein predicted complex.
The change in the electrostatic interaction surface of MSH2 upon variance might result in different association equilibrium with MLH1. The DNA repair mechanism is a multistep process that includes cooperation between different proteins. Electrostatic forces that act at distances of 5–10 Å can be affected by the p.R243Q variant [71]. More recently, it has been shown that the electrostatic potential disruption between MLH1 and MSH2 can lead to the accumulation of DNA errors and an impact on the protein–protein complex [72]. The electrostatic properties of MLH1 seem to be highly regulated via acetylation/deacetylation by the Histone deacetylase 6 (HDAC6). Moreover, the results from [72] also imply that a dynamic of association/dissociation between MutLα, MutSα and HDAC6 is required for DNA repair. In this regard, electrostatic forces, disrupted by the p.R243Q, would play an important role and could result in a significant effect.
Beyond the MSH2 variant, another putative probably deleterious variant in the FANCD2 gene (p.A627S) was identified. FANCD2 is required for maintenance of chromosomal stability. It also promotes accurate and efficient pairing of homologs during meiosis. It is involved in the repair of DNA double-strand breaks, both by homologous recombination and single-strand annealing. It may participate in S phase and G2 phase checkpoint activation upon DNA damage playing a role in preventing breakage. This gene encodes the protein for complementation group D2.
The A627 amino acid is located at the opposite helices facing the protein–protein interface with FANCI that also contains an ubiquitination site at residue K561. FANCD2 ubiquitination stabilizes a conformational change of the protein–protein complex [48] that is required to increase the affinity for the double stranded DNA [49]. The perturbation of the flexibility by p.A627S variant of FANCD2 may result in two effects. First, the formation of the complex FANCD2 and the ubiquitin-conjugating enzyme E2-UBE2T would have to overcome the energy barrier induced by p.A627S local destabilization since the variation occurs at a close distance to K561. The second effect may result in the impairment of the association between FANCD2 and FANCI induced by destabilization of the local interface near the variant’s site. Moreover, since monoubiquitination occurs on the evolutionarily conserved lysine residues, variants affecting FANCD2 at K561 residue seem to result in major molecular defects in response to DNA damage agents in the Fanconi Anemia cells [73,74].
Our index case comes from a consanguineous marriage with accumulation of two potentially deleterious variants besides potential deficiency in the cell adhesion system, suggesting a putative increasing risk of several cancers in this family. Unfortunately, we were not able to screen the index case’s brother with CCR or her two affected sisters (breast and ovarian cancers) for this variant since they were dead. Susceptibility to cancer in offspring of consanguineous marriages has been largely studied. For countries such as Tunisia with common consanguineous marriage, the association between consanguinity and mortality due to cancers is highly important for public health programs [75,76,77]. Moreover, it has been reported that consanguinity and inbreeding plausibly led to the accumulation of population-specific founder pathogenic/or likely pathogenic sequence variants (PSVs) [78]. In fact, two reported studies have observed that inbreeding and ROH result in an increased risk of CCR [79] and developing leukemia, lymphoma, colorectal and prostate cancer [80]. Supplementary genetic screenings of this variant in larger cohorts in this region as in Tunisia are needed to confirm these hypotheses.
5. Conclusions
Our findings highlight the genetic heterogeneity of cancer predisposition in Tunisia and the importance of the use of target NGS to identify clinically actionable genetic variants for disease management. To our best knowledge, no study has investigated the molecular basis of GC in LS context in either Tunisia or North Africa. Our findings suggest that the MSH2 c.728G>A (p.R243Q) rare variant could represent a candidate marker for LS II screening. We suggested this variant be shared with Mediterranean countries. Nevertheless, genetic screening of a larger cohort of GC is needed to confirm its association with LSII syndrome in Tunisia. Our structural model prediction showed that this variant is likely pathogenic and is involved in the MSH2-MLH1 complex stability suggesting the revision of its status from VUS to likely pathogenic. The novel FANCD2 variant appears to have putative functional effects as well. The identification of such likely deleterious variants will help in the setting up of specific clinical surveillance protocol for individuals at high risk of developing this disease in terms of yearly endoscopy and colonoscopy as well as a mammography biannually for women at risk. We acknowledge however the importance of functional validation for our results of both identified variants (the VUS and the novel FANCD2 variant) which would be the next focus of our study.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13081355/s1, Figure S1: FANCD2 p.A627S variant within the ubiquitinated complex FANCD2/FANCI; Table S1: List of primers and settings for sanger sequencing of MSH2, CDH1 and CTNNA1 genes; Table S2: List of identified genes after applying filtering strategy.
Author Contributions
M.K. and J.B.A.-H. led the study and the writing of the paper; M.K., H.O., J.B.A.-H., S.E. and M.S.B. contributed to the experimental design and conception of the study; Sample and clinical data acquisition were performed by M.M. (Mouna Medhioub), M.M. (Moufida Mahmoudi), A.K., H.B., H.T.K., L.H., E.C. and M.M.A.; M.K., J.B.A.-H. and H.O. contributed to data analysis and result interpretation; M.K., J.B.A.-H. and H.O. provided the bioinformatics study analysis; M.K., J.B.A.-H., S.L., A.J.-G., H.T. and A.M. provided the technical support for the experimental study; Involvement in the drafting of the manuscript: J.B.A.-H. and H.O.; S.A. and M.S.B. provided the critical analysis of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Tunisian Ministry of Public Health, the Tunisian Ministry of Higher Education and Scientific Research (LR16IPT05) and MOBIDOC device funded by European Union in the frame EMORI program (EMORI-Horizon2020TN) and administered by the National Agency of scientific research support (ANPR).
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki Principles and was obtained from the biomedical ethics committee of Institut Pasteur de Tunis (IRB) (2017/6F/I/Gastric Cancer on 22 May 2018).
Informed Consent Statement
Informed consents were obtained from subjects involved in the current study.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its additional files.
Conflicts of Interest
The authors declare that they have no competing interest. All authors have approved the submission of this article in its current version.
Abbreviations
| HDGC | Hereditary Diffuse Gastric Carcinoma |
| MLPA | Multiplex Ligation Probe Amplification |
| GC/DGC | Gastric Carcinoma/Diffuse Gastric Carcinoma |
| LS | Lynch Syndrome |
| BC/LBC | Breast Cancer/Lobular Breast Cancer |
| IGCLC | International Gastric Cancer Linkage Consortium |
| gDNA | Genomic DNA |
| sDNA | Somatic DNA |
| HP | Helicobacter Pylori |
| PDB | Protein Data Bank |
| FFPE | Formalin Fixed Paraffin Embedded |
| CCR | Colorectal Carcinoma |
| OC | Ovarian Carcinoma |
| B/LB | Benign/Likely Benign |
| VUS | Variant of Uncertain Significance |
| NR | Not Reported |
| NI | Non Indicated |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Gastroenterol. Rev./Przegląd Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Moussa, S.A.-B.; Moussa, A.; Kourda, N.; Mezlini, A.; Abdelli, N.; Zerimech, F.; Najjar, T.; Jilani, S.B.; Porchet, N.; Ayed, F.B. Lynch syndrome in Tunisia: First description of clinical features and germline mutations. Int. J. Colorectal Dis. 2011, 26, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Setia, N.; Clark, J.W.; Duda, D.G.; Hong, T.S.; Kwak, E.L.; Mullen, J.T.; Lauwers, G.Y. Familial Gastric Cancers. Oncologist 2015, 20, 1365–1377. [Google Scholar] [CrossRef] [PubMed]
- Vecchia, C.L.; Negri, E.; Gentile, A.; Franceschi, S. Family history and the risk of stomach and colorectal cancer. Cancer 1992, 70, 50–55. [Google Scholar] [CrossRef]
- Fitzgerald, R.C.; Hardwick, R.; Huntsman, D.; Carneiro, F.; Guilford, P.; Blair, V.; Chung, D.C.; Norton, J.; Ragunath, K.; Van Krieken, J.H. International Gastric Cancer Linkage Consortium: Hereditary diffuse gastric cancer: Updated consensus guidelines for clinical management and directions for future research. J. Med. Genet. 2010, 47, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Peltomäki, P. Update on Lynch syndrome genomics. Fam. Cancer 2016, 15, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Rahner, N.; Steinke, V.; Schlegelberger, B.; Olschwang, S.; Eisinger, F.; Hutter, P. Clinical utility gene card for: Lynch syndrome (MLH1, MSH2, MSH6, PMS2). Eur. J. Hum. Genet. 2010, 18, 1071. [Google Scholar] [CrossRef] [PubMed]
- Lorans, M.; Dow, E.; Macrae, F.A.; Winship, I.M.; Buchanan, D.D. Update on Hereditary Colorectal Cancer: Improving the Clinical Utility of Multigene Panel Testing. Clin. Colorectal Cancer 2018, 17, e293–e305. [Google Scholar] [CrossRef]
- Lynch, H.T.; Smyrk, T.C.; Lynch, P.M.; Lanspa, S.J.; Boman, B.M.; Ens, J.; Lynch, J.F.; Strayhorn, P.; Carmody, T.; Cristofaro, G. Adenocarcinoma of the small bowel in Lynch syndrome II. Cancer 1989, 64, 2178–2183. [Google Scholar] [CrossRef]
- Dillon, J.L.; Gonzalez, J.L.; DeMars, L.; Bloch, K.J.; Tafe, L.J. Universal screening for Lynch syndrome in endometrial cancers: Frequency of germline mutations and identification of patients with Lynch-like syndrome. Hum. Pathol. 2017, 70, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Hampel, H.; Frankel, W.; Panescu, J.; Lockman, J.; Sotamaa, K.; Fix, D.; Comeras, I.; La Jeunesse, J.; Nakagawa, H.; Westman, J.A. Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res. 2006, 66, 7810–7817. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, E.; Mukherjee, B.; Raymond, V.M.; Tayob, N.; Kastrinos, F.; Sparr, J.; Wang, F.; Bandipalliam, P.; Syngal, S.; Gruber, S.B. Calculation of risk of colorectal and endometrial cancer among patients with Lynch syndrome. Gastroenterology 2009, 137, 1621–1627. [Google Scholar] [CrossRef] [PubMed]
- Capelle, L.G.; Grieken, N.C.T.V.; Lingsma, H.F.; Steyerberg, E.W.; Klokman, W.J.; Bruno, M.J.; Vasen, H.F.A.; Kuipers, E.J. Risk and Epidemiological Time Trends of Gastric Cancer in Lynch Syndrome Carriers in The Netherlands. Gastroenterology 2010, 138, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Gylling, A.; Abdel-Rahman, W.M.; Juhola, M.; Nuorva, K.; Hautala, E.; Järvinen, H.J.; Mecklin, J.-P.; Aarnio, M.; Peltomäki, P. Is gastric cancer part of the tumour spectrum of hereditary non-polyposis colorectal cancer? A molecular genetic study. Gut 2007, 56, 926–933. [Google Scholar] [CrossRef]
- Park, Y.J.; Shin, K.H.; Park, J.G. Risk of gastric cancer in hereditary nonpolyposis colorectal cancer in Korea. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 2994–2998. [Google Scholar]
- Sheehan, M.; Heald, B.; Yanda, C.; Kelly, E.D.; Grobmyer, S.; Eng, C.; Kalady, M.; Pederson, H. Investigating the Link between Lynch Syndrome and Breast Cancer. Eur. J. Breast Health 2020, 16, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Gylling, A.H.S.; Nieminen, T.T.; Abdel-Rahman, W.M.; Nuorva, K.; Juhola, M.; Joensuu, E.I.; Järvinen, H.J.; Mecklin, J.-P.; Aarnio, M.; Peltomäki, P.T. Differential cancer predisposition in Lynch syndrome: Insights from molecular analysis of brain and urinary tract tumors. Carcinogenesis 2008, 29, 1351–1359. [Google Scholar] [CrossRef]
- Rouprêt, M.; Yates, D.R.; Comperat, E.; Cussenot, O. Upper urinary tract urothelial cell carcinomas and other urological malignancies involved in the hereditary nonpolyposis colorectal cancer (lynch syndrome) tumor spectrum. Eur. Urol. 2008, 54, 1226–1236. [Google Scholar] [CrossRef]
- Lu, K.H.; Daniels, M. Endometrial and ovarian cancer in women with Lynch syndrome: Update in screening and prevention. Fam. Cancer 2013, 12, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Randall, L.M.; Pothuri, B. The genetic prediction of risk for gynecologic cancers. Gynecol. Oncol. 2016, 141, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Schmeler, K.M.; Lynch, H.T.; Chen, L.; Munsell, M.F.; Soliman, P.T.; Clark, M.B.; Daniels, M.S.; White, K.G.; Boyd-Rogers, S.G.; Conrad, P.G. Prophylactic surgery to reduce the risk of gynecologic cancers in the Lynch syndrome. N. Engl. J. Med. 2006, 354, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Lynch, H.T.; Lanspa, S.J.; Boman, B.M.; Smyrk, T.; Watson, P.; Lynch, J.F.; Lynch, P.M.; Cristofaro, G.; Bufo, P.; Tauro, A.V. Hereditary nonpolyposis colorectal cancer–Lynch syndromes I and II. Gastroenterol. Clin. N. Am. 1988, 17, 679–712. [Google Scholar] [CrossRef]
- Lynch, H.T.; Lynch, P.M.; Lanspa, S.J.; Snyder, C.L.; Lynch, J.F.; Boland, C.R. Review of the Lynch syndrome: History, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin. Genet. 2009, 76, 1–18. [Google Scholar] [CrossRef]
- Watson, P.; Vasen, H.F.; Mecklin, J.-P.; Bernstein, I.; Aarnio, M.; Järvinen, H.J.; Myrhøj, T.; Sunde, L.; Wijnen, J.T.; Lynch, H.T. The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int. J. Cancer 2008, 123, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Kanth, P.; Grimmett, J.; Champine, M.; Burt, R.; Samadder, N.J. Hereditary Colorectal Polyposis and Cancer Syndromes: A Primer on Diagnosis and Management. Am. J. Gastroenterol. 2017, 112, 1509–1525. [Google Scholar] [CrossRef] [PubMed]
- Møller, P.; Seppälä, T.T.; Bernstein, I.; Holinski-Feder, E.; Sala, P.; Gareth Evans, D.; Lindblom, A.; Macrae, F.; Blanco, I.; Sijmons, R.H.; et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: A report from the Prospective Lynch Syndrome Database. Gut 2018, 67, 1306–1316. [Google Scholar] [CrossRef]
- Ben Sghaier, R.; Jansen, A.M.L.; Bdioui, A.; Van Wezel, T.; Ksiaa, M.; Elgolli, L.; Ben Fatma, L.; Ben Ahmed, S.; Azzouz, M.M.; Hellara, O.; et al. Targeted next generation sequencing screening of Lynch syndrome in Tunisian population. Fam. Cancer 2019, 18, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Jaballah-Gabteni, A.; Tounsi, H.; Kabbage, M.; Hamdi, Y.; Elouej, S.; Ben Ayed, I.; Medhioub, M.; Mahmoudi, M.; Dallali, H.; Yaiche, H.; et al. Identification of novel pathogenic MSH2 mutation and new DNA repair genes variants: Investigation of a Tunisian Lynch syndrome family with discordant twins. J. Transl. Med. 2019, 17, 212. [Google Scholar] [CrossRef] [PubMed]
- Hansford, S.; Kaurah, P.; Li-Chang, H.; Woo, M.; Senz, J.; Pinheiro, H.; Schrader, K.A.; Schaeffer, D.F.; Shumansky, K.; Zogopoulos, G.; et al. Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol. 2015, 1, 23. [Google Scholar] [CrossRef] [PubMed]
- Blair, V.R.; McLeod, M.; Carneiro, F.; Coit, D.G.; D’Addario, J.L.; van Dieren, J.M.; Harris, K.L.; Hoogerbrugge, N.; Oliveira, C.; van der Post, R.S.; et al. Hereditary diffuse gastric cancer: Updated clinical practice guidelines. Lancet Oncol. 2020, 21, e386–e397. [Google Scholar] [CrossRef]
- Singh, A.; Pandey, G.K. Primer Design Using Primer Express® for SYBR Green-Based Quantitative PCR. In PCR Primer Design; Basu, C., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; pp. 153–164. ISBN 978-1-4939-2365-6. [Google Scholar]
- Ben Aissa-Haj, J.; Kabbage, M.; Othmen, H.; Saulnier, P.; Kettiti, H.T.; Jaballah-Gabteni, A.; Ferah, A.; Medhioub, M.; Khsiba, A.; Mahmoudi, M.; et al. CDH1 Germline Variants in a Tunisian Cohort with Hereditary Diffuse Gastric Carcinoma. Genes 2022, 13, 400. [Google Scholar] [CrossRef] [PubMed]
- Chikhaoui, A.; Elouej, S.; Nabouli, I.; Jones, M.; Lagarde, A.; Ben Rekaya, M.; Messaoud, O.; Hamdi, Y.; Zghal, M.; Delague, V.; et al. Identification of a ERCC5 c.2333T>C (L778P) Variant in Two Tunisian Siblings With Mild Xeroderma Pigmentosum Phenotype. Front. Genet. 2019, 10, 111. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.J.; Pohlhaus, T.J.; Changela, A.; Iyer, R.R.; Modrich, P.L.; Beese, L.S. Structure of the Human MutSα DNA Lesion Recognition Complex. Mol. Cell 2007, 26, 579–592. [Google Scholar] [CrossRef]
- Gupta, S.; Gellert, M.; Yang, W. Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat. Struct. Mol. Biol. 2012, 19, 72–78. [Google Scholar] [CrossRef]
- Šali, A.; Blundell, T.L. Comparative Protein Modelling by Satisfaction of Spatial Restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, H.; Lam, R.; Tempel, W.; Kerr, I.D.; Min, J. Structure of the human MLH1 N-terminus: Implications for predisposition to Lynch syndrome. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 981–985. [Google Scholar] [CrossRef]
- Delgado, J.; Radusky, L.G.; Cianferoni, D.; Serrano, L. FoldX 5.0: Working with RNA, small molecules and a new graphical interface. Bioinformatics 2019, 35, 4168–4169. [Google Scholar] [CrossRef]
- Shahnazari, P.; Sayehmiri, K.; Minuchehr, Z.; Parhizkar, A.; Poustchi, H.; Montazeri, G.; Mohamadkhani, A. The Increased Level of Serum p53 in Hepatitis B-Associated Liver Cirrhosis. Iran. J. Med. Sci. 2014, 39, 446–451. [Google Scholar] [PubMed]
- Case, D.A.; Ben-Shalom, I.H.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Kovalenko, D.; Ghoreishi, G.; et al. AMBER 2019; University of California: San Francisco, CA, USA, 2019. [Google Scholar]
- Vascon, F.; Gasparotto, M.; Giacomello, M.; Cendron, L.; Bergantino, E.; Filippini, F.; Righetto, I. Protein electrostatics: From computational and structural analysis to discovery of functional fingerprints and biotechnological design. Comput. Struct. Biotechnol. J. 2020, 18, 1774–1789. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids. Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef]
- Rodriguez, N.J.; Reineke, P.; LaDuca, H.; Xicola, R.M.; Llor, X. 1066-Phenotypic Expression of Cdh1 Germline Mutations Unselected for Hereditary Diffuse Gastric Cancer. Gastroenterology 2018, 154, S-204. [Google Scholar] [CrossRef]
- Schouten, J.P.; McElgunn, C.J.; Waaijer, R.; Zwijnenburg, D.; Diepvens, F.; Pals, G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids. Res. 2002, 30, e57. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef] [PubMed]
- Goellner, E.M.; Putnam, C.D.; Graham, W.J.; Rahal, C.M.; Li, B.-Z.; Kolodner, R.D. Identification of Exo1-Msh2 interaction motifs in DNA mismatch repair and new Msh2-binding partners. Nat. Struct. Mol. Biol. 2018, 25, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Murphy, V.J.; Charron, A.; van Twest, S.; Sharp, M.; Constantinou, A.; Parker, M.W.; Crismani, W.; Bythell-Douglas, R.; Deans, A.J. Preparation and purification of mono-ubiquitinated proteins using Avi-tagged ubiquitin. PLoS ONE 2020, 15, e0229000. [Google Scholar] [CrossRef]
- Wang, R.; Wang, S.; Dhar, A.; Peralta, C.; Pavletich, N.P. DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex. Nature 2020, 580, 278–282. [Google Scholar] [CrossRef] [PubMed]
- Aarnio, M.; Sankila, R.; Pukkala, E.; Salovaara, R.; Aaltonen, L.A.; de la Chapelle, A.; Peltomäki, P.; Mecklin, J.P.; Järvinen, H.J. Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int. J. Cancer 1999, 81, 214–218. [Google Scholar] [CrossRef]
- Goecke, T.; Schulmann, K.; Engel, C.; Holinski-Feder, E.; Pagenstecher, C.; Schackert, H.K.; Kloor, M.; Kunstmann, E.; Vogelsang, H.; Keller, G.; et al. Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: A report by the German HNPCC Consortium. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2006, 24, 4285–4292. [Google Scholar] [CrossRef] [PubMed]
- Vasen, H.F.; Stormorken, A.; Menko, F.H.; Nagengast, F.M.; Kleibeuker, J.H.; Griffioen, G.; Taal, B.G.; Moller, P.; Wijnen, J.T. MSH2 mutation carriers are at higher risk of cancer than MLH1 mutation carriers: A study of hereditary nonpolyposis colorectal cancer families. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2001, 19, 4074–4080. [Google Scholar] [CrossRef] [PubMed]
- Jakubowska, A.; Ławniczak, M.; Wojnarska, B.; Cybulski, C.; Huzarski, T.; Byrski, T.; Tołoczko-Grabarek, A.; Jaworska, K.; Durda, K.; Starzyńska, T.; et al. CDH1 gene mutations do not contribute in hereditary diffuse gastric cancer in Poland. Fam. Cancer 2010, 9, 605–608. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wang, W.; Lee, S.; Nafa, K.; Lee, J.; Romans, K.; Watson, P.; Gruber, S.B.; Euhus, D.; Kinzler, K.W. Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 2006, 296, 1479–1487. [Google Scholar] [CrossRef] [PubMed]
- Drost, M.; Lützen, A.; Van Hees, S.; Ferreira, D.; Calléja, F.; Zonneveld, J.B.; Nielsen, F.C.; Rasmussen, L.J.; de Wind, N. Genetic screens to identify pathogenic gene variants in the common cancer predisposition Lynch syndrome. Proc. Natl. Acad. Sci. USA 2013, 110, 9403–9408. [Google Scholar] [CrossRef] [PubMed]
- Pande, M.; Wei, C.; Chen, J.; Amos, C.I.; Lynch, P.M.; Lu, K.H.; Lucio, L.A.; Boyd-Rogers, S.G.; Bannon, S.A.; Mork, M.E. Cancer spectrum in DNA mismatch repair gene mutation carriers: Results from a hospital based Lynch syndrome registry. Fam. Cancer 2012, 11, 441–447. [Google Scholar] [CrossRef]
- Pearlman, R.; Frankel, W.L.; Swanson, B.; Zhao, W.; Yilmaz, A.; Miller, K.; Bacher, J.; Bigley, C.; Nelsen, L.; Goodfellow, P.J.; et al. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients With Early-Onset Colorectal Cancer. JAMA Oncol. 2017, 3, 464–471. [Google Scholar] [CrossRef]
- Pino, M.S.; Chung, D.C. Application of molecular diagnostics for the detection of Lynch syndrome. Expert Rev. Mol. Diagn. 2010, 10, 651–665. [Google Scholar] [CrossRef][Green Version]
- Tamura, G. Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J. Gastroenterol. 2006, 12, 192. [Google Scholar] [CrossRef]
- Watson, P.; Lynch, H.T. Cancer risk in mismatch repair gene mutation carriers. Fam. Cancer 2001, 1, 57–60. [Google Scholar] [CrossRef] [PubMed]
- Yurgelun, M.B.; Kulke, M.H.; Fuchs, C.S.; Allen, B.A.; Uno, H.; Hornick, J.L.; Ukaegbu, C.I.; Brais, L.K.; McNamara, P.G.; Mayer, R.J.; et al. Cancer Susceptibility Gene Mutations in Individuals With Colorectal Cancer. J. Clin. Oncol. 2017, 35, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Ziada-Bouchaar, H.; Sifi, K.; Filali, T.; Hammada, T.; Satta, D.; Abadi, N. First description of mutational analysis of MLH1, MSH2 and MSH6 in Algerian families with suspected Lynch syndrome. Fam. Cancer 2017, 16, 57–66. [Google Scholar] [CrossRef]
- Romdhane, L.; Kefi, R.; Azaiez, H.; Halim, N.B.; Dellagi, K.; Abdelhak, S. Founder mutations in Tunisia: Implications for diagnosis in North Africa and Middle East. Orphanet J. Rare Dis. 2012, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Cherbal, F.; Bakour, R.; Adane, S.; Boualga, K.; Benais-Pont, G.; Maillet, P. BRCA1 and BRCA2 germline mutations screening in Algerian breast/ovarian cancer families. Dis. Markers 2010, 28, 377–384. [Google Scholar] [CrossRef]
- Kharrat, M.; Trabelsi, S.; Chaabouni, M.; Maazoul, F.; Kraoua, L.; Jemaa, L.B.; Gandoura, N.; Barsaoui, S.; Morel, Y.; M’rad, R.; et al. Only two mutations detected in 15 Tunisian patients with 11β-hydroxylase deficiency: The p.Q356X and the novel p.G379V. Clin. Genet. 2010, 78, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Troudi, W.; Uhrhammer, N.; Romdhane, K.B.; Sibille, C.; Amor, M.B.; El Khil, H.K.; Jalabert, T.; Mahfoudh, W.; Chouchane, L.; Ayed, F.B. Complete mutation screening and haplotype characterization of BRCA1 gene in Tunisian patients with familial breast cancer. Cancer Biomark. 2008, 4, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-S.; Cordero, J.J.; Can, S.; Cai, L.-Q.; You, X.; Herrera, C.; DeFillo-Ricart, M.; Shackleton, C.; Imperato-McGinley, J. Mutations in CYP11B1 gene: Phenotype–genotype correlations. Am. J. Med. Genet. A 2003, 122, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Zeinalian, M.; Hashemzadeh-Chaleshtori, M.; Salehi, R.; Kazemi, M.; Emami, M.H. Tumor microsatellite instability and clinicopathologic features in Iranian colorectal cancer patients at risk for Lynch syndrome. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2015, 20, 154–160. [Google Scholar]
- Vasen, H.F.; Wijnen, J.T.; Menko, F.H.; Kleibeuker, J.H.; Taal, B.G.; Griffioen, G.; Nagengast, F.M.; Meijers-Heijboer, E.H.; Bertario, L.; Varesco, L.; et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology 1996, 110, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Groothuizen, F.S.; Winkler, I.; Cristóvão, M.; Fish, A.; Winterwerp, H.H.K.; Reumer, A.; Marx, A.D.; Hermans, N.; Nicholls, R.A.; Murshudov, G.N.; et al. MutS/MutL Crystal Structure Reveals that the MutS Sliding Clamp Loads MutL onto DNA. Available online: https://elifesciences.org/articles/06744 (accessed on 29 September 2020).
- Zhou, H.-X.; Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, C.; Moses, N.; Haakenson, J.; Xiang, S.; Quan, D.; Fang, B.; Yang, Z.; Bai, W.; Bepler, G. HDAC6 regulates DNA damage response via deacetylating MLH1. J. Biol. Chem. 2019, 294, 5813–5826. [Google Scholar] [CrossRef]
- Romick-Rosendale, L.E.; Lui, V.W.Y.; Grandis, J.R.; Wells, S.I. The Fanconi anemia pathway: Repairing the link between DNA damage and squamous cell carcinoma. Mutat. Res. Mol. Mech. Mutagen. 2013, 743–744, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Nepal, M.; Che, R.; Ma, C.; Zhang, J.; Fei, P. FANCD2 and DNA Damage. Int. J. Mol. Sci. 2017, 18, 1804. [Google Scholar] [CrossRef] [PubMed]
- El-Kheshen, G.; Saadat, M. Prevalence of consanguineous marriages among Shi’a populations of Lebanon. J. Biosoc. Sci. 2013, 45, 675–682. [Google Scholar] [CrossRef] [PubMed]
- Rafiee, L.; Saadat, M. Prevalence of consanguineous marriages among Iranian Georgians. J. Biosoc. Sci. 2011, 43, 47. [Google Scholar] [CrossRef] [PubMed]
- Saadat, M.; Vakili-Ghartavol, R. Parental consanguinity and susceptibility to drug abuse among offspring, a case–control study. Psychiatry Res. 2010, 180, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Laitman, Y.; Friebel, T.M.; Yannoukakos, D.; Fostira, F.; Konstantopoulou, I.; Figlioli, G.; Bonanni, B.; Manoukian, S.; Zuradelli, M.; Tondini, C.; et al. The spectrum of BRCA1 and BRCA2 pathogenic sequence variants in Middle Eastern, North African, and South European countries. Hum. Mutat. 2019, 40, e1–e23. [Google Scholar] [CrossRef] [PubMed]
- Rich, J.N. Cancer stem cells in radiation resistance. Cancer Res. 2007, 67, 8980–8984. [Google Scholar] [CrossRef] [PubMed]
- Bener, A.; El Ayoubi, H.R.; Chouchane, L.; Ali, A.I.; Al-Kubaisi, A.; Al-Sulaiti, H.; Teebi, A.S. Impact of consanguinity on cancer in a highly endogamous population. Asian Pac. J. Cancer Prev. 2009, 10, 35–40. [Google Scholar] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).