Comparative Genomic Analysis of Antarctic Pseudomonas Isolates with 2,4,6-Trinitrotoluene Transformation Capabilities Reveals Their Unique Features for Xenobiotics Degradation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Species and Culture Conditions
2.2. Public Data Acquisition
2.3. Genome Sequencing and Assembly
2.4. Genome Annotation and Metabolic Potential Analysis
2.5. Pangenome Analysis
2.6. Phylogenetic and Phylogenomic Analyses
2.7. Prediction of Mobile Genetic Elements and Other Specific Genomic Features
2.8. Identification and Analysis of Putative TNT Metabolism-Related Enzymes
2.9. Reconstruction of TNT Metabolic Pathways
3. Results and Discussion
3.1. General Genomic Features of TNT Isolates
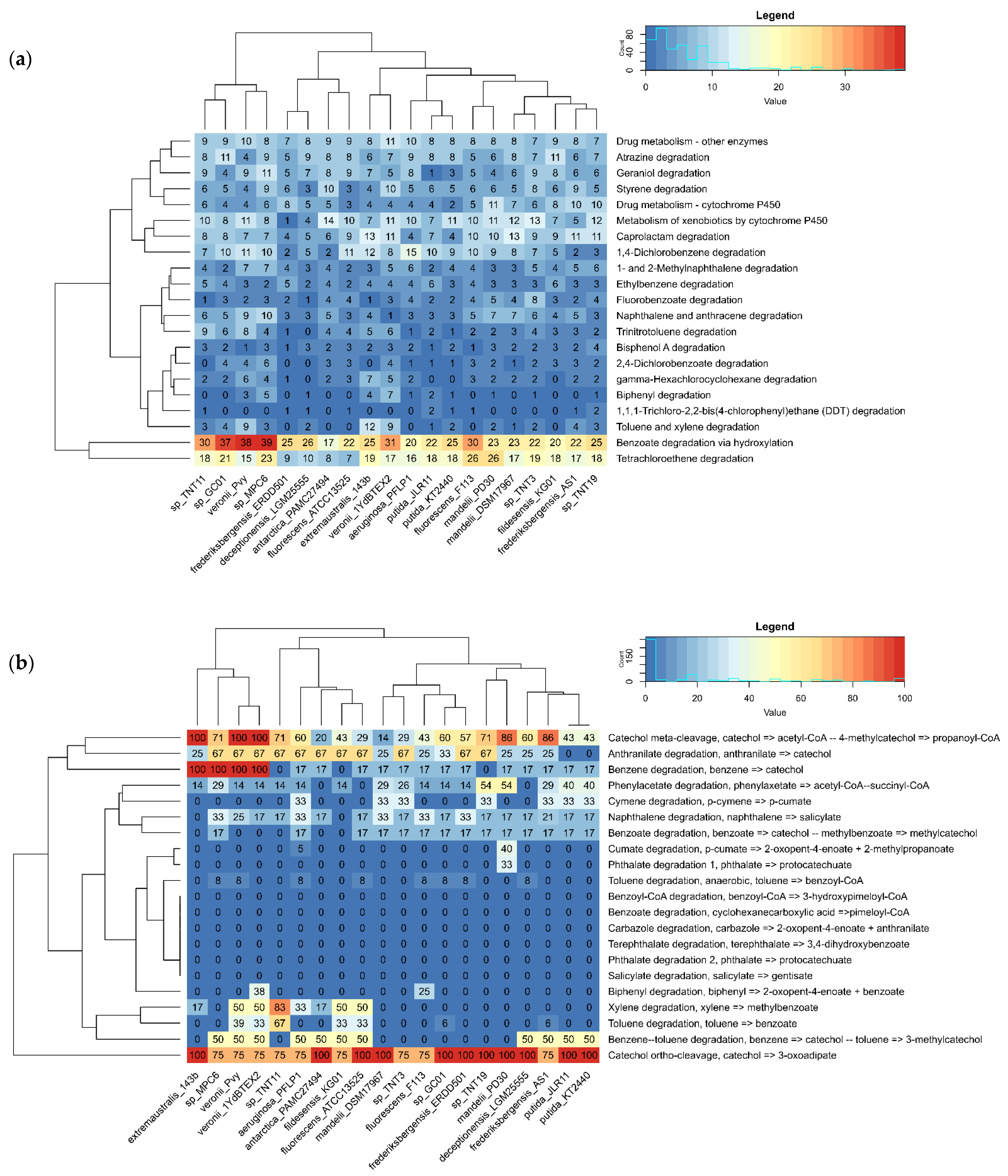
3.2. Annotation and Metabolic Potential Analysis
3.3. Pangenome Analysis and Unique Genes
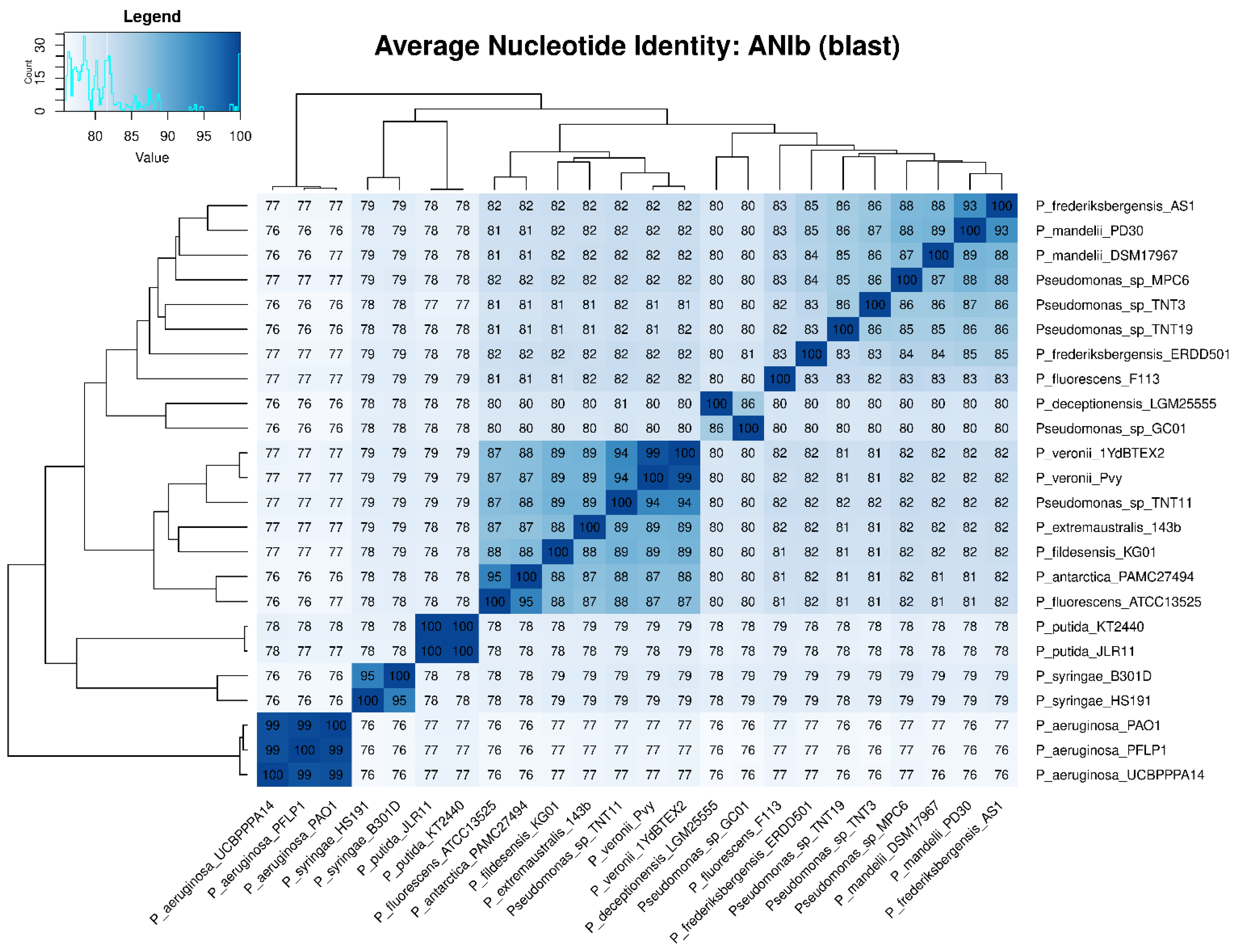
3.4. Phylogenetic and Phylogenomic Analyses
3.5. Mobile Genetic Elements (MGEs)
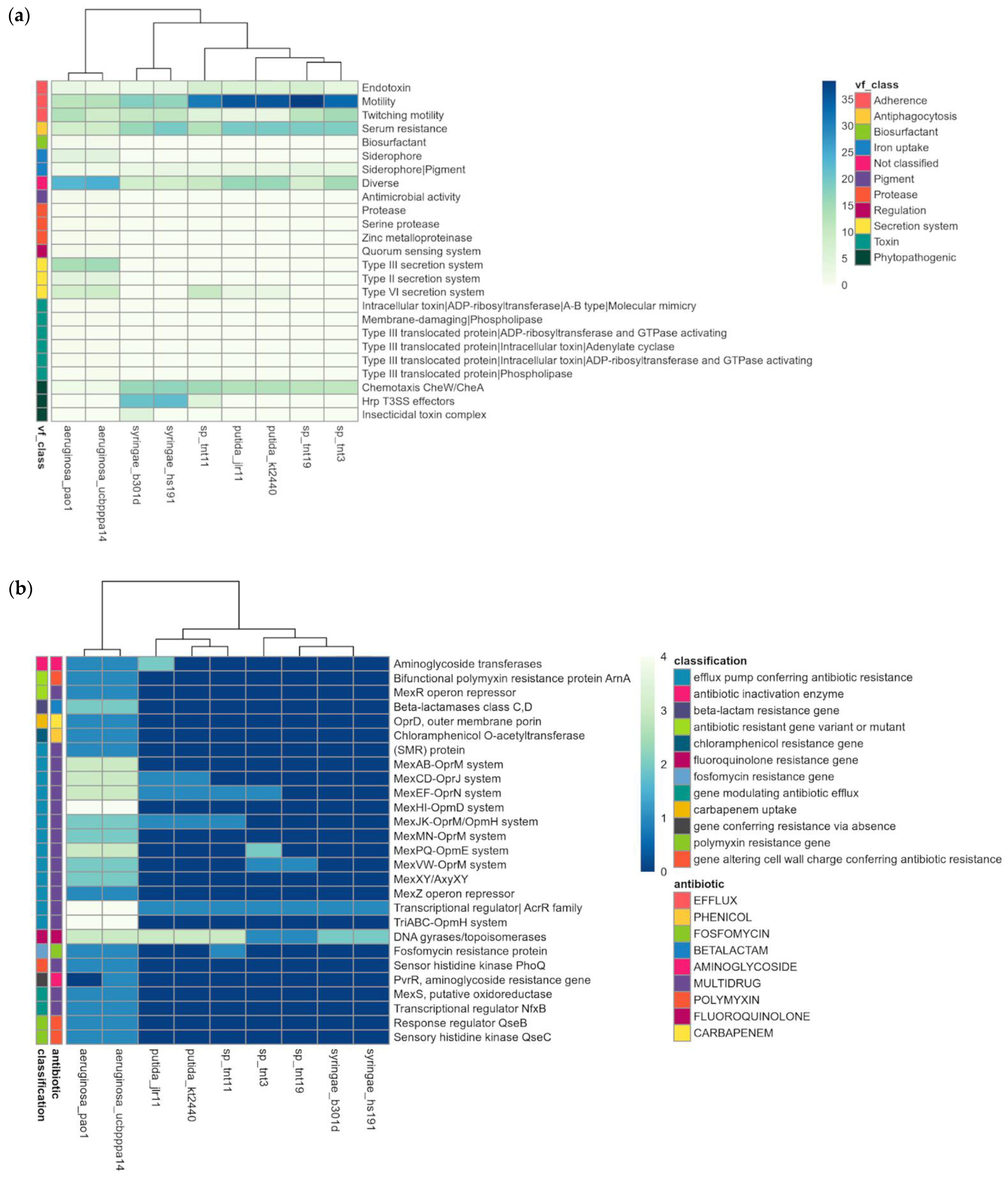
3.6. Pathogenic Profile and Other Specific Features of TNT Isolates
3.7. Analysis of Enzymes Involved in TNT Metabolism in TNT-Transforming Species
3.7.1. Nitroreductases
3.7.2. Xenobiotic Reductases
3.7.3. Azoreductases
3.7.4. Multidrug Efflux Pumps
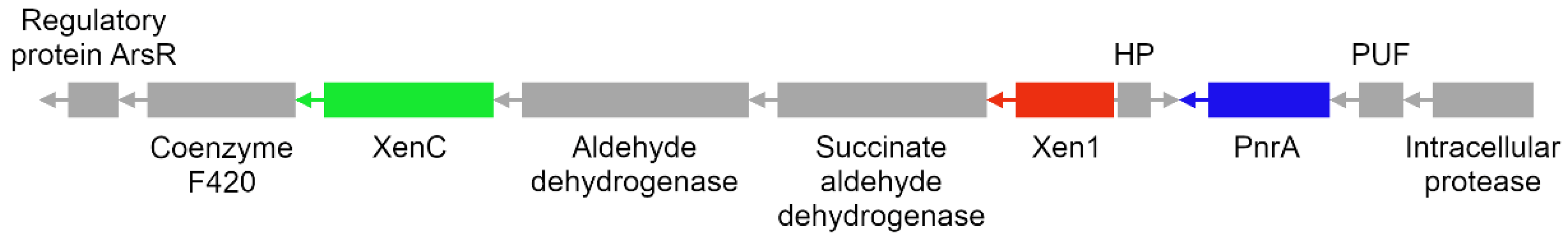
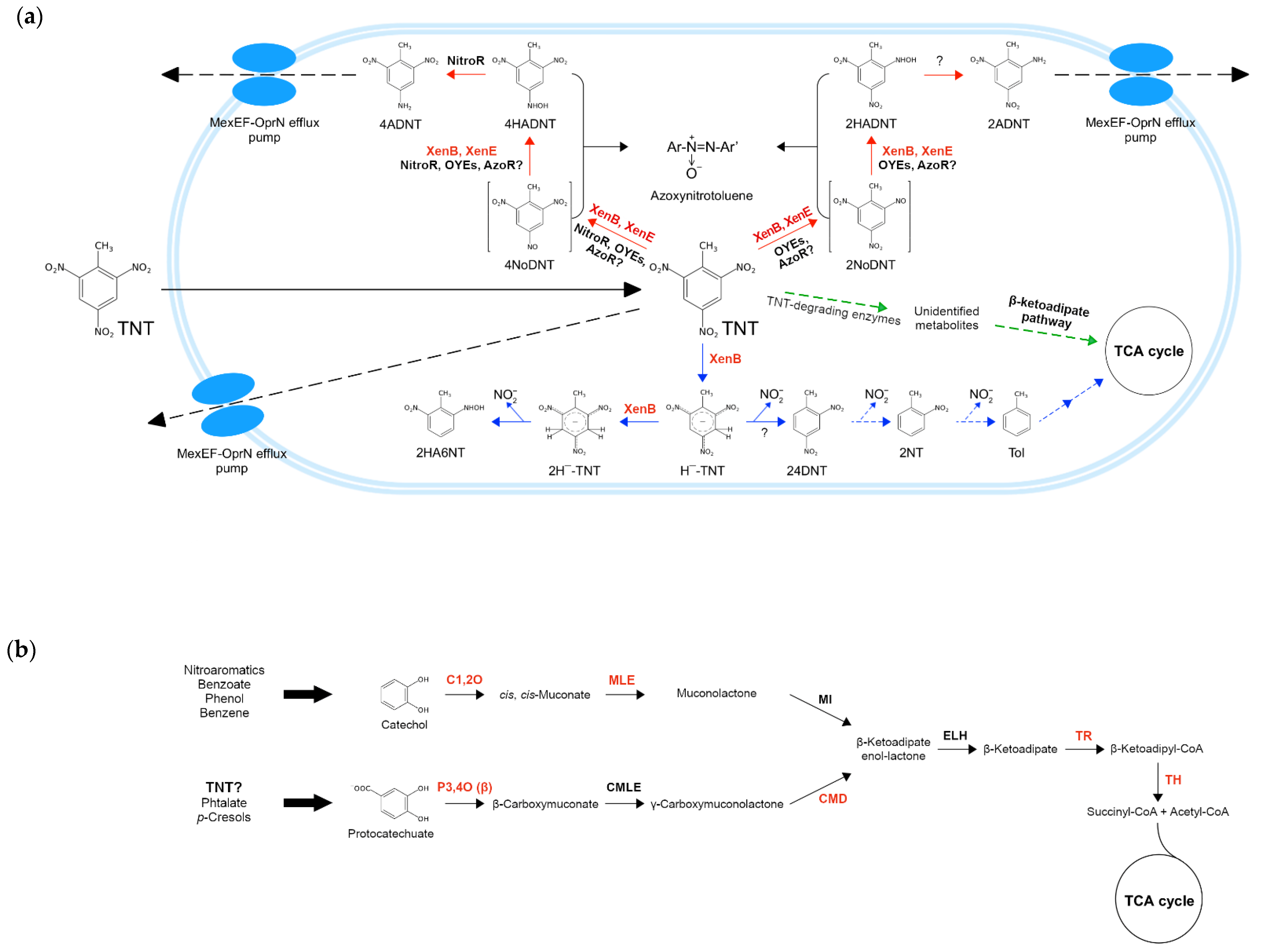
3.8. Reconstruction of TNT Degradation Pathways
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tiwari, J.; Tarale, P.; Sivanesan, S.; Bafana, A. Environmental Persistence, Hazard, and Mitigation Challenges of Nitroaromatic Compounds. Environ. Sci. Pollut. Res. 2019, 26, 28650–28667. [Google Scholar] [CrossRef]
- Khan, M.I.; Lee, J.; Park, J. A Toxicological Review on Potential Microbial Degradation Intermediates of 2,4,6-Trinitrotoluene, and Its Implications in Bioremediation. KSCE J. Civ. Eng. 2013, 17, 1223–1231. [Google Scholar] [CrossRef]
- Serrano-González, M.Y.; Chandra, R.; Castillo-Zacarias, C.; Robledo-Padilla, F.; Rostro-Alanis, M.D.J.; Parra-Saldivar, R. Biotransformation and Degradation of 2,4,6-Trinitrotoluene by Microbial Metabolism and Their Interaction. Def. Technol. 2018, 14, 151–164. [Google Scholar] [CrossRef]
- Honeycutt, M.E.; Jarvis, A.S.; McFarland, V.A. Cytotoxicity and Mutagenicity of 2,4,6-Trinitrotoluene and Its Metabolites. Ecotoxicol. Environ. Saf. 1996, 35, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Lata, K.; Kushwaha, A.; Ramanathan, G. Bacterial Enzymatic Degradation and Remediation of 2,4,6-Trinitrotoluene. In Microbial and Natural Macromolecules; Das, S., Dash, H.R., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 623–659. ISBN 9780128200841. [Google Scholar]
- EPA. Priority Pollutant List; United States Environmental Protection Agency: Washington, DC, USA, 2014.
- Ayoub, K.; van Hullebusch, E.D.; Cassir, M.; Bermond, A. Application of Advanced Oxidation Processes for TNT Removal: A Review. J. Hazard. Mater. 2010, 178, 10–28. [Google Scholar] [CrossRef] [PubMed]
- Maksimova, Y.G.; Maksimov, A.Y.; Demakov, V.A. Biotechnological Approaches to the Bioremediation of an Environment Polluted with Trinitrotoluene. Appl. Biochem. Microbiol. 2018, 54, 767–779. [Google Scholar] [CrossRef]
- Kalsi, A.; Celin, S.M.; Sahai, S. Agro Waste as Immobilization Carrier for in Situ Remediation of 2,4,6-Trinitrotoluene Contaminated Soil. Environ. Technol. Innov. 2022, 27, 102455. [Google Scholar] [CrossRef]
- Lamba, J.; Anand, S.; Dutta, J.; Chatterjee, S.; Nagar, S.; Celin, S.M.; Rai, P.K. Study on Aerobic Degradation of 2,4,6-Trinitrotoluene (TNT) Using Pseudarthrobacter chlorophenolicus Collected from the Contaminated Site. Environ. Monit. Assess. 2021, 193, 80. [Google Scholar] [CrossRef]
- Xu, M.; Liu, D.; Sun, P.; Li, Y.; Wu, M.; Liu, W.; Maser, E.; Xiong, G.; Guo, L. Degradation of 2,4,6-Trinitrotoluene (TNT): Involvement of Protocatechuate 3,4-Dioxygenase (P34O) in Buttiauxella sp. S19-1. Toxics 2021, 9, 231. [Google Scholar] [CrossRef] [PubMed]
- Barrows, S.E.; Cramer, C.J.; Truhlar, D.G.; Elovitz, M.S.; Weber, E.J. Factors Controlling Regioselectivity in the Reduction of Polynitroaromatics in Aqueous Solution. Environ. Sci. Technol. 1996, 30, 3028–3038. [Google Scholar] [CrossRef]
- Esteve-Núñez, A.; Caballero, A.; Ramos, J.L. Biological Degradation of 2,4,6-Trinitrotoluene. Microbiol. Mol. Biol. Rev. 2001, 65, 335–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blehert, D.S.; Fox, B.G.; Chambliss, G.H. Cloning and Sequence Analysis of Two Pseudomonas Flavoprotein Xenobiotic Reductases. J. Bacteriol. 1999, 181, 6254–6263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pak, J.W.; Knoke, K.L.; Noguera, D.R.; Fox, B.G.; Chambliss, G.H. Transformation of 2,4,6-Trinitrotoluene by Purified Xenobiotic Reductase B from Pseudomonas fluorescens I-C. Appl. Environ. Microbiol. 2000, 66, 4742–4750. [Google Scholar] [CrossRef] [Green Version]
- Orville, A.M.; Manning, L.; Blehert, D.S.; Fox, B.G.; Chambliss, G.H. Crystallization and Preliminary Analysis of Xenobiotic Reductase B from Pseudomonas fluorescens I-C. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 1289–1291. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Tomioka, Y.; Suzuki, H.; Yonezawa, M.; Hishinuma, T.; Mizugaki, M. Molecular Cloning of the NemA Gene Encoding N-Ethylmaleimide Reductase from Escherichia coli. Biol. Pharm. Bull. 1997, 20, 110–112. [Google Scholar] [CrossRef] [Green Version]
- González-Pérez, M.M.; Van Dillewijn, P.; Wittich, R.M.; Ramos, J.L. Escherichia coli has Multiple Enzymes that Attack TNT and Release Nitrogen for Growth. Environ. Microbiol. 2007, 9, 1535–1540. [Google Scholar] [CrossRef] [PubMed]
- French, C.E.; Nicklin, S.; Bruce, N.C. Aerobic Degradation of 2,4,6-Trinitrotoluene by Enterobacter cloacae PB2 and by Pentaerythritol Tetranitrate Reductase. Appl. Environ. Microbiol. 1998, 64, 2864–2868. [Google Scholar] [CrossRef] [Green Version]
- French, C.E.; Nicklin, S.; Bruce, N.C. Sequence and Properties of Pentaerythritol Tetranitrate Reductase from Enterobacter cloacae PB2. J. Bacteriol. 1996, 178, 6623. [Google Scholar] [CrossRef] [Green Version]
- Bryant, C.; Hubbard, L.; McElroy, W.D. Cloning, Nucleotide Sequence, and Expression of the Nitroreductase Gene from Enterobacter cloacae. J. Biol. Chem. 1991, 266, 4126–4130. [Google Scholar] [CrossRef]
- Van Dillewijn, P.; Wittich, R.M.; Caballero, A.; Ramos, J.L. Subfunctionality of Hydride Transferases of the Old Yellow Enzyme Family of Flavoproteins of Pseudomonas putida. Appl. Environ. Microbiol. 2008, 74, 6703–6708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caballero, A.; Lázaro, J.J.; Ramos, J.L.; Esteve-Núñez, A. PnrA, a New Nitroreductase-Family Enzyme in the TNT-Degrading Strain Pseudomonas putida JLR11. Environ. Microbiol. 2005, 7, 1211–1219. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas Genomes: Diverse and Adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef] [Green Version]
- Reddy, G.S.N.; Matsumoto, G.I.; Schumann, P.; Stackerbrandt, E.; Shivaji, S. Psychrophilic Pseudomonads from Antarctica: Pseudomonas antartica sp. Nov., Pseudomonas meridiana sp. Nov. and Pseudomonas proteolytica sp. Nov. Int. J. Syst. Evol. Microbiol. 2004, 54, 713–719. [Google Scholar] [CrossRef]
- Cabrera, M.Á.; Blamey, J.M. Cloning, Overexpression, and Characterization of a Thermostable Nitrilase from an Antarctic Pyrococcus sp. Extremophiles 2017, 21, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Márquez, S.L.; Atalah, J.; Blamey, J.M. Characterization of a Novel Thermostable (S)-Amine-Transaminase from an Antarctic Moderately-Thermophilic Bacterium Albidovulum sp. SLM16. Enzyme Microb. Technol. 2019, 131, 109423. [Google Scholar] [CrossRef] [PubMed]
- Orellana-Saez, M.; Pacheco, N.; Costa, J.I.; Mendez, K.N.; Miossec, M.J.; Meneses, C.; Castro-Nallar, E.; Marcoleta, A.E.; Poblete-Castro, I. In-Depth Genomic and Phenotypic Characterization of the Antarctic Psychrotolerant Strain Pseudomonas sp. MPC6 Reveals Unique Metabolic Features, Plasticity, and Biotechnological Potential. Front. Microbiol. 2019, 10, 1154. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, M.Á.; Márquez, S.L.; Quezada, C.P.; Osorio, M.I.; Castro-Nallar, E.; González-Nilo, F.D.; Pérez-Donoso, J.M. Biotransformation of 2,4,6-Trinitrotoluene by Pseudomonas sp. TNT3 Isolated from Deception Island, Antarctica. Environ. Pollut. 2020, 262, 113922. [Google Scholar] [CrossRef]
- Fernández, M.; Duque, E.; Pizarro-Tobías, P.; van Dillewijn, P.; Wittich, R.M.; Ramos, J.L. Microbial Responses to Xenobiotic Compounds. Identification of Genes That Allow Pseudomonas putida KT2440 to Cope with 2,4,6-Trinitrotoluene. Microb. Biotechnol. 2009, 2, 287–294. [Google Scholar] [CrossRef] [Green Version]
- Kalderis, D.; Juhasz, A.L.; Boopathy, R.; Comfort, S. Soils Contaminated with Explosives: Environmental Fate and Evaluation of State-Ofthe-Art Remediation Processes (IUPAC Technical Eport). Pure Appl. Chem. 2011, 83, 1407–1484. [Google Scholar] [CrossRef]
- Habineza, A.; Zhai, J.; Mai, T.; Mmereki, D.; Ntakirutimana, T. Biodegradation of 2, 4, 6-Trinitrotoluene (TNT) in Contaminated Soil and Microbial Remediation Options for Treatment. Period. Polytech. Chem. Eng. 2017, 61, 171–187. [Google Scholar] [CrossRef] [Green Version]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding Data and Analysis Capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Institute: England, UK, 2010. [Google Scholar]
- Bushnell, B. BBMap; Version 37.75; Joint Genome Institute: Walnut Creek, CA, USA, 2015.
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An Integrated Tool for Comprehensive Microbial Variant Detection and Genome Assembly Improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A Modular and Extensible Implementation of the RAST Algorithm for Building Custom Annotation Pipelines and Annotating Batches of Genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Perez, C.A.; Conrad, R.E.; Konstantinidis, K.T. MicrobeAnnotator: A User-Friendly, Comprehensive Functional Annotation Pipeline for Microbial Genomes. BMC Bioinform. 2021, 22, 1–16. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A New Software for Selection of Phylogenetic Informative Regions from Multiple Sequence Alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and Taxonomy in Diagnostics for Food Security: Soft-Rotting Enterobacterial Plant Pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Warnes, G.R. Gplots: Various R Programming Tools for Plotting Data. 2020. [Google Scholar]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Bertelli, C.; Laird, M.R.; Williams, K.P.; Lau, B.Y.; Hoad, G.; Winsor, G.L.; Brinkman, F.S.L. IslandViewer 4: Expanded Prediction of Genomic Islands for Larger-Scale Datasets. Nucleic Acids Res. 2017, 45, W30–W35. [Google Scholar] [CrossRef]
- Xie, Z.; Tang, H. ISEScan: Automated Identification of Insertion Sequence Elements in Prokaryotic Genomes. Bioinformatics 2017, 33, 3340–3347. [Google Scholar] [CrossRef] [PubMed]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an Update of CRISRFinder, Includes a Portable Version, Enhanced Performance and Integrates Search for Cas Proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A Comparative Pathogenomic Platform with an Interactive Web Interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic Resistome Surveillance with the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Martin, M.J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The Universal Protein Knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The Proteomics Server for in-Depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [Green Version]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated Protein Sequence and Structural Alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Mishra, K.; Ramanthan, G. Bioremediation of Nitroaromatic Compounds. Wastewater Treat. Eng. 2015, 2, 51–83. [Google Scholar] [CrossRef] [Green Version]
- Nogales, J.; García, J.L.; Díaz, E. Degradation of Aromatic Compounds in Pseudomonas: A Systems Biology View. In Aerobic Utilization of Hydrocarbons, Oils, and Lipids. Handbook of Hydrocarbon and Lipid Microbiology; Rojo, F., Ed.; Springer: Cham, Switzerland, 2017; pp. 1–49. ISBN 978-3-319-39782-5. [Google Scholar]
- Wells, T.; Ragauskas, A.J. Biotechnological Opportunities with the β-Ketoadipate Pathway. Trends Biotechnol. 2012, 30, 627–637. [Google Scholar] [CrossRef]
- Bhatt, P.; Pathak, V.M.; Joshi, S.; Bisht, T.S.; Singh, K.; Chandra, D. Major Metabolites after Degradation of Xenobiotics and Enzymes Involved in These Pathways. In Smart Bioremediation Technologies: Microbial Enzymes; Academic Press: Cambridge, MA, USA, 2019; ISBN 9780128183076. [Google Scholar]
- De Lima-Morales, D.; Chaves-Moreno, D.; Wos-Oxley, M.L.; Jáuregui, R.; Vilchez-Vargas, R.; Pieper, D.H. Degradation of Benzene by Pseudomonas veronii 1YdBTEX2 and 1YB2 Is Catalyzed by Enzymes Encoded in Distinct Catabolism Gene Clusters. Appl. Environ. Microbiol. 2016, 82, 167–173. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Echartea, E.; Suman, J.; Smrhova, T.; Ridl, J.; Pajer, P.; Strejcek, M.; Uhlik, O. Genomic Analysis of Dibenzofuran-Degrading Pseudomonas veronii Strain Pvy Reveals Its Biodegradative Versatility. G3 Genes 2021, 11, jkaa030. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Formusa, P.A.; Hsiang, T.; Habash, M.B.; Lee, H.; Trevors, J.T. Genome Sequence of Pseudomonas mandelii PD30. Genome Announc. 2014, 2, e00713–e00714. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Park, W. Genome Analysis of Naphthalene-Degrading Pseudomonas sp. AS1 Harboring the Megaplasmid PAS1. J. Microbiol. Biotechnol. 2018, 28, 330–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortier, L.C.; Sekulovic, O. Importance of Prophages to Evolution and Virulence of Bacterial Pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef]
- Bertelli, C.; Tilley, K.E.; Brinkman, F.S.L. Microbial Genomic Island Discovery, Visualization and Analysis. Brief. Bioinform. 2019, 20, 1685–1698. [Google Scholar] [CrossRef] [Green Version]
- Feil, H.; Feil, W.S.; Chain, P.; Larimer, F.; DiBartolo, G.; Copeland, A.; Lykidis, A.; Trong, S.; Nolan, M.; Goltsman, E.; et al. Comparison of the Complete Genome Sequences of Pseudomonas syringae Pv. Syringae B728a and Pv. Tomato DC3000. Proc. Natl. Acad. Sci. USA 2005, 102, 11064–11069. [Google Scholar] [CrossRef] [Green Version]
- Dziewit, L.; Radlinska, M. Two Inducible Prophages of an Antarctic Pseudomonas sp. ANT_H14 Use the Same Capsid for Packaging Their Genomes—Characterization of a Novel Phage Helper-Satellite System. PLoS ONE 2016, 11, e0158889. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial Insertion Sequences: Their Genomic Impact and Diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef] [Green Version]
- Golubov, A. Genome Instability in Bacteria: Causes and Consequences. Genome Stab. 2021, 73–90. [Google Scholar] [CrossRef]
- Kallastu, A.; Hõrak, R.; Kivisaar, M. Identification and Characterization of IS1411, a New Insertion Sequence Which Causes Transcriptional Activation of the Phenol Degradation Genes in Pseudomonas putida. J. Bacteriol. 1998, 180, 5306–5312. [Google Scholar] [CrossRef] [Green Version]
- Christie-Oleza, J.A.; Nogales, B.; Martín-Cardona, C.; Lanfranconi, M.P.; Albertí, S.; Lalucat, J.; Bosch, R. ISPst9, an ISL3-like Insertion Sequence from Pseudomonas stutzeri AN 10 Involved in Catabolic Gene Inactivation. Int. Microbiol. 2008, 11, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Springael, D.; Top, E.M. Horizontal Gene Transfer and Microbial Adaptation to Xenobiotics: New Types of Mobile Genetic Elements and Lessons from Ecological Studies. Trends Microbiol. 2004, 12, 53–58. [Google Scholar] [CrossRef]
- Mohapatra, B.; Phale, P.S. Microbial Degradation of Naphthalene and Substituted Naphthalenes: Metabolic Diversity and Genomic Insight for Bioremediation. Front. Bioeng. Biotechnol. 2021, 9, 602445. [Google Scholar] [CrossRef]
- Xu, A.; Wang, D.; Ding, Y.; Zheng, Y.; Wang, B.; Wei, Q.; Wang, S.; Yang, L.; Ma, L.Z. Integrated Comparative Genomic Analysis and Phenotypic Profiling of Pseudomonas aeruginosa Isolates from Crude Oil. Front. Microbiol. 2020, 11, 519. [Google Scholar] [CrossRef]
- He, C.; Li, Y.; Huang, C.; Chen, F.; Ma, Y. Genome Sequence and Metabolic Analysis of a Fluoranthene-Degrading Strain Pseudomonas aeruginosa DN1. Front. Microbiol. 2018, 9, 2595. [Google Scholar] [CrossRef]
- Das, D.; Baruah, R.; Sarma Roy, A.; Singh, A.K.; Deka Boruah, H.P.; Kalita, J.; Bora, T.C. Complete Genome Sequence Analysis of Pseudomonas aeruginosa N002 Reveals Its Genetic Adaptation for Crude Oil Degradation. Genomics 2015, 105, 182–190. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete Genome Sequence of Pseudomonas aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Rahme, L.G.; Ausubel, F.M.; Cao, H.; Drenkard, E.; Goumnerov, B.C.; Lau, G.W.; Mahajan-Miklos, S.; Plotnikova, J.; Tan, M.W.; Tsongalis, J.; et al. Plants and Animals Share Functionally Common Bacterial Virulence Factors. Proc. Natl. Acad. Sci. USA 2000, 97, 8815–8821. [Google Scholar] [CrossRef] [Green Version]
- Ravindran, A.; Jalan, N.; Yuan, J.S.; Wang, N.; Gross, D.C. Comparative Genomics of Pseudomonas syringae Pv. Syringae Strains B301D and HS191 and Insights into Intrapathovar Traits Associated with Plant Pathogenesis. Microbiologyopen 2015, 4, 553–573. [Google Scholar] [CrossRef] [PubMed]
- Ichinose, Y.; Taguchi, F.; Mukaihara, T. Pathogenicity and Virulence Factors of Pseudomonas syringae. J. Gen. Plant Pathol. 2013, 79, 285–296. [Google Scholar] [CrossRef]
- Nelson, K.E.; Weinel, C.; Paulsen, I.T.; Dodson, R.J.; Hilbert, H.; Fouts, D.E.; Gill, S.R.; Pop, M.; Holmes, M.; Brinkac, L.; et al. Complete Genome Sequence and Comparative Analysis of the Metabolically Versatile Pseudomonas putida KT2440. Environ. Microbiol. 2002, 4, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Poole, K. Multidrug Efflux Pumps and Antimicrobial Resistance in Pseudomonas aeruginosa and Related Organisms. J. Mol. Microbiol. Biotechnol. 2001, 3, 255–264. [Google Scholar] [PubMed]
- Sharrar, A.M.; Crits-Christoph, A.; Méheust, R.; Diamond, S.; Starr, E.P.; Banfield, J.F. Bacterial Secondary Metabolite Biosynthetic Potential in Soil Varies with Phylum, Depth, and Vegetation Type. MBio 2020, 11, 1–17. [Google Scholar] [CrossRef]
- Pandey, S.S.; Patnana, P.K.; Rai, R.; Chatterjee, S. Xanthoferrin, the α-Hydroxycarboxylate-Type Siderophore of Xanthomonas campestris Pv. Campestris, Is Required for Optimum Virulence and Growth inside Cabbage. Mol. Plant Pathol. 2017, 18, 949–962. [Google Scholar] [CrossRef]
- Roldán, M.D.; Pérez-Reinado, E.; Castillo, F.; Moreno-Vivián, C. Reduction of Polynitroaromatic Compounds: The Bacterial Nitroreductases. FEMS Microbiol. Rev. 2008, 32, 474–500. [Google Scholar] [CrossRef] [Green Version]
- Wittich, R.M.; Haïdour, A.; Van Dillewijn, P.; Ramos, J.L. OYE Flavoprotein Reductases Initiate the Condensation of TNT-Derived Intermediates to Secondary Diarylamines and Nitrite. Environ. Sci. Technol. 2008, 42, 734–739. [Google Scholar] [CrossRef]
- Spiegelhauer, O.; Werther, T.; Mende, S.; Knauer, S.H.; Dobbek, H. Determinants of Substrate Binding and Protonation in the Flavoenzyme Xenobiotic Reductase A. J. Mol. Biol. 2010, 403, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Spiegelhauer, O.; Dickert, F.; Mende, S.; Niks, D.; Hille, R.; Ullmann, M.; Dobbek, H. Kinetic Characterization of Xenobiotic Reductase A from Pseudomonas putida 86. Biochemistry 2009, 48, 11412–11420. [Google Scholar] [CrossRef]
- Griese, J.J.; Jakob, R.P.; Schwarzinger, S.; Dobbek, H. Xenobiotic Reductase A in the Degradation of Quinoline by Pseudomonas putida 86: Physiological Function, Structure and Mechanism of 8-Hydroxycoumarin Reduction. J. Mol. Biol. 2006, 361, 140–152. [Google Scholar] [CrossRef]
- Kumagai, Y.; Shimojo, N. Possible Mechanisms for Induction of Oxidative Stress and Suppression of Systemic Nitric Oxide Production Caused by Exposure to Environmental Chemicals. Environ. Health Prev. Med. 2002, 7, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Stenuit, B.A.; Agathos, S.N. Microbial 2,4,6-Trinitrotoluene Degradation: Could We Learn from (Bio)Chemistry for Bioremediation and Vice Versa? Appl. Microbiol. Biotechnol. 2010, 88, 1043–1064. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, W.; Bhatt, P.; Chen, S. Recent Advanced Technologies for the Characterization of Xenobiotic-Degrading Microorganisms and Microbial Communities. Front. Bioeng. Biotechnol. 2021, 9, 632059. [Google Scholar] [CrossRef] [PubMed]
- Ridl, J.; Suman, J.; Fraraccio, S.; Hradilova, M.; Strejcek, M.; Cajthaml, T.; Zubrova, A.; Macek, T.; Strnad, H.; Uhlik, O. Complete Genome Sequence of Pseudomonas alcaliphila JAB1 (=DSM 26533), a Versatile Degrader of Organic Pollutants. Stand. Genomic Sci. 2018, 13, 3. [Google Scholar] [CrossRef] [PubMed]
- Heiss, G.; Knackmuss, H.-J. Bioelimination of Trinitroaromatic Compounds: Immobilization versus Mineralization. Curr. Opin. Microbiol. 2002, 5, 282–287. [Google Scholar] [CrossRef]
- Misal, S.A.; Gawai, K.R. Azoreductase: A Key Player of Xenobiotic Metabolism. Bioresour. Bioprocess. 2018, 5, 1–9. [Google Scholar] [CrossRef]
- Misal, S.A. Biotransformation of Nitro Aromatic Compounds by Flavin-Free NADHAzoreductase. J. Bioremediation Biodegrad. 2015, 6, 272. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Zhou, J.; Lv, H.; Xiang, X.; Wang, J.; Zhou, M.; Qv, Y. Azoreductase from Rhodobacter sphaeroides AS1.1737 Is a Flavodoxin That Also Functions as Nitroreductase and Flavin Mononucleotide Reductase. Appl. Microbiol. Biotechnol. 2007, 76, 1271–1279. [Google Scholar] [CrossRef]
- Gonçalves, A.M.D.; Mendes, S.; De Sanctis, D.; Martins, L.O.; Bento, I. The Crystal Structure of Pseudomonas putida Azoreductase—The Active Site Revisited. FEBS J. 2013, 280, 6643–6657. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.J.; Hagemeier, C.; Rahman, N.; Lowe, E.; Noble, M.; Coughtrie, M.; Sim, E.; Westwood, I. Molecular Cloning, Characterisation and Ligand-Bound Structure of an Azoreductase from Pseudomonas aeruginosa. J. Mol. Biol. 2007, 373, 1213–1228. [Google Scholar] [CrossRef]
- Díaz, E.; Jiménez, J.I.; Nogales, J. Aerobic Degradation of Aromatic Compounds. Curr. Opin. Biotechnol. 2013, 24, 431–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, J.; Das, S. Molecular Perspectives and Recent Advances in Microbial Remediation of Persistent Organic Pollutants. Environ. Sci. Pollut. Res. 2016, 23, 16883–16903. [Google Scholar] [CrossRef]
- Mutanda, I.; Sun, J.; Jiang, J.; Zhu, D. Bacterial Membrane Transporter Systems for Aromatic Compounds: Regulation, Engineering, and Biotechnological Applications. Biotechnol. Adv. 2022, 59, 107952. [Google Scholar] [CrossRef]
- Vermaas, J.V.; Dixon, R.A.; Chen, F.; Mansfield, S.D.; Boerjan, W.; Ralph, J.; Crowley, M.F.; Beckham, G.T. Passive Membrane Transport of Lignin-Related Compounds. Proc. Natl. Acad. Sci. USA 2019, 116, 23117–23123. [Google Scholar] [CrossRef]
- Van der Meer, J.R.; de Vos, W.M.; Harayama, S.; Zehnder, A.J. Molecular Mechanisms of Genetic Adaptation to Xenobiotic Compounds. Microbiol. Rev. 1992, 56, 677–694. [Google Scholar] [CrossRef]
- Oh, B.T.; Shea, P.J.; Drijber, R.A.; Vasilyeva, G.K.; Sarath, G. TNT Biotransformation and Detoxification by a Pseudomonas aeruginosa Strain. Biodegradation 2003, 14, 309–319. [Google Scholar] [CrossRef]
- Pascual, J.; Udaondo, Z.; Molina, L.; Segura, A.; Esteve-Núñez, A.; Caballero, A.; Duque, E.; Ramos, J.L.; van Dillewijn, P. Draft Genome Sequence of Pseudomonas putida JLR11, a Facultative Anaerobic 2,4,6-Trinitrotoluene Biotransforming Bacterium. Genome Announc. 2015, 3, 10–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteve-Nuñez, A.; Lucchesi, G.; Philipp, B.; Schink, B.; Ramos, J.L. Respiration of 2,4,6-Trinitrotoluene by Pseudomonas sp. Strain JLR11. J. Bacteriol. 2000, 182, 1352–1355. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R.; Acharya, V.; Mukhia, S.; Singh, D.; Kumar, S. Complete Genome Sequence of Pseudomonas frederiksbergensis ERDD5:01 Revealed Genetic Bases for Survivability at High Altitude Ecosystem and Bioprospection Potential. Genomics 2019, 111, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Redondo-Nieto, M.; Barret, M.; Morrisey, J.P.; Germaine, K.; Martínez-Granero, F.; Barahona, E.; Navazo, A.; Sánchez-Contreras, M.; Moynihan, J.A.; Giddens, S.R.; et al. Genome Sequence of the Biocontrol Strain Pseudomonas fluorescens F113. J. Bacteriol. 2012, 194, 1273–1274. [Google Scholar] [CrossRef] [PubMed]
- ATCC. Pseudomonas Fluorescens Migula 13525; ATCC: Manassas, VA, USA, 2021. [Google Scholar]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.; Saucier, M.; Déziel, E.; et al. Genomic Analysis Reveals That Pseudomonas aeruginosa Virulence Is Combinatorial. Genome Biol. 2006, 7, R90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahto, K.U.; Das, S. Whole Genome Characterization and Phenanthrene Catabolic Pathway of a Biofilm Forming Marine Bacterium Pseudomonas aeruginosa PFL-P1. Ecotoxicol. Environ. Saf. 2020, 206, 111087. [Google Scholar] [CrossRef] [PubMed]
- Andersen, S.M.; Johnsen, K.; Sørensen, J.; Nielsen, P.; Jacobsen, C.S. Pseudomonas frederiksbergensis sp. Nov., Isolated from Soil at a Coal Gasification Site. Int. J. Syst. Evol. Microbiol. 2000, 50, 1957–1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallardo-Benavente, C.; Campo-Giraldo, J.L.; Castro-Severyn, J.; Quiroz, A.; Pérez-Donoso, J.M. Genomics Insights into Pseudomonas sp. CG01: An Antarctic Cadmium-Resistant Strain Capable of Biosynthesizing CdS Nanoparticles Using Methionine as S-Source. Genes 2021, 12, 187. [Google Scholar] [CrossRef]
- Carrión, O.; Miñana-Galbis, D.; Montes, M.; Mercadé, E. Pseudomonas deceptionensis sp. Nov., a Psychrotolerant Bacterium from the Antarctic. Int. J. Syst. Evol. Microbiol. 2011, 61, 2401–2405. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Cho, Y.; JY, Y.; YJ, J.; SG, H.; OS, K. Complete Genome Sequence of Pseudomonas antarctica PAMC 27494, a Bacteriocin-Producing Psychrophile Isolated from Antarctica. J. Biotechnol. 2017, 259, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, M.S.; Lira, F.; Martínez, J.L.; Olivares, J.; Marshall, S.H. Draft Genome Sequence of Antarctic Pseudomonas sp. Strain KG01 with Full Potential for Biotechnological Applications. Genome Announc. 2015, 3, e00906–e00915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tribelli, P.M.; Iustman, L.J.R.; Catone, M.V.; Di Martino, C.; Revale, S.; Méndez, B.S.; López, N.I. Genome Sequence of the Polyhydroxybutyrate Producer Pseudomonas extremaustralis, a Highly Stress-Resistant Antarctic Bacterium. J. Bacteriol. 2012, 194, 2381–2382. [Google Scholar] [CrossRef] [Green Version]
- Verhille, S.; Baida, N.; Dabboussi, F.; Izard, D.; Leclerc, H. Taxonomic Study of Bacteria Isolated from Natural Mineral Waters: Proposal of Pseudomonas jessenii sp. Nov. and Pseudomonas mandelii sp. Nov. Syst. Appl. Microbiol. 1999, 22, 45–58. [Google Scholar] [CrossRef]

| Genome Feature | Isolate | ||
|---|---|---|---|
| TNT3 | TNT11 | TNT19 | |
| Contig count | 138 | 718 | 92 |
| L50 value | 22 | 126 | 11 |
| Contigs N50 (bp) | 93,061 | 13,749 | 166,334 |
| Completeness (%) | 99.2 | 96.8 | 99.2 |
| Genome size (bp) | 6,458,871 | 5,861,354 | 6,454,788 |
| GC content (%) | 58.55 | 60.44 | 58.60 |
| Total genes | 6185 | 6345 | 6385 |
| Total number CDSs | 6120 | 6280 | 6322 |
| rRNA genes | 5 | 6 | 5 |
| tRNA genes | 60 | 59 | 58 |
| Proteins with functional assignment | 4751 | 4943 | 4644 |
| Hypothetical proteins | 1369 | 1337 | 1678 |
| Isolate | ||||||
|---|---|---|---|---|---|---|
| Route | Enzyme | TNT3 | TNT11 | TNT19 | ||
| Central metabolic pathway | β-ketoadipate pathway | Protocatechuate | P3,4O (β-chain) | Yes | Yes (2 isoenzymes) | Yes |
| CMD | Yes (isoenzyme 1) | Yes (isoenzyme 1) | Yes (isoenzyme 1) | |||
| Yes (isoenzyme 2) | Yes (isoenzyme 2) | Yes (isoenzyme 2) | ||||
| Ortho-catechol | C1,2O | Yes | Yes | Yes | ||
| MLE | Yes | Yes | Yes | |||
| Ortho-catechol and protocatechuate | TR | Yes (A chain) | Yes (A chain) | Yes (A chain) | ||
| Yes (B chain) | Yes (B chain) | Yes (B chain) | ||||
| TH | Yes | Yes | Yes | |||
| Peripheral metabolic pathway | TNT degradation pathway | Nitroreduction | NitroR4 | Yes | Yes | Yes |
| NitroR5 | Yes | Yes | Yes | |||
| NitroR6 | - | Yes | - | |||
| AzoR-a | Yes | Yes | Yes | |||
| AzoR-b | Yes | Yes | - | |||
| AzoR-c | Yes | - | - | |||
| Nitroreduction and denitration | XenA | - | Yes | - | ||
| XenB | Yes | Yes | Yes | |||
| XenC | - | Yes | - | |||
| XenE | Yes | Yes | Yes | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabrera, M.Á.; Márquez, S.L.; Pérez-Donoso, J.M. Comparative Genomic Analysis of Antarctic Pseudomonas Isolates with 2,4,6-Trinitrotoluene Transformation Capabilities Reveals Their Unique Features for Xenobiotics Degradation. Genes 2022, 13, 1354. https://doi.org/10.3390/genes13081354
Cabrera MÁ, Márquez SL, Pérez-Donoso JM. Comparative Genomic Analysis of Antarctic Pseudomonas Isolates with 2,4,6-Trinitrotoluene Transformation Capabilities Reveals Their Unique Features for Xenobiotics Degradation. Genes. 2022; 13(8):1354. https://doi.org/10.3390/genes13081354
Chicago/Turabian StyleCabrera, Ma. Ángeles, Sebastián L. Márquez, and José M. Pérez-Donoso. 2022. "Comparative Genomic Analysis of Antarctic Pseudomonas Isolates with 2,4,6-Trinitrotoluene Transformation Capabilities Reveals Their Unique Features for Xenobiotics Degradation" Genes 13, no. 8: 1354. https://doi.org/10.3390/genes13081354
APA StyleCabrera, M. Á., Márquez, S. L., & Pérez-Donoso, J. M. (2022). Comparative Genomic Analysis of Antarctic Pseudomonas Isolates with 2,4,6-Trinitrotoluene Transformation Capabilities Reveals Their Unique Features for Xenobiotics Degradation. Genes, 13(8), 1354. https://doi.org/10.3390/genes13081354






