Source-To-Sink Transport of Sugar and Its Role in Male Reproductive Development
Abstract
1. Introduction
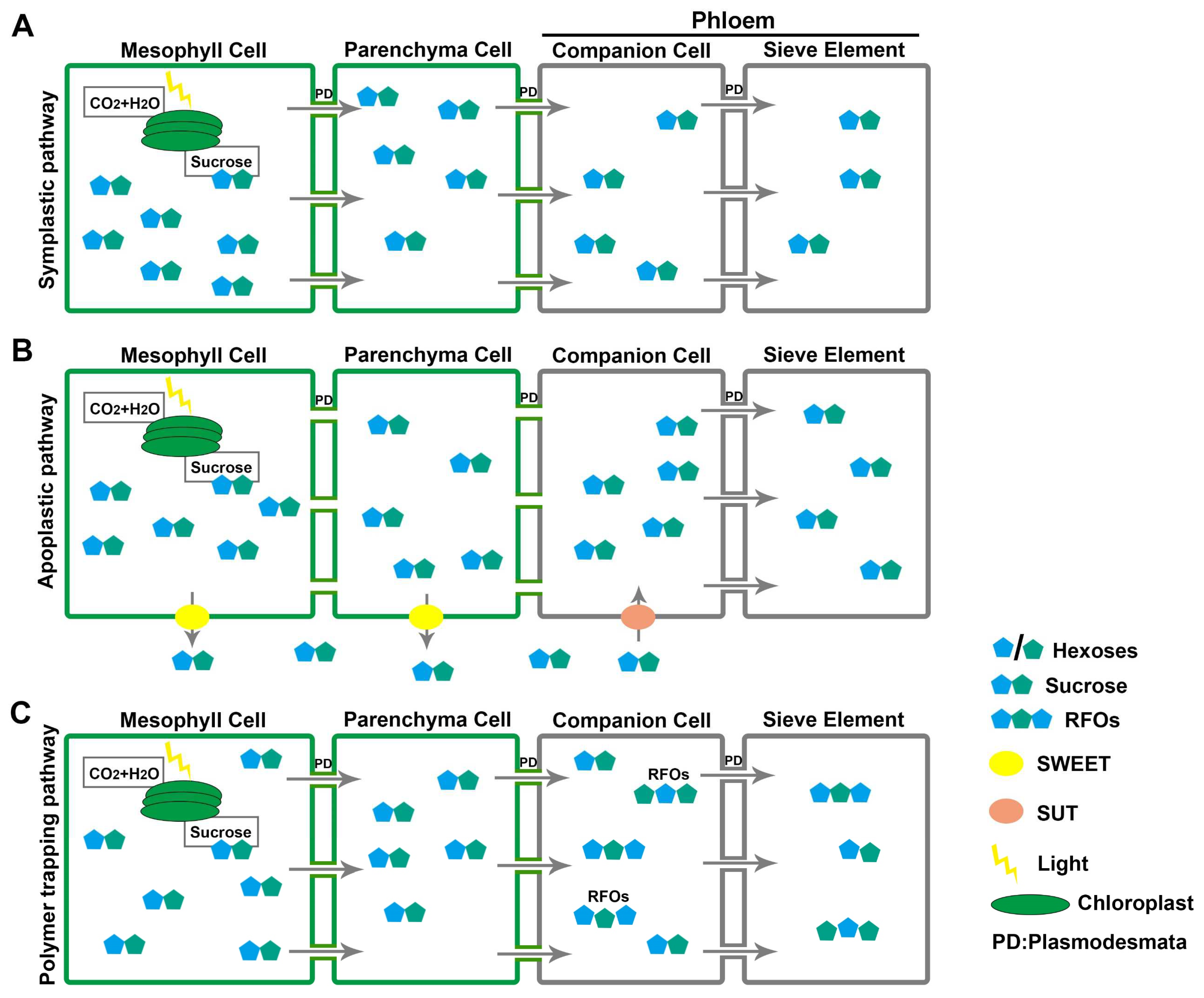
2. Strategies of Source-to-Sink Sugar Partitioning
3. Proteins Involved in Sugar Partitioning
3.1. Sucrose Transporters (SUTs)
3.2. Sugars Will Eventually Be Exported Transporters (SWEETs)
3.3. Invertases (INVs)
3.4. Monosaccharide Transporters (MSTs)
4. Roles of Sugar Transporters in Phloem Loading and Unloading
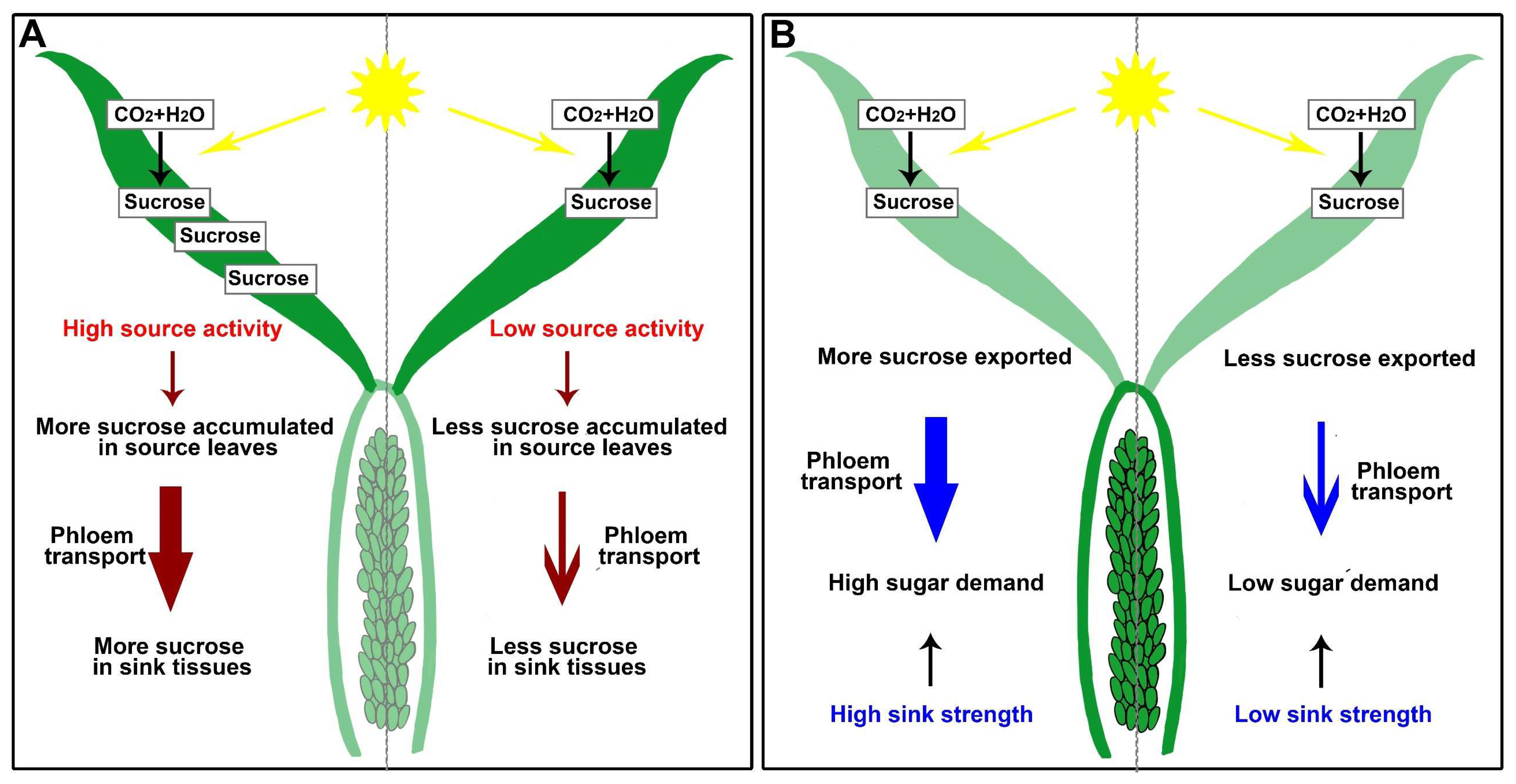
5. Sugar Balance and Signaling Transduction
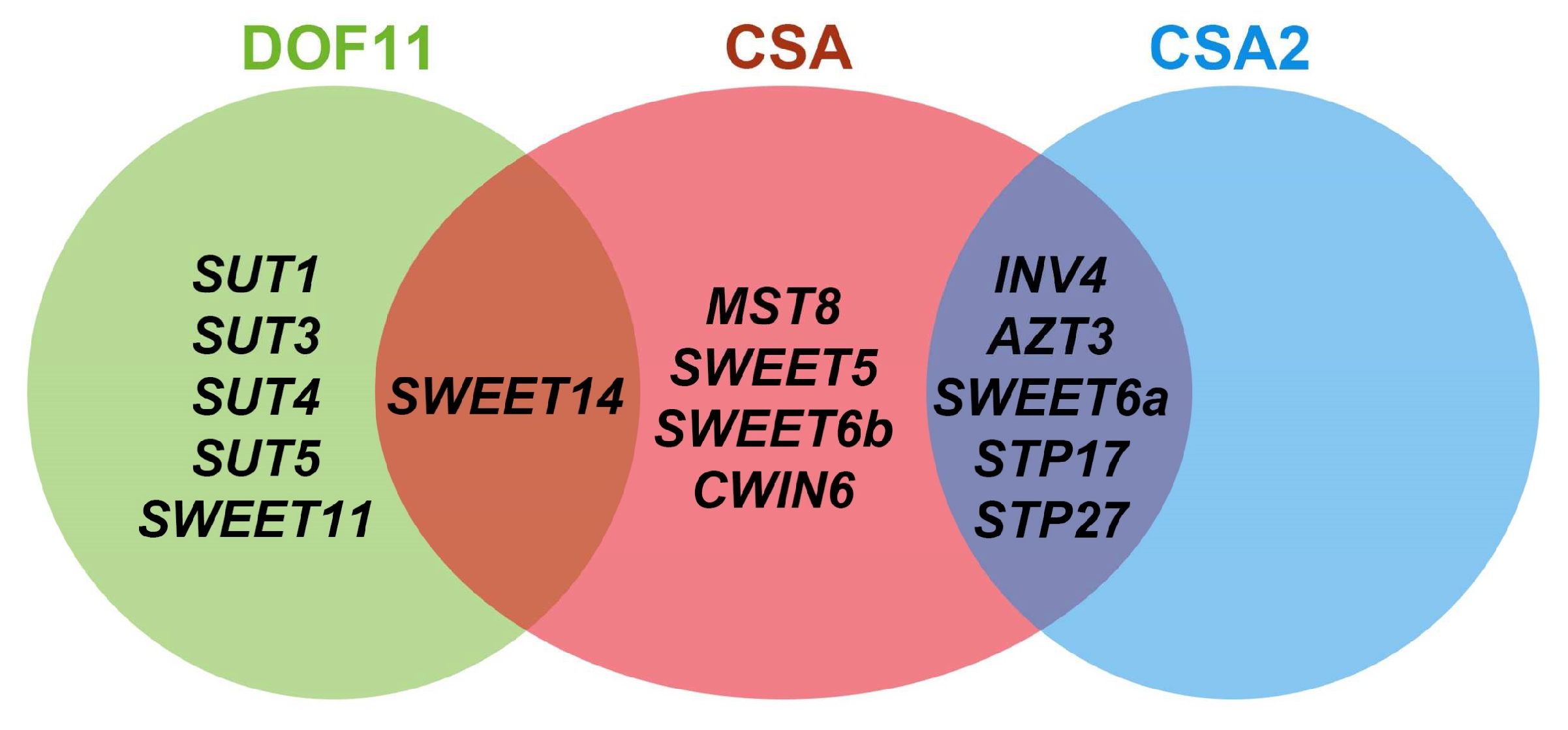
6. Sugar Regulatory Network
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Izawa, T.; Shimamoto, K. Becoming a model plant: The importance of rice to plant science. Trends Plant Sci. 1996, 1, 95–99. [Google Scholar] [CrossRef]
- Delseny, M.; Salses, J.; Cooke, R.; Sallaud, C.; Regad, F.; Lagoda, P.; Guiderdoni, E.; Ventelon, M.; Brugidou, C.; Ghesquière, A. Rice genomics: Present and future. Plant Physiol. Biochem. 2001, 39, 323–334. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, L. Specification of tapetum and microsporocyte cells within the anther. Curr. Opin. Plant Biol. 2014, 17, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Luo, X.; Zhu, L. Cytological analysis and genetic control of rice anther development. J. Genet. Genom. 2011, 38, 379–390. [Google Scholar] [CrossRef]
- Zhang, D.; Wilson, Z.A. Stamen specification and anther development in rice. Chin. Sci. Bull. 2009, 54, 2342–2353. [Google Scholar] [CrossRef]
- Scott, R.J.; Spielman, M.; Dickinson, H. Stamen structure and function. Plant Cell 2004, 16 (Suppl. S1), S46–S60. [Google Scholar] [CrossRef]
- Ma, H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 2005, 56, 393–434. [Google Scholar] [CrossRef]
- Mercier, R.; Grelon, M. Meiosis in plants: Ten years of gene discovery. Cytogenet. Genome Res. 2008, 120, 281–290. [Google Scholar] [CrossRef]
- Stanley, R.G. Pollen Chemistry and Tube Growth. Pollen 1971, 131–155. [Google Scholar] [CrossRef]
- Oliver, S.N.; van Dongen, J.; Alfred, S.C.; Mamun, E.A.; Zhao, X.; Saini, H.S.; Fernandes, S.F.; Blanchard, C.; Sutton, B.G.; Geigenberger, P.; et al. Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant, Cell Environ. 2005, 28, 1534–1551. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, W.; Yang, X.; Luo, X.; Jiang, N.; Ma, H.; Zhang, D. Carbon Starved Anther Encodes a MYB Domain Protein That Regulates Sugar Partitioning Required for Rice Pollen Development. Plant Cell 2010, 22, 672–689. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, C.; He, Y.; Zong, J.; Yang, X.; Si, H.; Sun, Z.; Hu, J.; Liang, W.; Zhang, D. Mutation in CSA creates a new photoperiod-sensitive genic male sterile line applicable for hybrid rice seed production. Proc. Natl. Acad. Sci. USA 2013, 110, 76–81. [Google Scholar] [CrossRef]
- Wang, D.; Li, J.; Sun, L.; Hu, Y.; Yu, J.; Wang, C.; Zhang, F.; Hou, H.; Liang, W.; Zhang, D. Two rice MYB transcription factors maintain male fertility in response to photoperiod by modulating sugar partitioning. New Phytol. 2021, 231, 1612–1629. [Google Scholar] [CrossRef]
- Sun, L.; Yuan, Z.; Wang, D.; Li, J.; Shi, J.; Hu, Y.; Yu, J.; Chen, X.; Chen, S.; Liang, W.; et al. Carbon Starved Anther modulates sugar and ABA metabolism to protect rice seed germination and seedling fitness. Plant Physiol. 2021, 187, 2405–2418. [Google Scholar] [CrossRef]
- Chu, Z.; Yuan, M.; Yao, J.; Ge, X.; Yuan, B.; Xu, C.; Li, X.; Fu, B.; Li, Z.; Bennetzen, J.L.; et al. Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 2006, 20, 1250–1255. [Google Scholar] [CrossRef]
- Antony, G.; Zhou, J.; Huang, S.; Li, T.; Liu, B.; White, F.; Yang, B. Rice xa13 Recessive Resistance to Bacterial Blight Is Defeated by Induction of the Disease Susceptibility Gene Os-11N3. Plant Cell 2010, 22, 3864–3876. [Google Scholar] [CrossRef]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef]
- Kalt-Torres, W.; Kerr, P.S.; Usuda, H.; Huber, S.C. Diurnal changes in maize leaf photosynthesis: I. Carbon exchange rate, assimilate export rate, and enzyme activities. Plant Physiol. 1987, 83, 283–288. [Google Scholar] [CrossRef]
- Turgeon, R.; Wolf, S. Phloem Transport: Cellular Pathways and Molecular Trafficking. Annu. Rev. Plant Biol. 2009, 60, 207–221. [Google Scholar] [CrossRef]
- Osorio, S.; Ruan, Y.-L.; Fernie, A.R. An update on source-to-sink carbon partitioning in tomato. Front. Plant Sci. 2014, 5, 516. [Google Scholar] [CrossRef]
- White, P.J. Long-distance transport in the xylem and phloem. In Marschner’s Mineral Nutrition of Higher Plants; Academic Press: Cambridge, MA, USA, 2012; pp. 49–70. [Google Scholar]
- Zhang, C.; Turgeon, R. Mechanisms of phloem loading. Curr. Opin. Plant Biol. 2018, 43, 71–75. [Google Scholar] [CrossRef]
- Lalonde, S.; Tegeder, M.; Throne-Holst, M.; Frommer, W.B.; Patrick, J.W. Phloem loading and unloading of sugars and amino acids. Plant, Cell Environ. 2003, 26, 37–56. [Google Scholar] [CrossRef]
- Turgeon, R.; Ayre, B.G. Pathways and Mechanisms of Phloem Loading. In Vascular Transport in Plants; Academic Press: Cambridge, MA, USA, 2005; pp. 45–67. [Google Scholar]
- McCaskill, A.; Turgeon, R. Phloem loading in Verbascum phoeniceum L. depends on the synthesis of raffinose-family oligosaccharides. Proc. Natl. Acad. Sci. USA 2007, 104, 19619–19624. [Google Scholar] [CrossRef]
- Rennie, E.A.; Turgeon, R. A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. USA 2009, 106, 14162–14167. [Google Scholar] [CrossRef]
- Turgeon, R.; Medville, R. The absence of phloem loading in willow leaves. Proc. Natl. Acad. Sci. USA 1998, 95, 12055–12060. [Google Scholar] [CrossRef]
- Bush, D.R. Proton-coupled sugar and amino acid transporters in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993, 44, 513–542. [Google Scholar] [CrossRef]
- Turgeon, R. Phloem loading and plasmodesmata. Trends Plant Sci. 1996, 1, 418–423. [Google Scholar] [CrossRef]
- Haritatos, E.; Medville, R.; Turgeon, R. Minor vein structure and sugar transport in Arabidopsis thaliana. Planta 2000, 211, 105–111. [Google Scholar] [CrossRef]
- Comtet, J.; Turgeon, R.; Stroock, A.D. Phloem Loading through Plasmodesmata: A Biophysical Analysis. Plant Physiol. 2017, 175, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.M.; Slewinski, T.L. Genetic Control of Carbon Partitioning in Grasses: Roles of Sucrose Transporters and Tie-dyed Loci in Phloem Loading. Plant Physiol. 2009, 149, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Riesmeier, J.W.; Willmitzer, L.; Frommer, W.B. Isolation and characterization of a sucrose carrier cDNA from spinach by functional expression in yeast. EMBO J. 1992, 11, 4705–4713. [Google Scholar] [CrossRef] [PubMed]
- Aoki, N.; Hirose, T.; Scofield, G.N.; Whitfeld, P.R.; Furbank, R.T. The Sucrose Transporter Gene Family in Rice. Plant Cell Physiol. 2003, 44, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Kühn, C. A Comparison of the Sucrose Transporter Systems of Different Plant Species. Plant Biol. 2003, 5, 215–232. [Google Scholar] [CrossRef]
- Lim, J.D.; Cho, J.-I.; Park, Y.-I.; Hahn, T.-R.; Choi, S.-B.; Jeon, J.-S. Sucrose transport from source to sink seeds in rice. Physiol. Plant. 2006, 126, 572–584. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirose, T.; Aoki, N.; Takahashi, S.; Ono, K.; Yamamoto, S.; Wu, J.; Saji, S.; Baba, T.; Ugaki, M.; et al. Antisense Expression of a Rice Sucrose Transporter OsSUT1 in Rice (Oryza sativa L.). Plant Cell Physiol. 2001, 42, 1181–1185. [Google Scholar] [CrossRef]
- Scofield, G.N.; Hirose, T.; Aoki, N.; Furbank, R.T. Involvement of the sucrose transporter, OsSUT1, in the long-distance pathway for assimilate transport in rice. J. Exp. Bot. 2007, 58, 3155–3169. [Google Scholar] [CrossRef]
- Eom, J.S.; Cho, J.I.; Reinders, A.; Lee, S.W.; Yoo, Y.; Tuan, P.Q.; Choi, S.B.; Bang, G.; Park, Y.I.; Cho, M.H.; et al. Impaired function of the tonoplast-localized sucrose transporter in rice, OsSUT2, limits the transport of vacuolar reserve sucrose and affects plant growth. Plant Physiol. 2011, 157, 109–119. [Google Scholar] [CrossRef]
- Eom, J.-S.; Choi, S.-B.; Ward, J.; Jeon, J.-S. The mechanism of phloem loading in rice (Oryza sativa). Mol. Cells 2012, 33, 431–438. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, L.; Huang, W.; Yuan, M.; Zhou, F.; Li, X.; Lin, Y. Overexpression of OsSWEET5 in Rice Causes Growth Retardation and Precocious Senescence. PLoS ONE 2014, 9, e94210. [Google Scholar] [CrossRef]
- Wang, G.; Wu, Y.; Ma, L.; Lin, Y.; Hu, Y.; Li, M.; Li, W.; Ding, Y.; Chen, L. Phloem loading in rice leaves depends strongly on the apoplastic pathway. J. Exp. Bot. 2021, 72, 3723–3738. [Google Scholar] [CrossRef]
- Li, J.; Wang, D.; Sun, S.; Sun, L.; Zong, J.; Lei, Y.; Yu, J.; Liang, W.; Zhang, D. The regulatory role of CARBON STARVED ANTHER-mediated photoperiod-dependent male fertility in rice. Plant Physiol. 2022, 189, 955–971. [Google Scholar] [CrossRef]
- Yang, J.; Luo, D.; Yang, B.; Frommer, W.B.; Eom, J. SWEET 11 and 15 as key players in seed filling in rice. New Phytol. 2018, 218, 604–615. [Google Scholar] [CrossRef]
- Toyofuku, K.; Kasahara, M.; Yamaguchi, J. Characterization and Expression of Monosaccharide Transporters (OsMSTs) in Rice. Plant Cell Physiol. 2000, 41, 940–947. [Google Scholar] [CrossRef]
- Ngampanya, B.; Sobolewska, A.; Takeda, T.; Toyofuku, K.; Narangajavana, J.; Ikeda, A.; Yamaguchi, J. Characterization of rice functional monosaccharide transporter, OsMST5. Biosci. Biotechnol. Biochem. 2003, 67, 556–562. [Google Scholar] [CrossRef]
- Monfared, H.H.; Chew, J.K.; Azizi, P.; Xue, G.-P.; Ee, S.-F.; Kadkhodaei, S.; Hedayati, P.; Ismail, I.; Zainal, Z. Overexpression of a Rice Monosaccharide Transporter Gene (OsMST6) Confers Enhanced Tolerance to Drought and Salinity Stress in Arabidopsis thaliana. Plant Mol. Biol. Rep. 2020, 38, 151–164. [Google Scholar] [CrossRef]
- Xu, X.; Ren, Y.; Wang, C.; Zhang, H.; Wang, F.; Chen, J.; Liu, X.; Zheng, T.; Cai, M.; Zeng, Z.; et al. OsVIN2 encodes a vacuolar acid invertase that affects grain size by altering sugar metabolism in rice. Plant Cell Rep. 2019, 38, 1273–1290. [Google Scholar] [CrossRef]
- Jia, L.; Zhang, B.; Mao, C.; Li, J.; Wu, Y.; Wu, P.; Wu, Z. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.). Planta 2008, 228, 51–59. [Google Scholar] [CrossRef]
- Wang, E.; Xu, X.; Zhang, L.; Zhang, H.; Lin, L.; Wang, Q.; Li, Q.; Ge, S.; Lu, B.R.; Wang, W.; et al. Research article Duplication and independent selection of cell-wall invertase genes GIF1 and OsCIN1 during rice evolution and domestication. BMC Evol. Biol. 2010, 10, 108. [Google Scholar] [CrossRef]
- Saier, M.H. TCDB: The Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006, 34, D181–D186. [Google Scholar] [CrossRef]
- Hamada, M.; Wada, S.; Kobayashi, K.; Satoh, N. Ci-Rga, a gene encoding an MtN3/saliva family transmembrane protein, is essential for tissue differentiation during embryogenesis of the ascidian Ciona intestinalis. Differentiation 2005, 73, 364–376. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Hou, B.-H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.-Q.; Guo, W.-J.; Kim, J.-G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Qu, X.-Q.; Hou, B.-H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Lin, I.W.; Sosso, D.; Chen, L.-Q.; Gase, K.; Kim, S.-G.; Kessler, D.; Klinkenberg, P.M.; Gorder, M.K.; Hou, B.-H.; Qu, X.-Q.; et al. Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 2014, 508, 546–549. [Google Scholar] [CrossRef]
- Chen, L.-Q.; Lin, I.W.; Qu, X.-Q.; Sosso, D.; McFarlane, H.; Londoño, A.; Samuels, A.L.; Frommer, W.B. A Cascade of Sequentially Expressed Sucrose Transporters in the Seed Coat and Endosperm Provides Nutrition for the Arabidopsis Embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef]
- Yuan, M.; Zhao, J.; Huang, R.; Li, X.; Xiao, J.; Wang, S. Rice MtN3/saliva/SWEET gene family: Evolution, expression profiling, and sugar transport. J. Integr. Plant Biol. 2014, 56, 559–570. [Google Scholar] [CrossRef]
- Yuan, M.; Wang, S. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms. Mol. Plant 2013, 6, 665–674. [Google Scholar] [CrossRef]
- Ruan, Y.L.; Jin, Y.; Huang, J. Capping invertase activity by its inhibitor: Roles and implications in sugar signaling, carbon allocation, senescence and evolution. Plant Signal. Behav. 2009, 4, 983–985. [Google Scholar] [CrossRef]
- Ruan, Y.-L.; Jin, Y.; Yang, Y.-J.; Li, G.-J.; Boyer, J.S. Sugar Input, Metabolism, and Signaling Mediated by Invertase: Roles in Development, Yield Potential, and Response to Drought and Heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef]
- Shen, S.; Ma, S.; Liu, Y.; Liao, S.; Li, J.; Wu, L.; Kartika, D.; Mock, H.-P.; Ruan, Y.-L. Cell Wall Invertase and Sugar Transporters Are Differentially Activated in Tomato Styles and Ovaries During Pollination and Fertilization. Front. Plant Sci. 2019, 10, 506. [Google Scholar] [CrossRef]
- Ji, X.; Ende, W.V.D.; Van Laere, A.; Cheng, S.; Bennett, J. Structure, Evolution, and Expression of the Two Invertase Gene Families of Rice. J. Mol. Evol. 2005, 60, 615–634. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; An, B.; Zhong, H.; Yang, J.; Kong, W.; Li, Y. A Novel Insight into Functional Divergence of the MST Gene Family in Rice Based on Comprehensive Expression Patterns. Genes 2019, 10, 239. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.A.; Hill, J.P.; Thomas, M.A. The monosaccharide transporter gene family in land plants is ancient and shows differential subfamily expression and expansion across lineages. BMC Evol. Biol. 2006, 6, 64. [Google Scholar] [CrossRef][Green Version]
- Schulz, A.; Beyhl, D.; Marten, I.; Wormit, A.; Neuhaus, E.; Poschet, G.; Büttner, M.; Schneider, S.; Sauer, N.; Hedrich, R. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 2011, 68, 129–136. [Google Scholar] [CrossRef]
- Wormit, A.; Trentmann, O.; Feifer, I.; Lohr, C.; Tjaden, J.; Meyer, S.; Schmidt, U.; Martinoia, E.; Neuhaus, H.E. Molecular Identification and Physiological Characterization of a Novel Monosaccharide Transporter from Arabidopsis Involved in Vacuolar Sugar Transport. Plant Cell 2006, 18, 3476–3490. [Google Scholar] [CrossRef]
- Aluri, S.; Buttner, M. Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc. Natl. Acad. Sci. USA 2007, 104, 2537–2542. [Google Scholar] [CrossRef]
- Yamada, K.; Osakabe, Y.; Mizoi, J.; Nakashima, K.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional Analysis of an Arabidopsis thaliana Abiotic Stress-inducible Facilitated Diffusion Transporter for Monosaccharides. J. Biol. Chem. 2010, 285, 1138–1146. [Google Scholar] [CrossRef]
- Cho, M.-H.; Lim, H.; Shin, D.H.; Jeon, J.-S.; Bhoo, S.H.; Park, Y.-I.; Hahn, T. Role of the plastidic glucose translocator in the export of starch degradation products from the chloroplasts in Arabidopsis thaliana. New Phytol. 2011, 190, 101–112. [Google Scholar] [CrossRef]
- Klepek, Y.-S.; Volke, M.; Konrad, K.R.; Wippel, K.; Hoth, S.; Hedrich, R.; Sauer, N. Arabidopsis thaliana POLYOL/MONOSACCHARIDE TRANSPORTERS 1 and 2: Fructose and xylitol/H+ symporters in pollen and young xylem cells. J. Exp. Bot. 2010, 61, 537–550. [Google Scholar] [CrossRef]
- Schneider, S.; Schneidereit, A.; Konrad, K.; Hajirezaei, M.-R.; Gramann, M.; Hedrich, R.; Sauer, N. Arabidopsis INOSITOL TRANSPORTER4 Mediates High-Affinity H+ Symport of Myoinositol across the Plasma Membrane. Plant Physiol. 2006, 141, 565–577. [Google Scholar] [CrossRef]
- Kaschuk, G.; Hungria, M.; Leffelaar, P.A.; Giller, K.E.; Kuyper, T.W. Differences in photosynthetic behaviour and leaf senescence of soybean (Glycine max [L.] Merrill) dependent on N2 fixation or nitrate supply. Plant Biol. 2010, 12, 60–69. [Google Scholar] [CrossRef]
- Paul, M.J.; Pellny, T.K. Carbon metabolite feedback regulation of leaf photosynthesis and development. J. Exp. Bot. 2003, 54, 539–547. [Google Scholar] [CrossRef]
- Wright, K.M.; Roberts, A.G.; Martens, H.J.; Sauer, N.; Oparka, K.J. Structural and Functional Vein Maturation in Developing Tobacco Leaves in Relation to AtSUC2 Promoter Activity. Plant Physiol. 2003, 131, 1555–1565. [Google Scholar] [CrossRef]
- Truernit, E.; Sauer, N. The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 1995, 196, 564–570. [Google Scholar] [CrossRef]
- Gottwald, J.R.; Krysan, P.J.; Young, J.C.; Evert, R.F.; Sussman, M.R. Genetic evidence for the in planta role of phloem-specific plasma membrane sucrose transporters. Proc. Natl. Acad. Sci. USA 2000, 97, 13979–13984. [Google Scholar] [CrossRef]
- Han, L.; Zhu, Y.; Liu, M.; Zhou, Y.; Lu, G.; Lan, L.; Wang, X.; Zhao, Y.; Zhang, X.C. Molecular mechanism of substrate recognition and transport by the AtSWEET13 sugar transporter. Proc. Natl. Acad. Sci. USA 2017, 114, 10089–10094. [Google Scholar] [CrossRef]
- Sosso, D.; Luo, D.; Li, Q.-B.; Sasse, J.; Yang, J.; Gendrot, G.; Suzuki, M.; Koch, K.E.; Mccarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef]
- Cheng, W.H.; Taliercio, E.W.; Chourey, P.S. The Miniature1 Seed Locus of Maize Encodes a Cell Wall Invertase Required for Normal Development of Endosperm and Maternal Cells in the Pedicel. Plant Cell 1996, 8, 971–983. [Google Scholar] [CrossRef]
- Weber, H.; Borisjuk, L.; Wobus, U. Weber-Controlling seed development and seed size in Vicia faba a role for seed. Plant J. 1996, 10, 823–834. [Google Scholar] [CrossRef]
- Ruan, Y.L.; Chourey, P.S.; Delmer, D.P.; Perez-Grau, L. The Differential Expression of Sucrose Synthase in Relation to Diverse Patterns of Carbon Partitioning in Developing Cotton Seed. Plant Physiol. 1997, 115, 375–385. [Google Scholar] [CrossRef]
- Wang, E.; Wang, J.; Zhu, X.; Hao, W.; Wang, L.; Li, Q.; Zhang, L.; He, W.; Lu, B.; Lin, H.; et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat. Genet. 2008, 40, 1370–1374. [Google Scholar] [CrossRef]
- Slewinski, T.L. Diverse Functional Roles of Monosaccharide Transporters and their Homologs in Vascular Plants: A Physiological Perspective. Mol. Plant 2011, 4, 641–662. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, Y.; Zhang, Y.; Chai, C.; Wei, G.; Wei, X.; Xu, H.; Wang, M.; Ouwerkerk, P.B.F.; Zhu, Z. Molecular cloning, functional characterization and expression analysis of a novel monosaccharide transporter gene OsMST6 from rice (Oryza sativa L.). Planta 2008, 228, 525–535. [Google Scholar] [CrossRef]
- Nguyen, G.N.; Hailstones, D.L.; Wilkes, M.; Sutton, B.G. DROUGHT STRESS: Role of Carbohydrate Metabolism in Drought-Induced Male Sterility in Rice Anthers*. J. Agron. Crop Sci. 2010, 196, 346–357. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, H.; Wei, X.; Chai, C.; Xiao, Y.; Zhang, Y.; Chen, B.; Xiao, G.; Ouwerkerk, P.B.F.; Wang, M.; et al. Molecular cloning and expression analysis of a monosaccharide transporter gene OsMST4 from rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 439–451. [Google Scholar] [CrossRef]
- Mamun, E.; Alfred, S.; Cantrill, L.; Overall, R.; Sutton, B. Effects of chilling on male gametophyte development in rice. Cell Biol. Int. 2006, 30, 583–591. [Google Scholar] [CrossRef]
- Sharkey, T.D. Understanding carbon partitioning and its role in determining plant growth. Plant, Cell Environ. 2015, 38, 1963–1964. [Google Scholar] [CrossRef]
- Scialdone, A.; Mugford, S.; Feike, D.; Skeffington, A.; Borrill, P.; Graf, A.; Smith, A.M.; Howard, M. Arabidopsis plants perform arithmetic division to prevent starvation at night. eLife 2013, 2, e00669. [Google Scholar] [CrossRef]
- Graf, A.; Smith, A.M. Starch and the clock: The dark side of plant productivity. Trends Plant Sci. 2011, 16, 169–175. [Google Scholar] [CrossRef]
- Zeeman, S.C.; Rees, T.A. Changes in carbohydrate metabolism and assimilate export in starch-excess mutants of Arabidopsis. Plant Cell Environ. 1999, 22, 1445–1453. [Google Scholar] [CrossRef]
- Smith, A.M.; Stitt, M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007, 30, 1126–1149. [Google Scholar] [CrossRef] [PubMed]
- Soy, J.; Leivar, P.; González-Schain, N.; Sentandreu, M.; Prat, S.; Quail, P.H.; Monte, E. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J. 2012, 71, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Zeeman, S.C. Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 2012, 15, 282–292. [Google Scholar] [CrossRef]
- Noordally, Z.B.; Ishii, K.; Atkins, K.A.; Wetherill, S.J.; Kusakina, J.; Walton, E.J.; Kato, M.; Azuma, M.; Tanaka, K.; Hanaoka, M.; et al. Circadian Control of Chloroplast Transcription by a Nuclear-Encoded Timing Signal. Science 2013, 339, 1316–1319. [Google Scholar] [CrossRef]
- Haydon, M.; Mielczarek, O.; Robertson, F.C.; Hubbard, K.; Webb, A. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature 2013, 502, 689–692. [Google Scholar] [CrossRef]
- Weber, H.; Borisjuk, L.; Wobus, U. Molecular physiology of legume seed development. Annu. Rev. Plant Biol. 2005, 56, 253–279. [Google Scholar] [CrossRef]
- Rolland, F.; Moore, B.; Sheen, J. Sugar Sensing and Signaling in Plants. Plant Cell 2002, 14, S185–S205. [Google Scholar] [CrossRef]
- Leon, P.; Sheen, J. Sugar and hormone connections. Trends Plant Sci. 2003, 8, 110–116. [Google Scholar] [CrossRef]
- Mason, M.G.; Ross, J.J.; Babst, B.A.; Wienclaw, B.N.; Beveridge, C.A. Sugar demand, not auxin, is the initial regulator of apical dominance. Proc. Natl. Acad. Sci. USA 2014, 111, 6092–6097. [Google Scholar] [CrossRef]
- Hyun, Y.; Richter, R.; Coupland, G. Competence to Flower: Age-Controlled Sensitivity to Environmental Cues. Plant Physiol. 2017, 173, 36–46. [Google Scholar] [CrossRef]
- Choudhary, A.; Kumar, A.; Kaur, N.; Kaur, H. Molecular cues of sugar signaling in plants. Physiol. Plant. 2022, 174, e13630. [Google Scholar] [CrossRef]
- Botha, C.E.; Aoki, N.; Scofield, G.N.; Liu, L.; Furbank, R.T.; White, R.G. A xylem sap retrieval pathway in rice leaf blades: Evidence of a role for endocytosis? J. Exp. Bot. 2008, 59, 2945–2954. [Google Scholar] [CrossRef]
- Wang, L.; Lu, Q.; Wen, X.; Lu, C. Enhanced sucrose loading improves rice yield by increasing grain size. Plant Physiol. 2015, 169, 2848–2862. [Google Scholar] [CrossRef]
- Wu, Y.; Lee, S.-K.; Yoo, Y.; Wei, J.; Kwon, S.-Y.; Lee, S.-W.; Jeon, J.-S.; An, G. Rice Transcription Factor OsDOF11 Modulates Sugar Transport by Promoting Expression of Sucrose Transporter and SWEET Genes. Mol. Plant 2018, 11, 833–845. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT Protein Movement Contributes to Long-Distance Signaling in Floral Induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Yano, M.; Kojima, S.; Takahashi, Y.; Lin, H.; Sasaki, T. Genetic Control of Flowering Time in Rice, a Short-Day Plant. Plant Physiol. 2001, 127, 1425–1429. [Google Scholar] [CrossRef]
- Lee, Y.-S.; An, G. Regulation of flowering time in rice. J. Plant Biol. 2015, 58, 353–360. [Google Scholar] [CrossRef]
- Yuan, L.P. Hybrid rice technology for food security in the world. Int. Rice Comm. Newsl. 2004, 53, 24–25. [Google Scholar]
- Rich-Griffin, C.; Stechemesser, A.; Finch, J.; Lucas, E.; Ott, S.; Schäfer, P. Single-Cell Transcriptomics: A High-Resolution Avenue for Plant Functional Genomics. Trends Plant Sci. 2020, 25, 186–197. [Google Scholar] [CrossRef]
- Balcerowicz, M.; Shetty, K.N.; Jones, A.M. Fluorescent biosensors illuminating plant hormone research. Plant Physiol. 2021, 187, 590–602. [Google Scholar] [CrossRef]
- Wang, B.; Zhu, L.; Zhao, B.; Zhao, Y.; Xie, Y.; Zheng, Z.; Li, Y.; Sun, J.; Wang, H. Development of a Haploid-Inducer Mediated Genome Editing System for Accelerating Maize Breeding. Mol. Plant 2019, 12, 597–602. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
| Gene Family | Number of Genes | Reported Genes/Reference |
|---|---|---|
| SUT | 5 | SUT1 [38,39]; SUT2 [40]; SUT3 [41]; SUT4, SUT5 [35] |
| SWEET | 21 | SWEET5 [42]; SWEET6a [43,44]; SWEET6b [44]; SWEET11 [15]; SWEET14 [16]; SWEET15 [45] |
| MST | 64 | |
| AZT subfamily | 6 | AZT3 [13] |
| ERD subfamily | 6 | |
| pGlcT subfamily | 4 | |
| Xylose subfamily | 2 | |
| STP subfamily | 15 | MST1, MST2, MST3 [46]; MST5 [47]; MST6 [48]; MST8 [11]; SPT17, SPT27 [43,44] |
| PLT subfamily | 28 | |
| INT subfamily | 3 | |
| Invertases | 18 | |
| VIN | 2 | VIN2 [49] |
| CIN | 8 | CIN8/Cyt-INV1 [50] |
| CWIN | 8 | GIF1 [51]; INV4 [10]; CWIN6 [44] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Kim, Y.-J.; Zhang, D. Source-To-Sink Transport of Sugar and Its Role in Male Reproductive Development. Genes 2022, 13, 1323. https://doi.org/10.3390/genes13081323
Li J, Kim Y-J, Zhang D. Source-To-Sink Transport of Sugar and Its Role in Male Reproductive Development. Genes. 2022; 13(8):1323. https://doi.org/10.3390/genes13081323
Chicago/Turabian StyleLi, Jingbin, Yu-Jin Kim, and Dabing Zhang. 2022. "Source-To-Sink Transport of Sugar and Its Role in Male Reproductive Development" Genes 13, no. 8: 1323. https://doi.org/10.3390/genes13081323
APA StyleLi, J., Kim, Y.-J., & Zhang, D. (2022). Source-To-Sink Transport of Sugar and Its Role in Male Reproductive Development. Genes, 13(8), 1323. https://doi.org/10.3390/genes13081323






